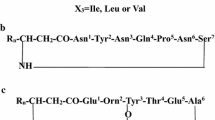Abstract
Bacterial lipopeptides have become a research focus of many studies owing to their industrial and pharmaceutical importance. Although such studies focused on researching purification procedures and qualitative analysis, much remains to be explored and developed to improve the current methods. To enable thorough studies of lipopeptides, this paper describes a new method for purification and characterization of in-gel anionic lipopeptides. Specifically, lipopeptides attributed to the anti-staphylococcal activity of Bacillus mojavensis HF were separated using SDS-PAGE (sodium dodecyl sulphate–polyacrylamide gel electrophoresis) and subsequently characterized using mass spectrometry. Lipopeptide band obtained by gel electrophoresis was first visualized using three different staining methods. Next, the lipopeptide isomers were efficiently recovered from the gel band and structural characterization of the extracted lipopeptides was carried out by matrix-assisted laser desorption ionization time-of-flight mass spectrometry (MALDI-TOF MS). MS analysis revealed that Bacillus mojavensis HF produced three types of lipopeptides including surfactin, fengycin, and kurstakin. 14 clusters of ion peaks were identified as fengycin A with fatty acid of C15-C17, fengycin B (C16, C17), surfactin (C13-C16), and kurstakin (C9-C12). Moreover, tandem mass spectrometric analysis (MS/MS) revealed the sequences of fengycin A and surfactin. In this study, we identified a high variety and number of surfactin and fengycin isomers, which previous reports lacked. To the best of our knowledge, we are the first to report the presence of kurstakin in Bacillus mojavensis species. Finally, we demonstrated that our gel-based study of lipopeptides allowed for a precise and reproducible investigation of these molecules.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
A number of bacterial and fungal species are capable of producing lipopeptides such as the members of genus Bacillus. Cyclic lipopeptides are amphiphilic molecules containing a lipid tail attached to a peptidic part. Amongst various types of lipopeptides, surfactin, fengycin, iturin, and kurstakin are abundant and well-known extracellular lipopeptides with molecular weights of approximately 1.5 kDa (Roy et al. 2013; Meena & Kanwar 2015). The unique physicochemical properties and antimicrobial activity make bacterial lipopeptides attractive compounds for biotechnological and pharmaceutical applications (Naughton et al. 2019). Surfactin consists of a cyclic heptapeptide, which is bonded to a β-hydroxy fatty acid (C12 to C16) resulting in a cyclic lactone ring structure. Natural surfactin is a mixture of isoforms differing in fatty acid chain length and amino acid composition (Chen et al. 2015). Fengycin consists of a decapeptide linked to a saturated or unsaturated fatty acid (C14 to C18). The ring structure of its cyclic peptide is formed by linking the residue at position 3 to the C-terminal –COOH group of the amino acid at position 10, such that eight amino acids are involved in the cyclic structure. The members of iturin family consisting of iturin A, mycosubtilin, bacillomycin, and mojavensin exhibit strong antifungal activity (Ma et al. 2012; Pathak & Keharia 2013; Liu et al. 2020). The peptidyl part of these lipopeptides is a cyclic heptapeptide connected to a β-hydroxy fatty acid chain, which contains from 14 to 17 carbon groups (Meena & Kanwar 2015). Kurstakin is a new family of lipopeptides, which was discovered in 2000. This lipopeptide is a biomarker of Bacillus thuringiensis species and has antifungal activity against Stachybotrys charatum. Although this molecule is a secreted lipopeptide, it is normally associated with the bacterial cells and often not recovered in their cell free culture supernatant. It seems that the high affinity of kurstakin to the cell membrane of kurstakin producing bacteria is due to the cationic charge of histidine on the peptidyl part and its interaction with anionic phospholipid membranes (Béchet et al. 2012). All of the mentioned lipopeptides are produced via non-ribosomal synthesis pathway and contain unusual building blocks. For example, the cyclic structure of the peptide part and the presence of D-amino acids play a unique structural role in bacterial lipopeptides.
Many approaches have been proposed for the purification of lipopeptides; yet, current purification protocols are application specific and often produce fairly crude extracts. The selection and optimization of methods for lipopeptides purification depend on the properties of the molecules such as solubility in water, ionic charge, molecular weight, size, and hydrophilicity. Amphiphilic nature of bacterial lipopeptides is a critical characteristic mainly considered in the selection of purification processes. Examples of previously reported recovery methods for lipopeptide purification are foam fractionation, phase separation, solvent extraction, adsorption onto polymeric resins/ activated carbon, and ultrafiltration (Chen et al. 2008). Use of a sequence of purification steps may be required for an efficient recovery of the lipopeptides. The basic process mainly used for downstream purification of lipopeptides consists of three steps: phase separation using centrifuge or filters (to remove insoluble substances such as cells from cultures), acid precipitation (to reduce the solubility of lipopeptides in water), and solvent extraction (to select lipopeptides based on their solubility in solvents) (Dlamini 2017). Higher purity could be achieved by applying other methods such as thin-layer chromatography (Waewthongrak et al. 2014), size-exclusion chromatography, and reversed-phase high-performance liquid chromatography (Kim et al. 2010; Sharma et al. 2014). In general, gel-free study of lipopeptides has been widely used to investigate a wide range of bacterial samples while there is no report regarding the gel-based identifications of these molecules. Characterization of lipopeptides from acrylamide gel offers various advantages. For example, allowing lipopeptide molecules to separate on the gel results in removal of impurities. In addition, acrylamide gel is a safe matrix with fixed pores commonly used for transport of proteins as well as for protein in-gel digestion and may be suitable for a long-term storage of lipopeptides.
In the present study polyacrylamide gel electrophoresis was employed for purification of anionic lipopeptides. Furthermore, we applied mass spectrometry as a powerful tool to characterize a number of lipopeptide isoforms originating from complex biological extracts of Bacillus mojavensis HF. The new protocol for gel extraction of lipopeptides presented in this work allowed structural determination of in-gel lipopeptides by MALDI-TOF MS analysis.
Materials and methods
Bacterial strain and culture conditions
Lipopeptide-producing bacterium, Bacillus mojavensis HF (GenBank accession number: KX611851.1) which we previously isolated from the water of Iran’s Lake Urmia (Fanaei and Emtiazi 2018) was investigated in this study. The strain was grown in nutrient broth at 30 °C, overnight, and inoculated (in 0.5 McFarland's standard concentration) in productive medium with the following composition (g l−1): glycerol, 15; yeast extract, 15; peptone, 5; MgSO4, 0.5; K2HPO4, 1; pH 7.0 ± 0.2; growth temperature, 37 °C; rotate, 250 rpm. Lipopeptides were harvested in stationary phase.
Lipopeptide extraction
The bacterial cells were removed at the stationary phase of growth (70 h culture broth) by centrifuging at 7750 g for 20 min. Concentrated HCl was added to the supernatant to achieve a final pH of 2.0 and the sample was kept at 4 °C overnight to precipitate formation. The pellet was obtained by centrifugation (7750 g, 20 min), dissolved in chloroform: methanol solvent (2:1 v/v), stirred for 30 min, and centrifuged at 7750 g for 20 min. Soluble fraction was completely evaporated at room temperature to remove the solvent (Smyth et al. 2010). The dried extracted lipopeptides (Gel-free lipopeptides) were analyzed by mass spectrometry. Additionally, these bacterial lipopeptide extracts were separated by SDS-PAGE (Gel-based lipopeptides) and subsequent mass spectrometric analyses of gel extracted lipopeptides were carried out.
SDS-PAGE and identification of lipopeptide band on the gel
The extracted lipopeptides were dissolved in SDS-PAGE loading buffer and SDS-PAGE was carried out with 15% (w/v) separating and 5% (w/v) stacking gels. The voltage was adjusted to 80 V until samples reached running gel afterwards it was set at 120 V (Running time: 90 min). The gel was stained using 0.1% Coomassie Blue R250 in 50% methanol, 40% H2O, and 10% acetic acid for 30 min followed by exposure of the gel to a washing solution (H2O: acetic acid: methanol (8:1:1 v/v)) with several solvent changes to remove SDS and Coomassie stain (Sambrook et al. 1989). Finally, the gel was visualized on white and black backgrounds.
Staining of lipopeptide band with methylene blue and sudan black
The extracted lipopeptide mixture was loaded into two polyacrylamide gel electrophoresis wells. Following the completion of electrophoresis separation, the gel was split into two parts. The first lane was stained with methylene blue (0.1% (w/v) in 3% acetic acid (v/v)) (Kambourova et al. 2001) and the second lane was stained with sudan black (0.5% (w/v) sudan black in 15% acetic acid (v/v)) (Prat et al. 1969; Andrews 1986). Methylene blue and sudan black stained gels were destained with H2O: acetic acid: methanol (8:1:1 v/v) and H2O: acetone: acetic acid (65: 20: 15 v/v) respectively.
Antibacterial activity of lipopeptide band
SDS-PAGE technique was further applied for an investigation of the antibacterial activity of the lipopeptide band (Dehghanifar et al. 2019). The unstained gel was used to excise the lipopeptide band strips. Here, the position of the lipopeptide band on an unstained gel was determined using a stained gel with separated lipopeptide, which was run in parallel. The band strips were placed on the surface of a Mueller Hinton agar plate followed by spreading suspension of Staphylococcus aureus ATCC 6538 adjusted to 0.5 McFarland on the plate. The agar plate was incubated at 37 °C for 24 h and growth-inhibiting effect of the strips was investigated.
Lipopeptide extraction from acrylamide gel
After staining the gel with coomassie brilliant blue and visualization of the lipopeptide band on black background (point 2.3), the following sample preparation steps were carried out to extract the lipopeptides from the acrylamide gel for identification by mass spectrometry:
-
1.
Excise the bright band with a clean scalpel. Cut the excised bands into small pieces and transfer to a microcentrifuge tube.
-
2.
Add H2O: acetone: acetic acid (65: 20: 15 v/v) to completely cover gel pieces. Rock gently followed by centrifuge at 7750 g for 10 min to pellet gel pieces. Remove the liquid phase and discard.
-
3.
Repeat this step until the gel pieces become colorless (about 2 h). This removes stain and non-lipopeptide contaminants, which allows for subsequent lipopeptide mass spectrometric analysis.
-
4.
Wash pellet twice with dH2O.
-
5.
Add 1 mL of chloroform: methanol (2:1) and incubate the sample with shaking overnight at room temperature.
-
6.
Centrifuge at 7750 g for 10 min to pellet gel pieces. Transfer the liquid to a new microcentrifuge tube and allow sample to air dry completely.
-
7.
Add 1 mL of dH2O and lyophilize the sample.
The extracted substance was re-analyzed by SDS-PAGE to investigate the quality of the procedure.
MALDI-TOF mass spectrometry analysis
Gel-free extracted lipopeptides were re-suspended in double-distilled water (dd-H2O) and pH was adjusted to 7.0. The sample was centrifuged at 7750 g for 10 min and supernatant was transferred to a new microcentrifuge tube followed by freeze-drying. Both gel-free and gel-extracted lipopeptides (lipopeptides extracted from the bright band) were freeze dried and analyzed. For MALDI mass spectrometric analysis, lyophilized lipopeptide samples were re-dissolved in a 10% acetonitrile and 0.1% TFA (trifluoroacetic acid) aqueous solution. MALDI matrix, α–cyano–4–hydroxycinnamic acid (CHCA), was prepared as 5 mg mL−1 in 6 mM ammonium phosphate monobasic, 50% acetonitrile, 0.1% trifluoroacetic acid and mixed with the sample at 1:1 ratio (v/v).
Mass Spectrometry data were obtained using an AB Sciex 5800 TOF/TOF System, MALDI TOF TOF (Framingham, MA, USA). Data acquisition and data processing were respectively done using a TOF TOF Series Explorer and Data Explorer (both from AB Sciex). The instrument is equipped with a 349 nm Nd:YLF OptiBeam On-Axis laser. The laser pulse rate is 400 Hz. Reflectron positive and MS/MS positive modes were used. Reflectron and MS/MS modes were externally calibrated at 50 ppm mass tolerance. Each MS mass spectrum was collected as a sum of 500 shots and each MS/MS spectrum as a sum of 1000 shots.
Results
Identification of lipopeptide band on acrylamide gel
After destaining of the gel, a pale low molecular weight band (< 10 kDa) was identified (the following experiments showed that this band was related to lipopeptides) and the sharp blue bands at molecular weights over 10 kDa were identified as protein bands. Interestingly, the lipopeptide band appeared as a bright band against dark background of the gel (Fig. 1).
Staining of lipopeptide band and its antibacterial activity
The lipopeptides were detected as black and purple band on the gels stained with sudan black and methylene blue dyes respectively (Fig. 2, Lane 2 and 3 respectively). The positions of the bands on these gels corresponded to the bright band on the gel stained with Coomassie Brilliant Blu (Fig. 2, Lane 4). Staining of the gel with methylene blue and sudan black dyes demonstrated that the bright band had distinct characteristics that distinguished it from the other bands. Since sudan black dye is specific for lipids and methylene blue stains negatively charged substances, it was suggested that the bright band is an anionic compound containing a lipid moiety. Antibacterial assay of the bands in the sample lane demonstrated that a zone of inhibition against Staphylococcus aureus was observed around the bright band (Fig. 2, Lane 5).
Characterization of in-gel B. mojavensis HF lipopeptides using different staining methods and antibacterial activity. Lane 1: SDS-PAGE gel stained with coomassie blue against white background; lane 2: stained with sudan black; lane 3: methylene blue; and lane 4: coomassie blue against dark background. Lane 5: Antibacterial activity assay of the gel pieces against Staphylococcus aureus. The position of each slice was determined on the parallel gel lane (Lane 4). Slice 1 and 9 contain low and high molecular weight compounds respectively
Characterization of gel-free lipopeptides by MS analysis
Mass spectral analysis of the gel-free bacterial lipopeptide extracts revealed three clusters of peaks in the mass ranges from 600 to 1000 m/z, 1000–1200 m/z, and 1400–1600 m/z. Multiple replicates of spectra were acquired, including the spectra obtained by varying laser intensities, and the representative spectrum of each region is shown in Fig. 3. All of the peaks detected cumulatively are listed in Table 1.
Comparison of the obtained mass spectral data to literature reported masses of bacterial lipopeptides indicated that the ions in the range from 600 to 1000 m/z could be attributed to isoforms of kurstakin (Hathout et al. 2000; Li et al. 2016; Zhi et al. 2017). For example, molecular ions at 895.5, 901.5, and 961.5 were tentatively assigned to C9 [M + 2Na]+, C11 [M + Na]+, and C11 [M + K + 2Na]+ kurstakins respectively (Fig. 3a). The spectra generated by varying laser intensities as well as acquiring MS spectra from multiple spots on the MALDI plate cumulatively showed a variety of low intensity molecular ions similar to kurstakin. In this case, three homologs were detected as [M + H]+ at m/z 865.5, 879.5, and 893.5 differing by 14 Da (i.e. one –CH2 group), which corresponded to C10, C11, and C12 kurstakins respectively. Next, the cluster of peaks between 1000 and 1200 m/z was assigned to surfactin isomers (Li et al. 2016; Zhi et al. 2017). Herein, homologous series of ions differing by 14 Da were detected as sodium/ potassium adduct peaks of surfactin. For example, molecular ions at m/z 1016.7, 1030.7, 1044.7, and 1058.7 could be attributed to [M + Na]+ of C13, C13/C14, C14/C15, and C15 surfactins respectively. Moreover, ions at m/z 1074.7 and 1088.7 were tentatively assigned to [M + K]+ ions of C15 and C16 surfactins while ions at m/z 1066.7 and 1080.7 were assigned to [M + 2Na]+ ions of C14/C15 and C15 surfactin isomers respectively. Molecular ions at 1102.7, 1118.7, and 1134.7 were assigned to [M + 3Na]+, [M + K + 2Na]+, and [M + Na + 2 K]+ ions of C15 surfactin respectively (Fig. 3b). In mass spectra, five isomers of surfactin were detected at a high signal to noise ratio nearing the saturation of detector. Finally, the cluster of peaks in the range of 1400–1600 m/z was assigned to fengycin isomers (de Faria et al. 2011; Li et al. 2016; Dimkić et al. 2017). Examples of the mass spectral signals of fengycin isomers were m/z 1471.8, 1485.8, and 1499.8 (a homologous series differing by 14 Da), which were tentatively assigned to [M + Na]+ ions of C15, C16, and C17 fengycin A respectively. 1477.8 was assigned to [M + H]+ ion of C17 fengycin A. An ion at m/z 1499.8 was suggested to be [M + Na]+ of C17 fengycin A, indicated by the difference of 22 Da from [M + H]+, and an ion at m/z 1515.8 was suggested to be [M + K]+, indicated by the difference of 38 from [M + H]+. Moreover, the molecular ion peak at m/z 1529.8 could be related to [M + K]+ C16 fengycin B (Fig. 3c) (Pathak et al. 2014).
Characterization of gel-extracted lipopeptide band by MS analysis
The spectra obtained by MS analysis of the gel-extracted lipopeptide band indicated the presence of kurstakin (Fig. 4a), surfactin (Fig. 4b), and fengycin isomers (Fig. 4c), which corresponded to MS result obtained from the original gel-free lipopeptide bacterial extract. Molecular ions detected are listed in Table 1 and the representative spectrum of each region is shown in Fig. 4. The result showed that all the isomers of gel-free lipopeptides were detected in gel-extracted sample as well, confirming that the bright band contained kurstakin, fengycin, and surfactin lipopeptides. After reloading the gel-extracted lipopeptide sample for SDS-PAGE re-analysis, the bright band re-occurred, indicating a high quality of the lipopeptide gel extraction process (Fig. 5). As shown in Fig. 5, Lane 1 and 2, both a bright band (lipopeptides) as well as blue bands (proteins) were initially detected from the bacterial lipopeptide extract. However, the re-run of the gel-extracted sample exclusively contained the bright band (Fig. 5, Lane 1 and 3).
Characterization of in-gel lipopeptides purified using the protocol proposed herein. a Bacillus mojavensis HF lipopeptides separated by SDS-PAGE gel and visualized against white (Lane 1) and black (Lane 2) backgrounds. The bright band marked with an arrow was extracted from the gel. b The extracted lipopeptides reloaded on SDS-PAGE gel and lipopeptide free sample as negative control which were visualized on white (Lane 1 and 2 respectively) and black (Lane 3 and 4 respectively) backgrounds
Characterization of fengycin/surfactin lipopeptides by MS–MS analysis
Shown in Fig. 6a and b is the MS/MS spectrum of [M + H]+ fengycin ion at m/z 1477.8. Figure 6c demonstrates some of the major fengycin fragment ions (Sa et al. 2018). Here, the loss of 129.1 Da residue from the fragment at m/z 1209.8 produced m/z 1080.7. A mass of 129.1 Da corresponds to that of Glutamic acid, indicating that Glu is linked to hydroxy fatty acid chain. The ion at m/z 966.6 was related to the loss of Orn with residue mass of 114.1 Da from the fragment of m/z 1080.7. The ions at m/z 966.6 and 1080.7 are y-type fragment ions (y8 and y9 respectively) and they are considered fingerprints of fengycin A (Bie et al. 2009). The product ions b3, m/z 675.5, y4 + H2O, m/z 520.3, as well as y4, m/z 502.3, were also observed confirming the presence of Tyr (position 3) and the fragment containing Ile/Tyr/Gln/Pro (position 7 to 10). Moreover, the zoomed MS/MS spectrum of fengycin precursor at m/z 1477.8 in the mass range to 350 m/z (Fig. 6b) illustrated the ions of m/z 169.1, 226.1, and 292.2, corresponding to Ala/Pro, Pro/Gln and Gln/Tyr residues respectively. Here, additional fragments of Gln (m/z 129.1) and Orn (115.1) as well as immonium ions of Gln, Pro, and Ile (m/z 101.1, 70.1, and 72.1/86.1 respectively) were detected confirming the presence of these amino acids in the structure of the investigated fengycin. The suggested amino acid sequence of the studied fengycin A is shown in Fig. 6c (Sa et al. 2018). Furthermore, the product ions of m/z 1237.5, 1108.5, and 994.4 appeared in MS/MS spectra of the precursor ion of m/z 1477.8, confirming the presence of fengycin B as well (Li et al. 2012).
The MS/MS analysis of surfactin at m/z 1036.7 is demonstrated in Fig. 7. Two isobaric species of surfactin, C15 and C14 surfactins, with sequences C15 b-OH FA-Cyclo[E-I/L-I/L-V-D- I/L-I/L] and C14 b-OH FA-Cyclo[E-I/L-I/L-I/L-D-I/L-I/L] have been suggested based on the ions detected and assigned as illustrated in Fig. 7a (Pathak et al. 2014). A series of both y ions (y2 to y6) and y + H20 ions (y1 + H2O to y6 + H2O) was detected respectively at m/z 227.2, 342.3, 441.3, 554.4, and 667.5 and m/z 132.1, 245.2, 360.3, 459.3, 572.4, and 685.5 indicating the amino acid residues in the order I/L-I/L-D-V-I/L-I/L from the C terminus, as indicated in Figs. 7b and c. Analogue to y-series ions, b series ions were assigned as b (b1 to b6 at m/z 370.3, 483.4, 596.4, 695.6, 810.6, 923.7), b-CO or a ions (a1 to a6 at m/z 342.3, 455.4, 568.5, 667.5, 782.6, 895.7), and b-H2O (b1-H2O to b6-H2O at m/z 352.3, 465.4, 578.5, 677.5, 792.6, 905.7). These ions re-confirmed the C15 surfactin sequence as indicated in 7a. In addition, fragment ions at m/z 455.5, 568.5, and 681.5 were assigned to y4, y5, and y6 respectively. These ions were assigned to a C14 surfactin, where Val in C15 surfactin is replaced by I/L. These ions were further confirmed by complementary b1 and b3 ions at m/z 356.3 and 582.5 as well as a1 to a3 ions at m/z 328.2, 441.3, and 554.4. Moreover, the immonium ions of Leu (86.1 m/z) and Val (72.0 m/z) were observed confirming the presence of these amino acids in the structure of surfactin. The proposed surfactins in this study have been previously reported by Grangemard et al., (1997) and Kowall et al., (1998). The ion assignment done here closely followed the mass spectrometric analysis reported by Pathak et al., (2014). In fact, the presence of a low intensity signal at m/z 808.6 suggests the possibility of the presence of additional two isobaric sequences as identified by Pathak et al., (2014) C15 and C14 surfactins with their respective amino acid order being [E–I/L–I/L–V–I/L– D–I/L] and [E–I/L–I/L–I/L–I/L–D –I/L].
Tandem mass spectrometric analysis of surfactin precursor at m/z 1036.7. The proposed amino acid sequences of cyclic C15 and C14 surfactins are outlined and detected fragment ions are listed with the most intense ions indicated in bold (a). Full MS/MS spectrum (b) and zoomed MS/MS spectrum in the mass range from 100 to 500 Da (c) are shown
Discussion
This study introduces a new method for the extraction of lipopeptide band from acrylamide gel and demonstrates the presence of surfactin, fengycin, and kurstakin lipopeptides in the extract of Bacillus mojavensis HF. For the purpose of this study, methanol/chloroform protocol (as a common lipopeptide extraction method) was modified and was included as a gel extraction step. Methanol is an amphiphilic solvent, which dissolves the molecules containing a polar group attached to a water-insoluble hydrocarbon chain, and chloroform is a hydrophobic solvent, which dissolves the lipopeptides via the lipid moiety (Breil et al. 2017). However, some proteins, lipids, and other impurities dissolve in the organic solvents or exist at the chloroform–methanol interface resulting in inefficient separation and purification of lipopeptide molecules (Vaisar 2009). In this work, the crude lipopeptides were separated using SDS-PAGE gel, which was followed by their extraction from the gel. Although the appearance of the lipopeptide band on the gel has been previously reported (Moyne et al. 2001; Alajlani et al. 2016), to the best of our knowledge there is no clear and concrete information regarding the purification and identification of lipopeptides from the gel. Sudan black has a very high affinity for lipids and staining of the lipopeptide band with this dye confirms the existence of a lipid moiety. Coomassie blue is an anionic dye, which binds to proteins through interaction between sulfonic acid groups of the dye and positive amine groups of proteins. Accordingly, anionic peptides such as polyglutamic acid and polyaspartic acid will stain poorly with coomassie blue due to the absence of NH3 groups (de Moreno et al. 1986). In contrast, methylene blue is a basic dye which reacts with negatively charged molecules. As shown in Fig. 2, it is suggested that the band visualized by methylene blue but not with coomassie blue has anionic nature. To conclude, in the present study sudan black and methylene blue stains identified the lipopeptide band, while coomassie blue complemented the finding. To the best of our knowledge, this is the first report of the detection of anionic lipopeptides as a bright band on the gel.
Mass spectrometric analysis of the bacterial extracts and of the gel-extracted lipopeptides demonstrated the presence of surfactin, fengycin, and kurstakin lipopeptides. Specifically, surfactin produced the most intense signals in mass spectra, which might be attributed to a higher abundance of this lipopeptide in the extracts (Fig. S-1). Surfactins are negatively charged molecules (Uttlová et al. 2016) and the anionic nature of the extracted lipopeptides could be related to the dominance of surfactin isomers in the sample. Moreover, MS/MS analysis revealed that the amino acid sequence of the surfactin isomer at 1036 m/z was likely a mixture of [E-I/L-I/L-V-D-I/L-I/L] and [E-I/L-I/L-I/L-D-I/L-I/L] belonging to C15 and C14 surfactin molecules respectively. Ile/Leu/ Val are hydrophobic amino acids and likely appear in the core of the molecules, while the anionic amino acids (Glu/ Asp) might be present at the surface of surfactin isomers contributing to the overall negative charge of the molecules.
In the present study, a new protocol is proposed to extract lipopeptide band from acrylamide gel followed by characterization of the extracted lipopeptides using mass spectrometry. The protocol herein includes two general phases: gel washing and extraction of lipopeptides. During the washing process, washing solution (H2O: acetone: acetic acid) removes coomassie blue stain and contaminating components from the gel. This solution has been previously applied for destaining of acrylamide gel stained with sudan black (Prat et al. 1969; Andrews 1986). In addition to removing the contaminants, this step is essential for the efficient extraction of lipopeptides. Since lipopeptides are small molecules with few amino acids (< 20 residues), there is no need to in-gel digest the sample (using trypsin or similar protease) prior to extraction (Vaisar 2009). The absence of background peaks in MALDI-TOF mass spectra indicates the precise detection of the lipopeptide band and improved lipopeptide purity during the washing step. In this study, 14 lipopeptide isomers were detected in the gel-free lipopeptide sample extracted from culture supernatant as well as in the gel-extracted lipopeptide sample. Recovery of all the lipopeptide isomers from the gel indicates a high quality of the extraction and analysis process. Re-running the gel-extracted lipopeptide sample on SDS-PAGE resulted in a purified sample with removed protein bands. Here, we demonstrated that the antibacterial activity was in fact related to the lipopeptide band.
There are few reports regarding the production of surfactin, mojavensin (type of iturin), and fengycin by Bacillus mojavensis, but there is no report about the presence of kurstakin in the members of our species. Four isomers of surfactin in B. mojavensis RRC101 (Snook et al. 2009) as well as two isomers of fengycin and an iturin in B. mojavensis B0621 (Ma et al. 2012) have been reported so far. We are the first to report the presence of kurstakin isoforms in the culture supernatant of a Bacillus mogavensis strain. Moreover, the variety of surfactin and fengycin homologues identified in the present study is more than previously reported. These include five surfactin (C13-C16), five fengycin (C15-C17 fengycin A and C16-C17 fengycin B) homologues and four kurstakin isoforms (C9-C12). Finally, MS/MS analyses revealed the amino acid sequences of surfactin and fengycin A.
In addition to structural differences among the isolated lipopeptide isomers from the biosurfactant produced by Bacillus mojavensis HF, these isomers differ in their biological properties. Surfactin is an anionic antibacterial agent while fengycin as well as kurstakin are known antifungal molecules. In fact, Bacillus mojavensis HF and similar bacteria in genus Bacillus are capable of producing conformationally versatile isomers with a range of drug-binding affinities, which makes them candidates of great potential for use in biomedical applications (e.g. drug delivery). Given the necessity for purified lipopeptides in biomedicine, we presented a novel purification approach as a proper tool to improve the existing methods.
The method for purification and detection of lipopeptides described in this work is easy to use, the materials as well as equipment utilized in this process are readily available at low costs and the method is not complex to troubleshoot problems. The proposed method was successfully applied for the separation, gel-visualization, extraction, and mass spectrometric characterization of anionic lipopeptides without any matrix interference and with great precision. Although high-performance liquid chromatography (HPLC) is an efficient method for the separation of the molecules, it has limitations such as availability, cost of equipment and column capacity. The presented gel-based method allows easy identification and collection of anionic compounds and can be used for the pre-concentration of the sample to remove impurities as early in the process. Therefore, this method could be complementary to the existing methods such as HPLC. It is the first report of the detection of anionic lipopeptides as a bright band on polyacrylamide gel, in which the identification is founded on the physiochemical properties of the lipopeptides. With regard to the fact that the detection is not based on comparison with a standard sample, this technique could be an easy method for the screening of new lipopeptides and lipopeptide-producing strains. In addition, we propose the presented method to be readily applicable for isolation of anionic lipopeptides from various microorganisms, for separation of other anionic molecules, and for investigating the effects of different treatments on the lipopeptide production (along with protein profile investigation).
Data availability
The authors confirm that the data supporting the findings of this study are available within the article and its supplementary material.
References
Alajlani M, Shiekh A, Hasnain S, Brantner A (2016) Purification of bioactive lipopeptides produced by Bacillus subtilis strain BIA. Chromatographia 79:1527–1532
Andrews AT (1986) Electrophoresis: theory, techniques and biochemical and clinical applications. Clarendon Press, New York
Béchet M, Caradec T, Hussein W, Abderrahmani A, Chollet M, Leclère V, Dubois T, Lereclus D, Pupin M, Jacques P (2012) Structure, biosynthesis, and properties of kurstakins, nonribosomal lipopeptides from Bacillus spp. Appl Microbiol Biot 95:593–600
Bie X, Lu Z, Lu F (2009) Identification of fengycin homologues from Bacillus subtilis with ESI-MS/CID. J Microbiol Methods 79:272–278
Breil C, Abert Vian M, Zemb T, Kunz W, Chemat F (2017) “Bligh and Dyer” and Folch methods for solid–liquid–liquid extraction of lipids from microorganisms. Comprehension of solvatation mechanisms and towards substitution with alternative solvents. Int J Mol Sci. https://doi.org/10.3390/ijms18040708
Chen HL, Lee YS, Wei YH, Juang RS (2008) Purification of surfactin in pretreated fermentation broths by adsorptive removal of impurities. Biochem Eng J. https://doi.org/10.1016/j.bej.2008.01.020
Chen WC, Juang RS, Wei YH (2015) Applications of a lipopeptide biosurfactant, surfactin, produced by microorganisms. Biochem Eng J. https://doi.org/10.1016/j.bej.2015.07.009
de Faria AF, Stéfani D, Vaz BG, Silva ÍS, Garcia JS, Eberlin MN, Grossman MJ, Alves OL, Durrant LR (2011) Purification and structural characterization of fengycin homologues produced by Bacillus subtilis LSFM-05 grown on raw glycerol. J Ind Microbiol Biotechnol 38:863–871
de Moreno MR, Smith JF, Smith RV (1986) Mechanism studies of coomassie blue and silver staining of proteins. J Pharm Sci 75:907–911
Dehghanifar S, Keyhanfar M, Emtiazi G (2019) Production and partial purification of thermostable bacteriocins from Bacillus pumilus ZED17 and DFAR8 strains with antifungal activity Mol. Biol Res Commun 8:41
Dimkić I, Stanković S, Nišavić M, Petković M, Ristivojević P, Fira D, Berić T (2017) The profile and antimicrobial activity of Bacillus lipopeptide extracts of five potential biocontrol strains. Front Microbiol 8:925
Dlamini B (2017) Downstream purification of surfactin produced by Bacillus subtilis ATCC 21332. Stellenbosch University, Stellenbosch
Fanaei M, Emtiazi G (2018) Microbial assisted (Bacillus mojavensis) production of bio-surfactant lipopeptide with potential pharmaceutical applications and its characterization by MALDI-TOF-MS analysis. J Mol Liq. https://doi.org/10.1016/j.molliq.2018.07.103
Grangemard I, Peypoux F, Wallach J, Das BC, Labbé H, Caille A, Genest M, Maget-Dana R, Ptak M, Bonmatin JM (1997) Lipopeptides with improved properties: structure by NMR, purification by HPLC and structure–activity relationships of new isoleucyl-rich surfactins. J Pept Sci 3:145–154
Hathout Y, Ho YP, Ryzhov V, Demirev P, Fenselau C (2000) Kurstakins: a new class of Lipopeptides isolated from Bacillus thuringiensis. J Nat Prod 63:1492–1496
Kambourova M, Tangney M, Priest FG (2001) Regulation of polyglutamic acid synthesis by glutamate in Bacillus licheniformis and Bacillus subtilis. Appl Environ Microbiol 67:1004–1007
Kim PI, Ryu J, Kim YH, Chi YT (2010) Production of biosurfactant lipopeptides Iturin A, fengycin and surfactin A from Bacillus subtilis CMB32 for control of Colletotrichum gloeosporioides. J Microbiol Biotechnol 20:138–145
Kowall M, Vater J, Kluge B, Stein T, Franke P, Ziessow D (1998) Separation and characterization of surfactin isoforms produced by Bacillus subtilis OKB 105. J Colloid Interface Sci 204:1–8
Li XY, Mao ZC, Wang YH, Wu YX, He YQ, Long CL (2012) ESI LC-MS and MS/MS characterization of antifungal cyclic lipopeptides produced by Bacillus subtilis XF-1. J Mol Microbiol Biotechnol 22:83–93
Li X, Zhang Y, Wei Z, Guan Z, Cai Y, Liao X (2016) Antifungal activity of isolated Bacillus amyloliquefaciens SYBC H47 for the biocontrol of peach gummosis. PloS One 11:e0162125
Liu Y, Teng K, Wang T, Dong E, Zhang M, Tao Y, Zhong J (2020) Antimicrobial Bacillus velezensis HC6: production of three kinds of lipopeptides and biocontrol potential in maize. J Appl Microbiol 128:242–254
Ma Z, Wang N, Hu J, Wang S (2012) Isolation and characterization of a new iturinic lipopeptide, mojavensin A produced by a marine-derived bacterium Bacillus mojavensis B0621A. J Antibiot 65:317–322
Meena KR, Kanwar SS (2015) Lipopeptides as the antifungal and antibacterial agents: applications in food safety and therapeutics. Biomed Res Int. https://doi.org/10.1155/2015/473050
Moyne AL, Shelby R, Cleveland T, Tuzun S (2001) Bacillomycin D: an iturin with antifungal activity against Aspergillus flavus. J Appl Microbiol 90:622–629
Naughton P, Marchant R, Naughton V, Banat I (2019) Microbial biosurfactants: current trends and applications in agricultural and biomedical industries. J Appl Microbiol 127:12–28
Pathak KV, Keharia H (2013) Characterization of fungal antagonistic bacilli isolated from aerial roots of banyan (F icus benghalensis) using intact-cell MALDI-TOF mass spectrometry (ICMS). J Appl Microbiol 114:1300–1310
Pathak KV, Bose A, Keharia H (2014) Characterization of novel lipopeptides produced by Bacillus tequilensis P15 using liquid chromatography coupled electron spray ionization tandem mass spectrometry (LC–ESI–MS/MS). J Pept Res Ther 20:133–143
Prat J, Lamy J, Weill J (1969) Staining of lipoproteins after electrophoresis in polyacrylamide gel. Bulletin de la Societe de Chimie Biologique 51:1367–1367
Roy A, Mahata D, Paul D, Korpole S, Franco OL, Mandal SM (2013) Purification, biochemical characterization and self-assembled structure of a fengycin-like antifungal peptide from Bacillus thuringiensis strain SM1. Front Microbiol 4:332. https://doi.org/10.3389/fmicb.2013.00332
Sa RB, An X, Sui JK, Wang XH, Ji C, Wang CQ, Li Q, Hu YR, Liu X (2018) Purification and structural characterization of fengycin homologues produced by Bacillus subtilis from poplar wood bark. Australas Plant Path 47:259–268
Sambrook J, Fritsch EF, Maniatis T (1989) Molecular cloning: a laboratory manual. Cold spring harbor laboratory press
Sharma D, Mandal SM, Manhas RK (2014) Purification and characterization of a novel lipopeptide from Streptomyces amritsarensis sp. Nov. active against methicillin-resistant Staphylococcus aureus. AMB Express 4:1–9
Smyth T, Perfumo A, McClean S, Marchant R, Banat I (2010) Isolation and Analysis of Lipopeptides and High Molecular Weight Biosurfactants. In: Handbook of hydrocarbon and lipid microbiology. pp 3688–3704
Snook ME, Mitchell T, Hinton DM, Bacon CW (2009) Isolation and characterization of Leu7-surfactin from the endophytic bacterium Bacillus mojavensis RRC 101, a biocontrol agent for Fusarium verticillioides. J Agric Food Chem 57:4287–4292
Uttlová P, Pinkas D, Bechyňková O, Fišer R, Svobodová J, Seydlová G (2016) Bacillus subtilis alters the proportion of major membrane phospholipids in response to surfactin exposure. Biochimica et Biophysica Acta -Biomembranes 1858:2965–2971
Vaisar T (2009) Thematic review series: proteomics. Proteomic analysis of lipid-protein complexes. J Lipid Res 50:781–786
Waewthongrak W, Leelasuphakul W, McCollum G (2014) Cyclic lipopeptides from Bacillus subtilis ABS–S14 elicit defense-related gene expression in citrus fruit. PloS One. https://doi.org/10.1371/journal.pone.0109386
Zhi Y, Wu Q, Xu Y (2017) Genome and transcriptome analysis of surfactin biosynthesis in Bacillus amyloliquefaciens MT45. Sci Rep. https://doi.org/10.1038/srep40976
Acknowledgements
The authors wish to thank the University of Isfahan for financial support of this work.
Funding
The current study was supported by grant from the University of Isfahan to Maryam Fanaei for obtaining PhD degree.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The author declares that they have no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Fanaei, M., Jurcic, K. & Emtiazi, G. Detection of simultaneous production of kurstakin, fengycin and surfactin lipopeptides in Bacillus mojavensis using a novel gel-based method and MALDI-TOF spectrometry. World J Microbiol Biotechnol 37, 97 (2021). https://doi.org/10.1007/s11274-021-03064-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11274-021-03064-9