Abstract
Urban wastewater treatment plants (UWTPs) are among the major recipients of antibiotic-resistant bacteria (ARB), antibiotic resistance genes (ARGs), and antibiotic residues in urban environments. Although during treatment, bacteria of human and animal origin are removed, some are able to survive, persisting in the final effluent. The occurrence of these bacteria, especially those harboring ARGs, may have a direct impact on the quality of the treated wastewater that is returned to the environment. In this study, we aimed to assess if the final effluent bacterial communities of three UWTPs (PT1, PT2, and PT3) located next to each other were distinct and if such differences were related with the antibiotic resistance profiles.
It was observed that the bacterial community (16S rRNA gene Illumina sequencing) and load of selected ARGs of final effluent differed among the three UWTPs, irrespective of sampling time. Members of the families Aeromonadaceae, Campylobacteraceae, Veillonellaceae, [Weeksellaceae], and Porphyromonadaceae were observed to be positively correlated with some ARGs (blaCTX–M, blaOXA-A, blaSHV) and intI1 (p < 0.05), while Intrasporangiaceae were observed to be negatively correlated. While Aeromonadaceae are recognized relevant ARG harbors, the other bacterial families may represent bacteria that co-exist with the ARG hosts, which may belong to minor bacterial groups omitted in the analyses. These findings suggest the importance of bacterial dynamics during treatment to the ARB&ARGs removal, a rationale that may contribute to design new strategies to apply in the UWTPs to prevent the spread of antibiotic resistance.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Urban wastewater treatment plants (UWTPs) have received much attention as major recipients of antibiotic-resistant bacteria (ARB), antibiotic resistance genes (ARGs) (Rizzo et al. 2013), and antibiotic residues (Michael et al. 2013) in urban environments. Their capacity to remove ARB and ARGs has also been largely discussed and demonstrated (Manaia et al. 2016; Bengtsson-Palme et al. 2016; Wu et al. 2018). Nevertheless, most authors agree that ARB and ARG loads released by well-functioning UWTPs may still have a negative impact on the environment, with serious implications to the human health (Berendonk et al. 2015). While it is consensual that it is important to improve wastewater treatment processes in order to maximize the removal of ARB and ARGs, not much is known about the best strategies to achieve such a goal (Berendonk et al. 2015; Manaia et al. 2016; Vikesland et al. 2017). Although wastewater disinfection may promote the reduction of the total microbial loads and of the bacterial diversity, it can create the perfect conditions for the bacterial regrowth of certain groups of bacteria (Manaia et al. 2016; Becerra-Castro et al. 2017; Moreira et al. 2018).
The maintenance or propagation of antibiotic resistance during wastewater treatment may be due to two major driving forces, ARG horizontal gene transfer and/or ARB survival or selection (Berendonk et al. 2015; Vikesland et al. 2017). Despite the importance that horizontal gene transfer may have on ARG spread, it may be not the most relevant factor to dictate the fate of antibiotic resistance during wastewater treatment (Figueira et al. 2011a; Figueira et al. 2011b; Manaia et al. 2016). The survival or not of specific bacterial lineages during treatment may have consequences on the abundance of ARGs in the final effluent. In this study, we aimed to investigate if ARGs released in the final effluent of a UWTP are related with the diversity of bacteria present in that effluent, at least with some of the groups surviving the wastewater treatment, comprising bacteria frequently associated with humans and animals. In this rationale, it is assumed that the understanding of which bacterial groups are the major vehicles or the ecologic surrogates of ARGs could be a good strategy to identify suitable bacterial indicators of ARGs that survive treatment. This identification could be the basis to manage wastewater treatment processes in order to maximize the reduction or eliminate such bacterial groups. To test the aforementioned hypothesis, we compared the final effluent of three UWTPs, located in the same region (Northern Portugal), and explored whether it was possible to find significant correlations between the bacterial community composition and the profile of a selected group of ARGs.
Materials and methods
Sampling
Treated wastewater samples were collected in three UWTPs (PT1, PT2, and PT3) located, within a region of approximately 7 km, in Northern Portugal. These UWTPs, serve equivalent populations of 150,000–170,000 inhabitants, receive a daily flow of approximately 22,000 m3 (PT1), 33,000 m3 (PT2), and 24,000 m3 (PT3), and all have primary and secondary treatments operating with conventional activated sludge (CAS). PT2 is the only one reporting the reception of hospital effluents, and PT3 is the only one with UV disinfection of the final effluent. No information was provided regarding the reception of hospital effluents by PT1 or PT3. Soon after this study, PT1 was temporarily closed for improvement restoration.
Composite samples (24 h) were collected from PT1 and PT2 in four campaigns, in early Spring (average temperature of 9 °C) and early Autumn (average temperature of 17 °C), specifically in three consecutive days (Tuesday–Thursday) in March 2015 (M_2015), October 2015 (O_2015), March 2016 (M_2016), and September 2016 (S_2016). Due to technical problems at PT1, it was not possible to sample this UWTP in March 2016. Grab samples of PT3 were collected in June 2015 (J_2015), July 2015 (JL_2015), September 2015 (S_2015), and March 2016 (M_2016). Samples were stored in ice and transported to the laboratory to be processed within 12 h.
DNA extraction
Volumes of 100 to 250 mL of final effluent were filtered, in triplicate, through sterile polycarbonate membranes (0.22 μm porosity, Whatman, UK) that were stored at − 80 °C. DNA was extracted from each of the three replicates with the PowerWater® DNA Isolation Kit (MOBIO Laboratories Inc., CA, USA) according to the manufacturer’s instructions. DNA extract’s concentration was measured using the Qubit 3.0 Fluorometer (ThermoFisher Scientific, USA). A total of 33 DNA extracts, corresponding to 7 independent sampling dates, were stored at − 20 °C until further analysis.
Bacterial community composition
The bacterial community composition was analyzed in the 33 DNA extracts, based on MiSeq® Illumina DNA sequencing (Genoinseq, Cantanhede, Portugal) of the V3–V4 hypervariable region of the 16S rRNA gene using the Bakt_341F 5′-CCTACGGGNGGCWGCAG-3′ and Bakt_805R 5′-GACTACHVGGGTATCTAATCC-3′ primers set, according to manufacturer’s instructions (Illumina, San Diego, CA, USA). A filtering based on the reads size and quality trimming was performed using the software PRINSEQ (Schmieder and Edwards 2011). Reads shorter than 200 bp and with average quality scores lower than 25 were excluded. Sequence reads were demultiplexed automatically by the Illumina® Miseq® sequencer using the CASAVA package (Illumina, San Diego, CA, USA), and paired-end reads were merged using a QIIME script. Chimeric sequences were removed using USEARCH v6.1 (Edgar 2010). After the quality control, one of the three replicates of PT3 (September 2015) was removed from the analyses due to the low number of initial reads (n = 37,932). Sequences with nucleotide identity higher than 97% were assigned to operational taxonomic units (OTUs) (Edgar 2010) and to taxonomic groups using the Greengenes Database version 13_8 (updated: August 2013) (DeSantis et al. 2006). Singletons and chimeras as well as OTUs assigned to non-bacterial groups, such as chloroplasts or mitochondria, were removed from the data set. After this procedure, 3,109,382 good-quality sequences were obtained, grouped into 32,084 OTUs. The downstream analyses were performed after normalization with the cumulative sum scaling (CSS) (Paulson et al. 2014) of the data (except the computation of the alpha diversity metrics). The alpha diversity was evaluated based on bacterial richness (number of OTUs), diversity indices Shannon (Shannon and Weaver 1963), phylogenetic diversity (PD) whole tree (Faith 1992), and Simpson (Simpson 1949), and the richness estimator Chao1 (Chao 1984), using the QIIME 1 pipeline (Caporaso et al. 2010). Alpha diversity was represented as the mean value for each index and the respective standard deviation (SD) values and differences among UWTP effluents calculated by analysis of variance (ANOVA) with post hoc Tukey’s test (p < 0.01). Alpha rarefaction plot of observed OTUs was performed to verify the sequence coverage. All the plots reached a plateau phase meaning that the final data is a good representation of the bacterial diversity of the wastewater effluent. Beta diversity indices were assessed using the weighted UniFrac metric distances (Lozupone and Knight 2005) from QIIME pipeline (Caporaso et al. 2010) and the results presented as principal coordinates analysis (PCoA). This analysis was also performed by taking into consideration the UWTPs individually in order to assess possible seasonal variations. The relative abundance of the bacterial community composition at different taxonomic levels was compared based on the ANOVA and Tukey-Kramer post hoc tests (p < 0.01), and the p values were corrected for multiple testing using the Benjamini-Hochberg false discovery rate (FDR) (Benjamini and Hochberg 1995), using STAMP v2.1.3 software (Parks et al. 2014). Pairs of samples with an FDR value < 0.01 were considered significantly different.
The sequence data files that support the findings of this study have been deposited in GenBank within the BioProject PRJNA478220, Sequence Read Archive (SRA) SRS3472441, and with the accession numbers SRX4313251-SRX4313282.
Quantification of antibiotic resistance genes and class 1 integron-integrase
Quantitative polymerase chain reaction (qPCR, StepOneTM Real-Time PCR System, Life Technologies, Carlsbad, CA, USA) was used to measure the abundance (per milliliter of sample) of seven ARGs (blaCTX-M, blaOXA-A, blaSHV,blaTEM, sul1, sul2, and qnrS) and class 1 integron-integrase (intI1). The 16S rRNA gene was also quantified, as a measure of the total bacterial load, and used to determine ARGs and intI1 prevalence values (per 16S rRNA gene copy number). Calibration curves, built based on adequate dilutions of standards of each analyzed gene, were produced in parallel with the test samples. Each DNA extract was tested in duplicate for each run. The list of primers and protocols used, based on SYBR Green detection, has been described before (Narciso-da-Rocha et al. 2018). One-way analysis of variance (ANOVA) and Tukey’s post hoc tests were used to assess statistically significant differences (p < 0.01) of prevalence and/or abundance of 16S rRNA gene, ARGs, or intI1 using GraphPad Prism 6.00 software (GraphPad Software, San Diego, CA). Principal component analysis (PCA) based on the prevalence of ARGs was performed using Canoco 5.01 software (Leps and Smilauer 2014).
Comparative analyses of bacterial community composition and ARGs and intI1
A redundancy analysis (RDA) (van den Wollenberg, 1977) was performed to explore possible relationships between the bacterial community structure (families with relative abundance > 1%) and the ARGs and intI1 prevalence (target gene copy number/16S rRNA gene copy number) for all the UWTPs, using Canoco 5.01 software (Leps and Smilauer 2014). The significance of the environmental variables (ARGs and intI1) was tested using Monte Carlo permutation tests (999 unrestricted permutations, p < 0.05 and p < 0.01) after FDR adjustment. The same data set was used to assess Pearson’s correlations, testing each UWTP individually, to infer about possible relationships between the bacterial community composition and genes. Pearson’s correlation was performed using GraphPad Prism 6.00 software (GraphPad Software, San Diego, CA).
Results
Bacterial community
The analysis of the V3–V4 region of the 16S rRNA gene led to the identification of 32,084 OTUs in the final effluent of the three UWTPs. PT1 showed the highest number of OTUs (7238 ± 861), followed by PT2 (6732 ± 1114) and PT3 (5704 ± 996). These values were in agreement with the observation that PT3 displayed the lowest Chao1 richness estimator value (p < 0.01). However, this difference was not expressed in the Shannon’s and Simpson’s diversity indices, significantly higher in PT3 and PT2 than in PT1 (p < 0.01) (Table 1). The whole tree phylogenetic diversity (PD) index, which measures the shared phylogenetic distance among taxa in each sample, did not reveal significant differences of the bacterial communities (p > 0.01) in the final effluents of the three UWTPs.
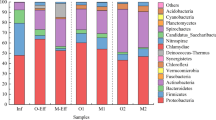
The phyla Proteobacteria and Bacteroidetes predominated in the final effluents of all UWTPs (PT1, 55.2–58.8% vs. 21.1–25.9%; PT2, 39.1–55.0% vs. 13.1–14.2%; and PT3, 42.9–44.8% vs. 9.4–11.3%) (Fig. 1). However, the candidate phyla, TM7 (renamed as Candidatus Saccharibacteria) and OD1 (renamed as Candidatus Parcubacteria), made the difference in PT2 and PT3, where they were also among the predominant, with relative abundance values ranging 9.2–18.3% and 2.7–6.6%, respectively, for PT2, and 5.9–12.7% and 8.4–14.0%, respectively, for PT3. These phyla were below 3.6% and 3.2%, respectively, in PT1. Predominant Proteobacteria classes were represented by OTUs affiliated to Gamma- (PT1, 9.3–11.8%; PT2, 9.3–12.5%; and PT3, 7.1–10.9%) and Betaproteobacteria (PT1, 31.2–38.4%; PT2, 16.1–26.6%; and PT3, 13.6–19.7%), in particular of the order Burkholderiales (PT1, 23.2–30.0%; PT2, 6.2–12.6 %; and PT3, 7.4–11.0%). Bacteroidales was the predominant order of Bacteroidetes (PT1, 10.0–17.0%; PT2, 3.9–6.9%; and PT3, 2.5–6.7%). It was also noticeable that, at all taxonomic levels, unclassified OTUs were more abundant in PT2 and PT3 than in PT1.
Relative abundance at bacterial phyla observed in the final effluent of the urban wastewater treatment plants (PT1, PT2, and PT3), in distinct sampling campaigns, each including three consecutive days, in March 2015 (M_2015), October 2015 (O_2015), March 2016 (M_2016), September 2016 (S_2016), June 2015 (J_2015), July 2015 (JL_2015), and September 2015 (S_2015). Taxa with abundance below 1% in all samples were designated as other phyla
Based on the PCoA analysis, the bacterial community observed in the final effluent of the three UWTPs formed different groups, which, according to this analysis, were not influenced by sampling date (Fig. 2a). PT1 displayed the most variable bacterial community composition (Fig. 2a), although PT2 and PT3 were also in separated groups (Fig. 2a). Given the fact that OTUs were defined at a sequence identity value of 97%, the number of OTUs shared by all UWTPs was surprisingly low, of only 22.0% (Fig. 2b). This number was nevertheless higher than that of unique OTUs, of 18.9%, 16.1%, and 10.8% in PT1, PT2, and PT3, respectively. Confirming the PCoA (Fig. 2a), PT3 and PT2 shared the highest number of OTUs (37.1%), while PT1 shared 34.3% with PT2 and 26.9% with PT3 (Fig. 2b). In PT1, unique OTUs were mainly Proteobacteria (58.4%) and Bacteroidetes (30.2%), of the families Comamonadaceae and Flavobacteriaceae, together with bacteria of the lineage GZKB119. The remaining bacterial phyla contributed less than 2.5% for the unique OTUs. In PT2 and PT3, unique OTUs were also mainly Proteobacteria (PT2, 43.8%; PT3, 37.8%) and Bacteroidetes (PT2, 9.1%; PT3, 7.2%), but other groups were observed, specifically TM7 (Candidatus Saccharibacteria; PT2, 19.6%; PT3, 7.4%) and OD1 (PT2, 7.7%; PT3, 16.5%). PT3 had also a relevant percentage of unique OTUs identified as Planctomycetes (> 5%). In PT2, unique OTUs identified at the family level belonged to Procabacteriaceae and Rhodocyclaceae, while for PT3, no unique bacterial families stood out.
Analysis of the wastewater bacterial diversity. a Principal coordinates analysis (PCoA) based on weighted UniFrac distances between different final effluents of the UWTPs, PT1 (pink), PT2 (green), and PT3 (blue), and the respective sampling campaigns, March 2015 (M_2015), October 2015 (O_2015), March 2016 (M_2016), September 2016 (S_2016), June 2015 (J_2015), July 2015 (JL_2015), and September 2015 (S_2015). b Venn diagram representation of the percentage of shared and exclusive bacterial OTUs between the different UWTPs
A more in-depth comparative analyses of the bacterial community composition in the final effluent of the three UWTPs considered all families with relative abundance higher than 1% (n = 24 families). This comparison showed that PT1 differed from the other two UWTPs by a higher relative abundance (p < 0.01) of [Weeksellaceae], Flavobacteriaceae, GZKB119, Porphyromonadaceae, Comamonadaceae, and Pseudomonadaceae, and a lower relative abundance (p < 0.01) of Saprospiraceae and Bdellovibrionaceae (Fig. 3). In PT2, the families Intrasporangiaceae, Chitinophagaceae, and Procabacteriaceae were more abundant than in the other two UWTPs (p < 0.01). PT3 differed from the other UWTPs on a higher relative abundance of Bdellovibrionaceae and lower relative abundance of Sphingomonadaceae and Verrucomicrobiaceae (Fig. 3). Curiously, PT2 and PT3 showed a higher abundance of unclassified families (PT2, 52.6%; PT3, 68.7%) than PT1 (14.7%).
Comparison of the relative abundance of families, accounting for more than 1%, identified in the final effluent of the different urban wastewater treatment plants (UWTPs), PT1, PT2, and PT3. The table below the bars represents the Pearson correlation between the relative abundance of a given family in a UWTP and each of the ARGs or intI1 gene. a, b, and c indicate significant (p < 0.01) different Tukey-Kramer groups after FDR correction. Positive significant correlations (ρ > 0.7; p < 0.01) are indicated in bold and negative significant correlations (ρ < − 0.7; p < 0.01) are plain text
Antibiotic resistance genes
A set of ARGs (blaCTX-M, blaTEM, blaOXA-A, blaSHV, sul1, sul2, and qnrS) and class 1 integron-integrase (intI1) gene were analyzed in terms of abundance and prevalence. The quantification of the 16S rRNA gene is a measure of the bacteria content in all samples and supported the estimation of prevalence values. Similar trends were observed for abundance and prevalence of ARGs and intI1, ranked as blaCTX-M < blaSHV < blaTEM < qnrS < sul2 < blaOXA-A < intI1 < sul1 (Fig. 4). Comparing the different UWTPs, it was observed that PT1 presented significantly higher (p < 0.01) abundance and prevalence of blaOXA-A and intI1 and higher abundance of 16S rRNA gene, blaTEM, and qnrS than PT2 and PT3. In PT3, the gene blaSHV was more abundant and sul1 was more prevalent than in the other plants. PT2 showed a higher prevalence of blaSHV and sul2 (p < 0.01) than the other two UWTPs (Fig. 4a). Despite these significant differences, the average abundance values of ARGs discharged by the different UWTPs differed less than 1.2 log-unit for blaCTX-M, blaSHV, blaTEM,sul1, sul2, qnrS, and intI1, being less than 2 log-units for the genes and blaOXA-A. These results show that the average abundance values of ARGs released by the three UWTPs analyzed are very similar, independently of the treatment used, suggesting that this may be associated with the region.
Quantification of ARGs and intI1 in the final effluent of the urban wastewater treatment plants (PT1, PT2, and PT3). a Gene abundance (gene copy number/mL of sample) in the upper part of the figure or prevalence (target gene copy number/16S rRNA gene copy number) in the bottom. a, b, and c indicate significant (p < 0.01) different Tukey’s groups. b Principal component analysis of the distribution of ARGs and intI1 gene prevalence
The bacterial community distribution seemed more stable than that of ARGs and intI1 prevalence values, even though the general pattern of distribution was similar with PT1 clustering apart from PT2 and PT3 (Figs. 2a and 4b). This observation motivated the search for possible relationships between the bacterial community composition and structure and the quantified genes in all UWTPs, using a redundancy analysis (RDA). According to this analysis, the ARGs and intI1 prevalence could explain approximately 84% of the variation observed (Fig. 5). Among the genes showing significant correlation with the bacterial community distribution were blaCTX–M, blaOXA-A, blaSHV, and intI1 (Monte Carlo permutations, p < 0.05 for blaSHV and intI1; p < 0.01 for blaCTX–M and blaOXA–A). The genes blaOXA-A and intI1 were mainly associated with PT1 samples and members of the families [Weeksellaceae], Comamonadaceae, Flavobacteriaceae, GZKB119, and Porphyromonadaceae predominant in that UWTP. In opposition, the genes blaCTX–M and blaSHV were mostly associated with PT2 and PT3 samples and with members of the families Bdellovibrionaceae, Gordoniaceae, and Saprospiraceae (Figs. 3 and 5).
Redundancy analysis (RDA) triplot of the bacterial community composition at the family level (relative abundance > 1 %) and environmental variables (ARGs and intI1 prevalence) in the 32 final effluent samples. Blue arrows indicate the members of the bacterial community. The gray arrows represent environmental variables (ARGs and intI1) with no significant correlation, based on the Monte Carlo permutation test after FDR correction. The pointed red and red arrows show the variables with significance lower than 0.05 and 0.01, respectively. PT1 (pink), PT2 (green), and PT3 (blue) regard the UWTPs in distinct sampling campaigns: March 2015 (M_2015), October 2015 (O_2015), March 2016 (M_2016), September 2016 (S_2016), June 2015 (J_2015), July 2015 (JL_2015), and September 2015 (S_2015)
For a further insight of the multivariate analysis provided by RDA, the comparative analyses presented in Fig. 3 were revisited for inferring about possible statistically significant correlations between specific bacterial families and ARGs and intI1 prevalence values. This analysis could hint bacterial groups putatively associated with ARGs, either because they harbor some of those genes or because they co-occur with the bacteria that harbor such genes (Ju et al. 2016). The identification of these bacteria is of interest, since being able to survive wastewater treatment, they may contribute to the antibiotic resistance spread. The same analysis could also suggest other groups whose presence might be associated with lower resistance prevalence, in the case of negative correlations. The analysis revealed that members of the families Aeromonadaceae and Campylobacteraceae (Proteobacteria), Veillonellaceae (Firmicutes), and [Weeksellaceae] and Porphyromonadaceae (Bacteroidetes) were significantly positively correlated with ARGs and intI1 in different UWTPs. None of these bacterial families were correlated with the sul1 gene. On the other hand, the Intrasporangiaceae were observed to be negatively correlated with most of the ARGs (Fig. 3).
Discussion
Some antibiotic resistance features in urban wastewater, such as the most common genes and predominant bacterial phyla, seem to follow a general pattern (Gatica et al. 2016; Manaia et al. 2016; Narciso-da-Rocha et al. 2018). However, it is recognized that each UWTP has specificities due not only to biogeographic factors but also to the composition of the influents received and the age of the plant, among others. These factors may surpass the effect of climate conditions and lead to final effluents with distinct resistance features and community composition. The bacterial community composition may be an important driver in determining the prevalence and patterns of resistance in the final effluent. Indeed, bacteria selection is suggested as an important biological process ruling the fate of ARGs during wastewater treatment, eventually with higher impact on the loads of resistance than horizontal gene transfer (Vaz-Moreira et al. 2014; Bengtsson-Palme et al. 2016; Manaia et al. 2016). In this study, we were interested in comparing the bacterial community and resistance profile in final effluents of three UWTPs. Based on such a comparison, we aimed at inferring about possible associations between antibiotic resistance and bacterial community members. Assuming that geographical and socioeconomic factors may have a strong effect on the bacterial communities and antibiotic resistance loads in the wastewater effluents, three UWTPs located within a distance of 7 km were selected for this comparative study. Interesting differences in these plants are the year of construction (more than 25 years ago for PT1, 15 years for PT2, and 18 years for PT3), the reception of hospital effluents (known only for PT2), and the existence of UV disinfection available at the time of sampling only in PT3. In all UWTPs effluents, the predominant bacterial phyla were the same, confirming previous reports that highlight that wastewater samples hold similar bacterial community compositions, at high taxonomic ranks (Munck et al. 2015). As in previous reports, Proteobacteria and Bacteroidetes were among the major phyla, although in Portugal wastewater samples, Actinobacteria are consistently poorly represented (Ye and Zhang 2013; Munck et al. 2015; Binh et al. 2018; Narciso-da-Rocha et al. 2018) (Fig. 1). However, eventually, as a result of the history and functioning differences of the three UWTPs, the bacterial community composition clustered into three distinct groups, each represented by a UWTP. Therefore, the current study evidenced that UWTPs located in close regions discharge treated effluents with a distinct profile of taxonomic groups (Figs. 1 and 2). Indeed, it was observed that in a total of 9 to 12 samples collected in each UWTP, in distinct seasons, with average temperatures of 9 °C (early Spring) and 17 °C (early Autumn), the final effluent of each plant fell into the same group, with the effluents of the three UWTPs forming three distinct groups. The UWTPs effect was more notorious than the season effect, for which no significant differences were observed. This suggests that the microbiota present in the final effluent is somehow characteristic of a given UWTP. This may be related to the previously noted functional stability of wastewater treatment bioreactors, which permit the maintenance of the system, buffering the occurrence of possible perturbations (LaPara et al. 2002).
UWTP PT1 was, among the three analyzed, the one with the most distinct bacterial community, with a significantly higher relative abundance of members of the families [Weeksellaceae], Flavobacteriaceae, GZKB119, Porphyromonadaceae, Comamonadaceae, and Pseudomonadaceae (Figs. 2a and 3). Also, of note was the fact that the phyla TM7 (Candidatus Saccharibacteria) and OD1 (Candidatus Parcubacteria) were more abundant in PT2 and PT3 than in PT1, eventually as a result of the higher bacterial diversity. Members of these phyla have small genomes and reduced metabolic capabilities (Albertsen et al. 2013; Nelson and Stegen 2015), although yielding several genes involved in complex carbon degradation or sulfate reduction (Kantor et al. 2013), which may explain their higher prevalence in the most recent UWTPs (Zhang et al. 2012; Ye and Zhang 2013).
Except for blaOXA-A, with the highest abundance and prevalence values in PT1, the average values of abundance or of prevalence of ARGs and intI1 did not differ by more than 1.2 log-unit in the different final effluents. This observation suggests that the average loads of ARGs may not differ much in different UWTPs located in the same region. However, the load of ARGs and intI1 suffered variations over the distinct sampling campaigns, and these were observed to be wider than those registered for the bacterial community composition (Fig. 2a vs. Fig. 4b). Interestingly, the distribution of the relative abundance of bacterial community members and of the ARGs and intI1 gene prevalence coincided, with the different UWTPs originating distinct groups (Fig. 2a vs. Fig. 4b). This observation seems to confirm the hypothesis that the selection promoted by wastewater treatment, rather than only horizontal gene transfer, may contribute to explain the occurrence of ARGs in the final effluent.
Bacterial families whose prevalence might be correlated with that of ARGs and intI1 included Aeromonadaceae and Campylobacteraceae (Proteobacteria), and Veillonellaceae (Firmicutes), whose relative abundance was significantly positively correlated with ARGs prevalence (Fig. 3). Surprisingly, none of these bacterial families were correlated with the sul1 gene, one of the most widespread genes in the environment. Except for Aeromonadaceae, these bacterial groups are not among the most probable harbors of the analyzed ARGs. However, these bacterial lineages may represent bacterial groups that co-exist with the ARGs hosts, in particular, because they share similar physiological properties or ecology traits. However, it must be argued that probably groups with relative abundance < 1%, whose tracking is difficult with the technique used to characterize the bacterial community, may, eventually, represent the most important carriers of ARGs in wastewater habitats. Further studies based on epicPCR or long-read sequence analyses may bring new insights into this issue (Spencer et al. 2016; Manaia et al. 2018).
Conclusions
The microbiota present in the final effluent is somehow characteristic of a given UWTP, and it is not strongly influenced by sampling date or season. In a UWTP, ARG prevalence presents higher variation than the relative abundance of bacterial families. The average ARG loads may not differ sharply in different UWTPs located in the same region. And, groups such as Aeromonadaceae, Campylobacteraceae, Veillonellaceae, [Weeksellaceae], and Porphyromonadaceae were observed to be significantly positively correlated with some ARGs.
References
Albertsen M, Hugenholtz P, Skarshewski A, Nielsen KL, Tyson GW, Nielsen PH (2013) Genome sequences of rare, uncultured bacteria obtained by differential coverage binning of multiple metagenomes. Nat Biotechnol 31:533–538. https://doi.org/10.1038/nbt.2579
Becerra-Castro C, Lopes AR, Teixeira S, Silva MEF, Pimenta E, Manaia CM, Nunes OC (2017) Characterization of bacterial communities from Masseiras, a unique Portuguese greenhouse agricultural system. Antonie van Leeuwenhoek, Int J Gen. Mol Microbiol 110:665–676. https://doi.org/10.1007/s10482-017-0833-7
Bengtsson-Palme J, Hammarén R, Pal C, Östman M, Björlenius B, Flach CF, Fick J, Kristiansson E, Tysklind M, Larsson DGJ (2016) Elucidating selection processes for antibiotic resistance in sewage treatment plants using metagenomics. Sci Total Environ 572:697–712. https://doi.org/10.1016/j.scitotenv.2016.06.228
Benjamini Y, Hochberg Y (1995) Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Ser B 57:289–300. https://doi.org/10.2307/2346101
Berendonk TU, Manaia CM, Merlin C, Fatta-Kassinos D, Cytryn E, Walsh F, Bürgmann H, Sørum H, Norström M, Pons MN, Kreuzinger N, Huovinen P, Stefani S, Schwartz T, Kisand V, Baquero F, Martinez JL (2015) Tackling antibiotic resistance: the environmental framework. Nat Rev Microbiol 13:310–317. https://doi.org/10.1038/nrmicro3439
Binh CTT, Petrovich ML, Chaudhary A, et al (2018) Metagenomics reveals the impact of wastewater treatment plants on the dispersal of microorganisms and genes in aquatic sediments. 84:1–15 https://doi.org/10.1128/AEM.02168-17
Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Peña AG, Goodrich JK, Gordon JI, Huttley GA, Kelley ST, Knights D, Koenig JE, Ley RE, Lozupone CA, McDonald D, Muegge BD, Pirrung M, Reeder J, Sevinsky JR, Turnbaugh PJ, Walters WA, Widmann J, Yatsunenko T, Zaneveld J, Knight R (2010) QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7:335–336. https://doi.org/10.1038/nmeth.f.303
Chao A (1984) Non-parametric estimation of the number of classes in a population 11:265–270
DeSantis TZ, Hugenholtz P, Larsen N et al (2006) Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl Environ Microbiol 72:5069–5072. https://doi.org/10.1128/AEM.03006-05
Edgar RC (2010) Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26:2460–2461
Faith DP (1992) Conservation evaluation and phylogenetic diversity. Biol Conserv 87:1–10. https://doi.org/10.1890/0012-9658(2006)87[1465:ATTFHF]2.0.CO;2
Figueira V, Serra E, Manaia CM (2011a) Differential patterns of antimicrobial resistance in population subsets of Escherichia coli isolated from waste- and surface waters. Sci Total Environ 409:1017–1023. https://doi.org/10.1016/j.scitotenv.2010.12.011
Figueira V, Vaz-Moreira I, Silva M, Manaia CM (2011b) Diversity and antibiotic resistance of Aeromonas spp. in drinking and waste water treatment plants. Water Res 45:5599–5611. https://doi.org/10.1016/j.watres.2011.08.021
Gatica J, Tripathi V, Green S, Manaia CM, Berendonk T, Cacace D, Merlin C, Kreuzinger N, Schwartz T, Fatta-Kassinos D, Rizzo L, Schwermer CU, Garelick H, Jurkevitch E, Cytryn E (2016) High throughput analysis of integron gene cassettes in wastewater environments. Environ Sci Technol 50:11825–11836. https://doi.org/10.1021/acs.est.6b03188
Ju F, Li B, Ma L, Wang Y, Huang D, Zhang T (2016) Antibiotic resistance genes and human bacterial pathogens: co-occurrence, removal, and enrichment in municipal sewage sludge digesters. Water Res 91:1–10. https://doi.org/10.1016/j.watres.2015.11.071
Kantor RS, Wrighton KC, Handley KM, Sharon I, Hug LA, Castelle CJ, Thomas BC, Banfield JF (2013) Small genomes and sparse metabolisms of sediment-associated bacteria from four candidate phyla. MBio 4:e00708–e00713. https://doi.org/10.1128/mBio.00708-13
LaPara TM, Nakatsu CH, Pantea LM, Alleman JE (2002) Stability of the bacterial communities supported by a seven-stage biological process treating pharmaceutical wastewater as revealed by PCR-DGGE. Water Res 36:638–646. https://doi.org/10.1016/S0043-1354(01)00277-9
Leps J, Smilauer P (2014) Multivariate analysis of ecological data using CANOCO 5
Lozupone C, Knight R (2005) UniFrac : a new phylogenetic method for comparing microbial communities. Appl Environ Microbiol 71:8228–8235. https://doi.org/10.1128/AEM.71.12.8228
Manaia CM, Macedo G, Fatta-Kassinos D, Nunes OC (2016) Antibiotic resistance in urban aquatic environments: can it be controlled? Appl Microbiol Biotechnol 100:1543–1557. https://doi.org/10.1007/s00253-015-7202-0
Manaia CM, Rocha J, Scaccia N, Marano R, Radu E, Biancullo F, Cerqueira F, Fortunato G, Iakovides IC, Zammit I, Kampouris I, Vaz-Moreira I, Nunes OC (2018) Antibiotic resistance in wastewater treatment plants: tackling the black box. Environ Int 115:312–324
Michael I, Rizzo L, McArdell CS et al (2013) Urban wastewater treatment plants as hotspots for the release of antibiotics in the environment: a review. Water Res 47:957–995. https://doi.org/10.1016/j.watres.2012.11.027
Moreira NFF, Narciso-da-Rocha C, Polo-López MI, Pastrana-Martínez LM, Faria JL, Manaia CM, Fernández-Ibáñez P, Nunes OC, Silva AMT (2018) Solar treatment (H 2 O 2, TiO 2-P25 and GO-TiO 2 photocatalysis, photo-Fenton) of organic micropollutants, human pathogen indicators, antibiotic resistant bacteria and related genes in urban wastewater. Water Res 135:195–206
Munck C, Albertsen M, Telke A, Ellabaan M, Nielsen PH, Sommer MOA (2015) Limited dissemination of the wastewater treatment plant core resistome. Nat Commun 6:8452. https://doi.org/10.1038/ncomms9452
Narciso-da-Rocha C, Rocha J, Vaz-Moreira I, Lira F, Tamames J, Henriques I, Martinez JL, Manaia CM (2018) Bacterial lineages putatively associated with the dissemination of antibiotic resistance genes in a full-scale urban wastewater treatment plant. Environ Int 118:179–188. https://doi.org/10.1016/j.envint.2018.05.040
Nelson WC, Stegen JC (2015) The reduced genomes of Parcubacteria (OD1) contain signatures of a symbiotic lifestyle. Front Microbiol 6. https://doi.org/10.3389/fmicb.2015.00713
Parks DH, Tyson GW, Hugenholtz P, Beiko RG (2014) STAMP: statistical analysis of taxonomic and functional profiles. Bioinformatics 30:3123–3124
Paulson JN, Stine CO, Bravo HC, Pop M (2014) Robust methods for differential abundance analysis in marker gene surveys. Nat Methods 10:1200–1202. https://doi.org/10.1038/nmeth.2658.Robust
Rizzo L, Manaia C, Merlin C, Schwartz T, Dagot C, Ploy MC, Michael I, Fatta-Kassinos D (2013) Urban wastewater treatment plants as hotspots for antibiotic resistant bacteria and genes spread into the environment: a review. Sci Total Environ 447:345–360. https://doi.org/10.1016/j.scitotenv.2013.01.032S0048-9697(13)00042-9 [pii]
Schmieder R, Edwards R (2011) Quality control and preprocessing of metagenomic datasets. Bioinformatics 27:863–864. https://doi.org/10.1093/bioinformatics/btr026
Shannon CE, Weaver W (1963) The mathematical theory of communication. University of Illinois Press, Urbana
Simpson EH (1949) Measurement of diversity. Nature
Spencer SJ, Tamminen MV, Preheim SP, Guo MT, Briggs AW, Brito IL, A Weitz D, Pitkänen LK, Vigneault F, Virta MPJ, Alm EJ (2016) Massively parallel sequencing of single cells by epicPCR links functional genes with phylogenetic markers. ISME J 10:427–436. https://doi.org/10.1038/ismej.2015.124
van den Wollenberg AL (1977) Redundancy analysis an alternative for canonical correlation analysis. Psychometrika 42:207–219
Vaz-Moreira I, Nunes OC, Manaia CM (2014) Bacterial diversity and antibiotic resistance in water habitats: searching the links with the human microbiome. FEMS Microbiol Rev 38:761–778
Vikesland PJ, Pruden A, Alvarez PJJ, Aga D, Bürgmann H, Li XD, Manaia CM, Nambi I, Wigginton K, Zhang T, Zhu YG (2017) Toward a comprehensive strategy to mitigate dissemination of environmental sources of antibiotic resistance. Environ Sci Technol 51:13061–13069. https://doi.org/10.1021/acs.est.7b03623
Wu D, Dolfing J, Xie B (2018) Bacterial perspectives on the dissemination of antibiotic resistance genes in domestic wastewater bio-treatment systems: beneficiary to victim. Appl Microbiol Biotechnol 102:597–604. https://doi.org/10.1007/s00253-017-8665-y
Ye L, Zhang T (2013) Bacterial communities in different sections of a municipal wastewater treatment plant revealed by 16S rDNA 454 pyrosequencing. Appl Microbiol Biotechnol 97:2681–2690. https://doi.org/10.1007/s00253-012-4082-4
Zhang T, Shao M-F, Ye L (2012) 454 Pyrosequencing reveals bacterial diversity of activated sludge from 14 sewage treatment plants. ISME J 6:1137–1147. https://doi.org/10.1038/ismej.2011.188
Acknowledgments
The authors gratefully acknowledge the support of the staff of the UWTPs and supplying entities that made this study possible by providing the water samples; Christophe Merlin that kindly provided the integrase gene qPCR protocol and Gonçalo Macedo and Jaqueline Rocha for technical assistance on sampling, DNA extraction, and qPCR analysis.
Funding
This work was funded by National Funds from FCT – Fundação para a Ciência e a Tecnologia through project WaterJPI/0001/2013 STARE – “Stopping Antibiotic Resistance Evolution,” and UID/Multi/50016/2013. IVM was supported by the FCT grant (SFRH/BPD/87360/2012).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest statement
The authors declare that they have no conflict of interest.
Additional information
Responsible editor: Diane Purchase
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Fernandes, T., Vaz-Moreira, I. & Manaia, C.M. Neighbor urban wastewater treatment plants display distinct profiles of bacterial community and antibiotic resistance genes. Environ Sci Pollut Res 26, 11269–11278 (2019). https://doi.org/10.1007/s11356-019-04546-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11356-019-04546-y