Abstract
In rice, moderate leaf rolling improves photosynthesis and crop yield. However, the molecular mechanisms underlying this important agronomic trait remain incompletely understood. Here, we investigated a dominant rolled leaf mutant (RL-D) developed from Nipponbare rice (WT). From the six-leaf stage, the leaves of the mutant rolled inward, and abnormal sclerenchyma tissues developed on the abaxial side of the leaf midribs. Additionally, leaf length, plant height, grain weight, and chlorophyll content were significantly greater in the mutant as compared to the WT. Genetic mapping analysis suggested that the leaf-rolling trait in the RL-D mutant was controlled by a single dominant gene, which was located in a 743-kb region on rice chromosome 3. Re-sequencing analysis showed that one gene in the mapped region encoding a protein phosphatase, Os03g0395100 (herein designated OsPP2C), had base mutations in the first exon. These mutations may have produced a truncated form of the OsPP2C protein in RL-D. Further transcriptomic analysis revealed that several biological processes, especially secondary cell wall formation and protein phosphorylation, were overrepresented among the differentially expressed genes (DEGs) between the mutant and the wild type. qRT-PCR verification also demonstrated that specific genes associated with leaf polarity and secondary cell wall formation were differentially expressed in the mutant. This study presents a novel dominant rolled-leaf germplasm that may help to improve rice leaf morphology in the future. The results also suggested that the RL-D phenotype might result from abnormal sclerenchyma tissue development, possibly regulated by OsPP2C via the dephosphorylation pathway. This may present a novel mechanism underlying leaf-rolling in rice.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Rice is a staple food feeding over half of the earth’s population (Muthayya et al. 2014). Changes in leaf morphology, especially moderate leaf rolling, improve photosynthesis and crop yield; thus, leaf rolling is usually considered a crucial agronomic trait in high-yield rice breeding (Xu et al. 2018).
During leaf maturation, the development of adaxial-abaxial polarity plays an important role (Bowman et al. 2002). Leaf adaxial development is mainly controlled by genes in the PHAN or homeodomain-leucine zipper III (HD-ZIPIII) families (Waites et al. 1998; Itoh et al. 2008), while leaf abaxial development is mainly controlled by genes in the YABBY or KANADI families (Dai et al. 2007; Liu et al. 2007a; Toriba et al. 2007; Eshed et al. 2004; Kerstetter et al. 2001; Zhang et al. 2009). Many of the leaf-rolling genes identified in rice to date regulate the development of leaf adaxial-abaxial polarity, including ADL1 (Hibara et al. 2009), ACL1 (Li et al. 2010; Fang et al. 2021), and OsAGO7 (Shi et al. 2007).
Several other genes have been shown to participate in the regulation of leaf rolling in rice, including genes associated with bulliform cells, such as REL1 (Chen et al. 2015), SRL1 (Xiang et al. 2012; Li et al. 2017), RL14 (Fang et al. 2012), Roc5 (Zou et al. 2011), NRL1 (Hu et al. 2010), OsLBD3-7 (Li et al. 2016a, b), REL2 (Yang et al. 2016), Hal1 (Matsumoto et al. 2018), and PSL1 (Zhang et al. 2021); genes associated with the development of sclerenchyma tissue around leaf vascular bundles, such as SLL1 (Zhang et al. 2009), SRL2 (Liu et al. 2016), and NRL2 (Zhao et al. 2016); and genes associated with cuticle development, such as CFL1 (Wu et al. 2011) and OsCHR4 (Guo et al. 2019). Other factors, such as the micro RNAs miR165/166 (Juarez et al. 2004; Mallory et al. 2004) and TAS3 (Fahlgren et al. 2006; Hunter et al. 2006), may indirectly control leaf morphology in plants by regulating downstream genes related to the development of leaf polarity. Some genes involved in the regulation of the auxin (IAA) response, such as miR160 (Mallory et al. 2005; Liu et al. 2007b), or in IAA biosynthesis, such as OsNAL7 (Fujino et al. 2008) and OsFMO(t) (Yi et al. 2013), may also participate in rolled leaf formation in rice, possibly by regulating the development of leaf polarity or vascular bundles.
Nevertheless, the molecular mechanisms underlying the leaf-rolling phenotype are still incompletely characterized in rice. Furthermore, in comparison with recessive mutants, dominant leaf-rolling mutants are advantageous for rice breeding because the hybrid progeny are easily identifiable by the rolled-leaf phenotype. However, to date, most identified rolled-leaf mutants are recessive; only a few dominant mutants have been identified in rice, including REL1 (Chen et al. 2015; Liang et al. 2018), CFL1 (Wu et al. 2011), R05 (Shi et al. 2007), and ACL-D (Xu et al. 2014), and the mechanisms controlling dominant leaf-rolling in rice remain unclear. In order to effectively utilize the dominant rolled-leaf phenotype in rice breeding, it is necessary to identify additional dominant leaf-rolling functional genes and to elucidate the underlying mechanisms.
In our preliminary study, a dominant rolled-leaf mutant (RL-D) was obtained via the ethyl methanesulfonate (EMS) mutagenesis of Nipponbare rice (Oryza sativa L. ssp. japonica) (Chen et al. 2013). However, it was unclear how this mutant differed from previously reported dominant leaf-rolling rice mutants. The underlying cause of this mutation in the RL-D mutant was also unknown. In the present study, the detailed phenotyping, histological alterations, and molecular mechanisms underlying leaf rolling in RL-D were investigated. Genetic mapping showed that the RL-D locus was defined in a 743-kb region on rice chromosome 3, while re-sequencing analysis showed that the first exon of a rice gene (Os03g0395100) encoding protein phosphatase (herein named OsPP2C) contained base mutations. These mutations may have produced a truncated form of the OsPP2C protein in the RL-D mutant. Our results also suggested that several metabolic pathways, especially secondary cell wall formation and protein phosphorylation, might participate in rolled-leaf formation in RL-D. Our study provided a novel dominant rolled-leaf germplasm, which may be valuable for high-yield rice breeding, and presented an analysis of the mechanisms underlying the RL-D mutation.
Materials and Methods
Plant Materials and Experimental Design
The rice RL-D mutant was obtained by performing EMS on suspension-cultured cells of Nipponbare rice (Oryza sativa L. ssp. japonica) (Chen et al. 2013). In the present study, the RL-D mutant was used for phenotypic, histological, transcriptomics, and qRT-PCR analyses, as compared to the wild-type Nipponbare rice variety (herein referred to as WT). In addition, we constructed two F2 populations: one F2 population (744 individuals in total), constructed by crossing RL-D (the male parent) with the wild type (WT), and the second F2 population (689 individuals in total), constructed by crossing RL-D (the male parent) with Jingxian 89 (a widely compatible flat-leaf rice variety), were used for genetic analysis. All 164 recessive (normal flat-leaf) individuals in the second F2 population were used for the molecular mapping of the rolled-leaf locus RL-D.
Phenotypic Investigation
At maturity, 30 RL-D or WT plants were randomly selected for the investigation of agronomic traits, and the rolled-leaf phenotypes were recorded for all plants from each F2 population. The top two leaves from each plant were used to calculate the leaf-rolling index (LRI) as previously described (Wang et al. 2017). Briefly, leaf width was measured in both a natural state (Ln) and an unfolded state (Lw), and then, LRI was calculated as (Lw − Ln)/Lw × 100%. The flag leaves at the maximum booting stage were used for the determination of chlorophyll content. Briefly, leaves from three randomly selected rice plants were ground to powder in liquid nitrogen, and chlorophyll was extracted by incubating 0.1 g of leaf powder in 95% ethanol for 12 h at room temperature in the dark. After incubation, the mixture was centrifuged (12,000 rpm, 10 min), and the chlorophyll absorbance of the supernatant was measured using a BioTek Epoch spectrophotometer at 649 nm and 665 nm. Total chlorophyll content (a + b) was calculated following Li et al. (2019). Three independent biological replicates of all experiments were performed.
Histological Observations of Leaves
Free-hand and semi-/ultra-thin transverse leaf sections were used for histological observation. To produce free-hand sections, the top leaf was harvested from a variety of WT and RL-D rice plants at different stages of growth. Each leaf was cut into slices (each about 100 μm thick) in a Petri dish with a small amount of ddH2O using a sharp blade, and the sections were observed under a normal light microscope. To produce semi-/ultra-thin sections, flag leaves at the boot stage were fixed in a solution of 4% paraformaldehyde-0.25% glutaraldehyde containing 0.1 mmol/L sodium phosphate (pH 7.0) and 0.1% Tween-20 for 24 h, and then transferred to a 1% osmic acid solution for 1.5 h. After fixation, the samples were dehydrated in a series of increasing concentrations of ethanol, infiltrated in acetone, and embedded in Eponate 12TM resin. The embedded samples were then polymerized for 24 h at 65 °C. Semi-thin sections (2 µm thick) and ultra-thin sections (80–90 nm thick) were prepared using a Leica UCT ultramicrotome. Semi-thin sections were stained with 0.1% toluidine blue solution, and then examined and photographed using a Zeiss Axio Observer D1 microscope. Ultra-thin sections were stained with uranyl acetate and lead citrate, and then examined and photographed using a Zeiss EVO MA 15 microscope.
Mapping of the RL-D Locus
The polymorphisms between RL-D (the dominant rolled-leaf mutant, male parent) and Jingxian 89 (female parent) were screened using simple sequence repeat (SSR) markers selected from the Gramene database (http://www.gramene.org/). The polymorphic markers were used for co-segregation analysis with 15 recessive (normal leaf) plants selected from the mapping population of the F2 generation. The identified roughly linked markers were used for an additional segregation analysis with all 164 recessive (normal leaf) individuals in the F2 generation. We used BLAST (https://blast.ncbi.nlm.nih.gov/Blast.cgi) to identify InDel variations between the reference genomic sequences of indica and japonica rice (downloaded from the nucleic acid database GenBank; https://www.ncbi.nlm.nih.gov/genbank/) in the genomic region anchored by the rough-mapped markers. New InDel markers, based on these sequence variations, were designed to fine map the RL-D locus.
Total genomic DNA was extracted from rice leaves for molecular mapping analysis using the CTAB method (Abdel-Latif and Osman 2017), and then used as templates for PCR amplification. To amplify SSR or insertion/deletion (InDel) markers, PCR amplifications were performed in 15.0 μL volumes, each containing 1.0 μL of DNA template, 1.5 μL of 10 × reaction buffer, 0.3 μL of 10 μM dNTPs mixture, 0.3 μL each of 10 μM forward and reverse primers, 0.15 μL (1.0 U) of Taq DNA polymerase, and 11.45 μL of ddH2O, using a MJ Research PTC-100 PCR instrument. The PCR cycling conditions were as follows: 95 °C for 5 min; 30 cycles of 95 °C for 30 s, 58 °C for 30 s, and 72 °C for 30 s; and a final extension of 72 °C for 10 min. The amplified products were subjected to 10% non-denaturing polyacrylamide gel electrophoresis (PAGE) and were visualized using the silver staining method (Xu et al. 2002).
Genetic maps were constructed using Mapmaker3.0 with a minimum likelihood of odd (LOD) threshold of 3.0 (Lander et al. 1987), and the Kosambi function was used to convert the recombinant rates (%) obtained from the molecular marker analysis into genetic distances (centiMorgan, cM). All coding sequences (CDSs) in the fine-mapped genomic DNA region were amplified by PCR and re-sequenced to identify sequence mutations between the RL-D mutant and the WT. Base positions in the mapped region and the locations of the base mutations in the tentative candidate genes were determined based on chromosome 3 of the Nipponbare genome, which is the reference genome for japonica rice varieties. This genome was obtained from the Rice Genome Annotation Project (RGAP, http://rice.plantbiology.msu.edu); we used BLAST against the RGAP database to perform sequence alignment and gene annotation.
Transcriptomic Analysis
We used RNA-seq to identify genes that were differently expressed during early leaf-rolling. As the leaf-rolling phenotype was first observed at the six-leaf stage in the RL-D mutant, we sampled the top (half-expanded) leaves of RL-D and WT Nipponbare plants at the early six-leaf stage. To ensure representative sampling, leaves from three different seedlings were mixed to generate a biological replicate, and three biological replicates were constructed per genotype (18 seedlings in total). Total RNA was extracted from each replicate using TRIzol reagent (Invitrogen, USA), following the manufacturer’s instructions, and then submitted to Novogene (http://www.novogene.com) for transcriptome sequencing. In brief, after RNA qualification, mRNA was enriched with Oligo(dT)-coupled magnetic beads, and then used as template for double-stranded cDNA (dscDNA) synthesis. Subsequently, the purified dscDNA samples were used to construct sequencing libraries and paired-end high-throughput sequencing was performed using a HiSeq 4000 sequencer (Illumina), with a read length of 150 bp (see Table S1 for RNA sequence statistics). After data filtration, the clean reads were compared to the reference genome (Rice Genome Annotation Project for Nipponbare, release 7; http://rice.plantbiology.msu.edu/) using TopHat2 (Kim et al. 2013). Gene expression levels were measured as using the expected number of fragments per kilobase of transcript sequence per millions base pairs sequenced (FPKM) method (Trapnell et al. 2010). DESeq (Anders and Huber 2010, 2013) was used to calculate P–values, and these P–values were adjusted using the Benjamini–Hochberg correction method. We identified differentially expressed genes (DEGs) between the mutant and the WT as those genes where |log2 fold change|≥ 1 and P–value < 0.05. We used GOseq (Young et al. 2010) to determine Gene Ontology (GO) enrichment, and KOBAS 2.0 (Mao et al. 2005) to determine pathway enrichment against the Kyoto Encyclopedia of Genes and Genomes (KEGG) database (https://www.kegg.jp/). Terms were considered significantly enriched in the DEGs when the P–value was < 0.05. Clean RNA-seq data were uploaded to the SRA database (http://www.ncbi.nlm.nih.gov/sra; accession no. PRJNA613143).
Quantitative Real-time RT-PCR (qRT-PCR)
Total RNA from rice leaves at different growth stages was extracted for gene expression analysis using TRIzol reagent (Invitrogen), and then treated with RNase-free DNase I (NEB), following the manufacturer’s instructions. Single-stranded cDNA was synthesized from 1.0 μg of total RNA using HiScript II Q Select RT SuperMix reverse transcriptase kits (Vazyme), and then used as templates for qRT-PCR. qRT-PCR amplifications were performed with specific primers (Table S2) using a Bio-Rad CFX96 and the 2 × SsoFast EvaGreen Supermix (Bio-Rad, USA), following the manufacturer’s instructions. The PCR cycling conditions were as follows: 95 °C for 3 min, followed by 38 cycles of 95 °C for 10 s, 58 °C for 10 s, and 72 °C for 10 s. The expression levels of the target genes were normalized against the expression of O. sativa actin (OsActin2, Os10g0510000) using the 2−ΔΔCt method (Livak and Schmittgen 2001). All PCRs were repeated three times.
Subcellular Localization of the Candidate Gene
Preliminary results identified an O. sativa protein phosphatase (OsPP2C) as the candidate gene for the RL-D locus. To determine whether mutations in this gene in the RL-D mutant altered the subcellular location of OsPP2C, we successively cloned the cauliflower mosaic virus (CaMV) 35S promoter, the green florescence protein (GFP) fragment, and the nopaline synthase (NOS) terminator of Agrobacterium tumefaciens into the pUC18 plasmid to construct the transient expression vector p35S-GFP, as described previously (Yang et al. 2014). The fragments containing the entire OsPP2C coding sequence without an end codon were amplified from the WT (Nipponbare) and the RL-D mutant using qRT-PCR. Each fragment was then inserted into separate p35S-GFP cloning vectors, upstream of GFP, to form WT and mutant p35S-OsPP2C-GFP fusion proteins. As described previously (Chen et al. 2006), recombinant vectors were transformed into rice protoplasts prepared from the leaf sheaths of 2-week-old seedlings of Zhonghua11 (ZH11, Oryza sativa L. ssp. japonica), and transient expression was analyzed.
Results
Phenotypic, Histological, and Physiological Characteristics of the RL-D Mutant
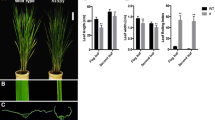
Until the five-leaf stage, the leaves of the RL-D mutant appeared similar to those of the WT (Fig. 1a). However, beginning at the six-leaf stage and continuing to maturity, the leaves of the RL-D mutant exhibited an obvious inward rolling (Fig. 1b–d). At maturity, the LRIs of the first two RL-D leaves (~49%) were significantly greater than those of the first two WT leaves (0%; Fig. 2a; P < 0.01). The first two RL-D leaves were significantly narrower and longer than those of the WT (P < 0.01): the first RL-D leaf was 41% narrower and 30% longer than the first WT leaf, while the second RL-D leaf was 41% narrower and 62% longer than the second WT leaf (Fig. 2b–c). The RL-D mutant was also significantly (19%) taller than the WT (P < 0.01; Fig. 2d). In addition, 100-grain weight and chlorophyll content were significantly greater than those of the WT: 100-grain weight increased by 8% (P < 0.01), and the chlorophyll content of the flag leaves increased by 9% (P < 0.05) (Fig. 2e–f). However, the seed setting rate and productive tiller number of the RL-D mutant were significantly lower than those of the WT (P < 0.01): seed setting rate decreased by 29% in the mutant, while productive tiller number decreased by 13% (Fig. 2g–h).
Quantitative agronomic traits of the rolled-leaf mutant rice (RL-D) as compared to the wild-type Nipponbare rice (WT). Data shown represent means ± SD (n = 30). a Leaf rolling index (LRI). b Leaf length. c Leaf width. d Plant height. e 100-grain weight. f Chlorophyll content. g Seed setting rate. h Productive tiller number. *P < 0.05, **P < 0.01; Student’s t test
Consistent with the observed changes in gross leaf morphology in the RL-D mutant, internal leaf anatomy, as revealed by the free-hand sections, did not differ between the mutant and the WT until the end of the five-leaf stage. However, from the beginning of the six-leaf stage, the sclerenchyma tissues appeared to develop abnormally on the abaxial side of the leaf midribs in the RL-D mutant, although no abnormalities were observed around the leaf lateral veins. No other obvious differences in leaf anatomy were observed between the RL-D and the WT (Fig. 3a–e). Semi-thin and ultra-thin sections confirmed that only the development of sclerenchyma tissues on the abaxial side of the midribs was defective in the RL-D mutant (Fig. 3f–m).
Histological observations of the leaves of wild-type Nipponbare rice (WT) and rolled-leaf mutant rice (RL-D) at the booting stage. b–e Free-hand cross-sections at the middle of the flag leaf as indicated by the red dashed-line box in a. Close-ups of free-hand cross sections of lateral veins (c) and midribs (d, e). Semi-thin cross sections of midribs (f, i) and lateral veins (l, m) of the flag leaf. g, j Close-ups of the solid-line boxed sections of f and i, respectively. h, k Ultra-thin cross sections of the midribs, corresponding to the circled areas in g and j, respectively. Red arrow indicates normal sclerenchyma tissues. sc, sclerenchyma cells; mc, mesophyll cells. Scale bar in b, 1 mm; scale bars c–e, f–g, i–j, and l–m, 100 µm; scale bars in h and k, 2 µm
Genetic Analysis and Molecular Mapping of the RL-D Locus
All plants in the F1 generation produced by crossing the mutant RL-D (male parent) with the WT Nipponbare variety or Jingxian 89 (a flat-leaf variety) expressed RL-D-like leaf rolling phenotypes. In the two F2 generations, the segregation ratios of rolled-leaf (mutant type) to flat-leaf (WT) plants were 563:181 = 3.11:1 (χ2 = 0.15 < χ20.05 = 3.84), and 525:164 = 3.20:1 (χ2 = 0.47 < χ20.05 = 3.84), respectively, consistent with the Mendelian ratio for one pair of alleles (3:1). This suggested that the RL-D locus was controlled by a single dominant gene. Using the F2 population constructed by crossing RL-D (male parent) with Jingxian 89, we located the rolled-leaf locus RL-D in a region about 743 kb long, anchored by InDel15402194 and InDel16145546 on chromosome 3 (Fig. 4). Notably, in the F1 and F2 offspring obtained by crossing RL-D with the WT or Jingxian 89 varieties, no obvious defects were observed in the RL-D phenotypes (e.g., lower seed setting rate and/or tiller number), implying that the RL-D locus may not be responsible for these phenotypical defects.
Molecular map of the RL-D locus and the tentative candidate genes in the mapped region. The numbers in parentheses indicate the recombination events detected by each marker in the F2 mapping population composed of 164 recessive (normal leaf) individuals (from Jingxian 89 × RL-D). Base positions in the mapped region and the locations of the base mutations in the tentative candidate genes are indicated based on chromosome 3 of the Nipponbare genome. Accession numbers in the format “LOC_Os × g × × × × × ” pertain to the Rice Genome Annotation Project database (http://rice.plantbiology.msu.edu), while those in the format “Os × g × × × × × × × ” pertain to the International Rice Annotation Project Database (http://rapdb.dna.affrc.go.jp)
Using the chromosome 3 sequence from the reference genome of the japonica Nipponbare variety (https://www.ncbi.nlm.nih.gov/nuccore/NC_029258.1), we identified 94 coding sequences (CDSs) in the mapped region. However, PCR amplification and re-sequencing of the RL-D and the WT identified only two genes in the mapped region with base mutations in the RL-D mutant: one substitution (G to A) was identified 1885 bp downstream of ATG in the sixth exon of Os03g0389900, corresponding to the conversion of serine to asparagine (Fig. 4 and Fig. S1), and two substitutions (G to T and G to A) were identified 13 bp and 63 bp, respectively, downstream of ATG in the first exon of Os03g0395100 (Fig. 4 and Fig. S1). The G-to-T substitution in Os03g0395100 introduced a termination codon (Fig. S1). Further analysis of the Os03g0395100 coding sequence in the mutant showed that the mutated CDS may be translated from ATG, which was identified 94 bp downstream of the original start codon, producing a truncated form of this protein. The re-sequencing results showed that the promoter regions of these two genes (i.e., the 2-kb region upstream of each start codon) were identical between RL-D and the WT. Os03g0389900 encodes OsFBO14, a protein containing a cyclin-like F-box domain, while Os03g0395100 encodes a protein herein named OsPP2C, containing a phosphatase 2C domain (Fig. 4).
Transcriptomic Analysis of the RL-D Mutant
To investigate the molecular mechanisms underlying the leaf-rolling phenotype of RL-D, we compared the whole genome transcriptomes of the RL-D mutant and the wild type (WT). In total, 551 differentially expressed genes (DEGs) (fold change ≥ 2.0, P < 0.05) were identified between the RL-D mutant and the WT at the early six-leaf stage, at which point the leaf-rolling phenotype is clearly visible in the mutant. Of these DEGs, 396 were upregulated in the RL-D mutant as compared to the WT, and 155 were downregulated (Table S3). Gene Ontology (GO) analysis showed that terms associated with programmed cell death (GO:0012501; GO:0008219), protein phosphorylation or modification (GO:0006468; GO:0016310; GO:0006796; GO:0043687), and the regulation of gene expression or various metabolic processes were overrepresented in the DEGs of RL-D (Table 1). KEGG pathway analysis indicated that pathways associated with the biosynthesis of secondary metabolites (ko01110), as well as various biomolecule biosynthesis and metabolic pathways, were most significantly enriched in the DEGs (Table 2).
Several previously described genes associated with leaf rolling were identified in the RNA-seq data (Table S4) and further verified using qRT-PCR (Fig. 5). The leaf polarity-related genes OsAGO7 (Shi et al. 2007) and OsYABBY1 (Toriba et al. 2007) were significantly upregulated in the RL-D leaves as compared with WT leaves, but OsYABBY5 (Toriba et al. 2007) was significantly downregulated (Fig. 5). SLL1, a gene associated with the sclerenchyma tissue (Zhang et al. 2009), was also significantly upregulated in the RL-D leaves as compared to the WT leaves (Fig. 5). Finally, several other genes related to the secondary cell wall were significantly differently expressed in the RL-D leaves as compared to the WT leaves: OsPAL7 (Tonnessen et al. 2015) and OsCAD7 (Li et al. 2009) were upregulated, while Os4CL3 (Gui et al. 2011), Dirigent1 (Davin and Lewis 2005), OsCesA4, OsCesA7, and OsCesA9 (Tanaka et al. 2003) were downregulated (Fig. 5).
qRT-PCR verification of the RNA-sequencing-detected genes associated with leaf development. Relative gene expression was measured in the top leaves. The 1st top (half-expanded) leaves from rice plants of the rolled-leaf mutant (RL-D) were compared to the wild-type Nipponbare (WT) at the early six-leaf stage. OsActin2 (Os10g0510000) was used as the internal standard, against which the expression levels of the analyzed genes were normalized. Expression levels in the WT were set to 1. Data shown represent means ± SD (n = 3). *P < 0.05, **P < 0.01; Student’s t test
Screening of Candidate Genes for the RL-D Locus
In RNA-seq analysis, Os03g0395100 in the mapped region was significantly upregulated in the RL-D leaves as compared to the WT leaves, corresponding to a 1.5-fold increase in relative expression level (P < 0.05) (Table S4). However, Os03g0389900 was not significantly differentially expressed in the RL-D mutant as compared to the WT (Table S4). To further screen the candidate gene for the RL-D locus, we quantified the expression levels of these two tentative candidate genes during early growth (just before and during the six-leaf stage, as well as during the seven-leaf stage) and during late growth (the tillering stage and the booting stage) using qRT-PCR. The expression levels of Os03g0389900 did not differ significantly between RL-D and the WT at any tested stages (Fig. 6a). However, Os03g0395100 was significantly upregulated in the RL-D mutant as compared to the WT at the five-leaf stage, just before the appearance of leaf rolling (Fig. 6b). This gene was also significantly upregulated compared to the WT at the six- and seven-leaf stages (Fig. 6b).
Relative expression of candidate genes for the RL-D locus in the top leaves from rolled-leaf mutant rice (RL-D) and wild-type Nipponbare rice (WT), as determined using qRT-PCR. (a) Os03g0389900. (b) Os03g0395100. OsActin2 (Os10g0510000) was used as the internal standard, against which the expression levels of the analyzed genes were normalized. Expression levels in the WT were set to 1. Data shown represent means ± SD (n = 3). *P < 0.05, **P < 0.01; Student’s t test
To explain the possible genetic causes of the RL-D mutation, which may be associated with mutations in Os03g0395100 (OsPP2C), we compared the subcellular locations of the WT and mutated proteins. The base mutation in the first exon of OsPP2C in the RL-D mutant may produce a truncated form of OsPP2C protein (as described above) and alter the N-terminal amino acid sequences of the OsPP2C protein in the RL-D mutant. This may shift the subcellular location of the protein. In the WT, the fluorescence signal of the p35S-OsPP2C-GFP fusion protein was observed only in the nucleus (Fig. 7b), in contrast to the GFP control (Fig. 7a). However, in the RL-D mutant, the fusion protein was expressed in both the nucleus and the cytoplasm (Fig. 7c).
Subcellular locations of the protein phosphatase-green florescence protein (OsPP2C-GFP) in leaf sheath protoplasts isolated from 2-week-old Zhonghua11 (ZH11, Oryza sativa L. ssp. japonica) rice seedlings. Brightfield, GFP, DAPI (4′,6-diamidino-2-phenylindole), and merged images of protoplasts are shown. a GFP control. b OsPP2C (WT)::GFP. c OsPP2C (RL-D)::GFP. Scale bars, 10 μm
Discussion
RL-D Represented a Novel Dominant Rolled-Leaf Germplasm Useful for Rice Breeding
In rice, moderate leaf rolling increases yield by improving photosynthetic efficiency, ventilation, and light transmission (Xu et al. 2018). Therefore, rolled-leaf germplasms are of great importance for the breeding of high-yield rice. At present, almost 50 leaf-rolling rice germplasms are available, but most of the reported alleles are recessive, which inhibits the effective large-scale application of the leaf-rolling trait in hybrid rice breeding. The few dominant rolled-leaf rice mutants available to date include REL1, in which the leaves roll from the seedling stage to maturity, but grain yield is reduced (Chen et al. 2015); CFL1, in which the leaves roll only from the late tillering stage (Wu et al. 2011); R05, in which leaf rolling increases only in the later vegetative stages, and chlorophyll content in the leaves is unchanged as compared to wild type (Shi et al. 2007); and ACL-D, which is clearly distinguishable at the three-leaf stage, but exhibits an abaxially rolled phenotype (Xu et al. 2014). Unlike these mutants, the leaves of RL-D rolled adaxially (inward) and were clearly identifiable beginning at the six-leaf stage. In the RL-D mutant, most of the cells in leaf cross sections were normal, except for the sclerenchymatous cells. Moreover, chlorophyll content, grain weight, leaf length, and plant height were all greater in the RL-D mutant as compared to the WT. The mutant also exhibited several negative characteristics, including a lower seed setting rate and a lower tiller number. However, hybrid breeding is usually a process of pyramiding favorable agronomic traits into one target cultivar (Wang et al. 2005). Therefore, RL-D not only represents a novel dominant rolled-leaf rice germplasm, but it also confers several beneficial agronomic traits that may support the future development of rice plants with improved phenotypes. In addition, because RL-D is a dominant moderate leaf-rolling mutation, it is easily identified in growing seedlings. Early identification is helpful when attempting to introduce favorable leaf traits into high-yield rice hybrids. Furthermore, moderate leaf-rolling helps to increase rice planting density and ventilation in the paddy field, which may increase rice yields in agricultural production.
Several Pathways, Including Leaf Polarity Regulation, Secondary Cell Wall Formation, and Protein Phosphorylation, May Participate in RL-D Formation
Previous studies have suggested that rolled-leaf formation in rice is influenced by several factors, including leaf polarity establishment (Xu et al. 2018), bulliform cell development (Xu et al. 2018), auxin (IAA) biosynthesis (Fujino et al. 2008), sclerenchyma tissue development (Zhang et al. 2009; Liu et al. 2016; Zhao et al. 2016), and leaf cuticle formation (Wu et al. 2011). In the RL-D mutant, we observed defective sclerenchyma tissues on the abaxial side of the leaf midribs (Fig. 3). Transcriptomic analysis identified many genes significantly differentially expressed between RL-D and WT. In particular, genes associated with transcription regulation were overrepresented in the DEGs (Table 1), suggesting that the regulation of gene expression is fundamental to the leaf-rolling phenotype in RL-D. Genes in the YABBY family mainly regulate the expression of genes that control the fate of leaf abaxial cells (Liu et al. 2007a; Toriba et al. 2007). Using qRT-PCR analysis, transcriptional changes in two YABBY genes were identified between the RL-D mutant and the WT: OsYABBY5 was downregulated in RL-D, while OsYABBY1 was upregulated (Fig. 5). This was in consistent with a previous study, in which the mRNA levels of the YABBY genes were differentially expressed in OsHox32-overexpressing rice plants with narrow- and rolled-leaf phenotypes (Li et al. 2016a, b). OsAGO7 belongs to the Argonaute (AGO) protein family; AGO proteins help to guide the cleavage of the mRNA of downstream genes, which are usually associated with leaf polarity development (Shi et al. 2007). Here, OsAGO7 was also upregulated in RL-D (Fig. 5). Notably, genes related to various relevant pathways, including programmed cell death, secondary cell wall formation, and protein phosphorylation or modification, were also overrepresented in the RL-D DEGs (Tables 1 and 2).
Sclerenchymatous cells are lignified dead cells with thickened secondary cell walls, and programmed cell death is necessary for the differentiation of mesophyll cells into sclerenchymatous cells (Zhang et al. 2009). Deficiencies in SLL1, SRL2, and NRL2, which directly regulate the development of sclerenchyma tissues, lead to the abnormal programmed cell death of the abaxial mesophyll cells, resulting in leaf rolling (Zhang et al. 2009; Liu et al. 2016; Zhao et al. 2016). Our histological observations identified defective abaxial sclerenchyma tissues around the leaf midrib in the RL-D mutant (Fig. 3). However, of the genes reported to be associated with sclerenchyma tissue development, only SLL1 was significantly upregulated in RL-D as compared to the WT (Fig. 5); the expression levels of the other genes did not differ between these lines (data not shown). This suggested that the mechanism of rolled-leaf formation in RL-D might differ from that previously proposed.
Transcriptional changes in genes associated with secondary cell wall formation were also identified in RL-D. Two genes related to lignin monomer synthesis (Os4CL3 and Dirigent1; Gui et al. 2011; Davin and Lewis 2005), and three genes encoding cellulose synthase catalytic subunits (OsCesA4, OsCesA7, and OsCesA9; Tanaka et al. 2003), which are necessary for the synthesis of secondary cell wall, were downregulated in RL-D as compared to the WT. This suggested that, in RL-D, the synthesis of lignin and cellulose might be reduced, inhibiting the formation of the secondary cell wall. These results were consistent with previous studies, which demonstrated that cell wall formation was repressed in rolled-leaf mutants (Fang et al. 2012). However, both OsPAL7 and OsCAD7 were upregulated in RL-D as compared to the WT. OsPAL7 encodes phenylalanine ammonia-lyase, a key enzyme in the phenylpropanoid pathway; lignin is one of the metabolites of this pathway (Tonnessen et al. 2015). OsCAD7, which encodes cinnamyl alcohol dehydrogenase, also plays an important role in lignin biosynthesis (Li et al. 2009). The upregulation of these genes in RL-D as compared to the WT suggested that lignin biosynthesis in RL-D might be increased, reinforcing the plant cell wall despite the downregulation of Os4CL3, Dirigent1, OsCesA4, OsCesA7, and OsCesA9. Thus, various genes previously shown to be associated with leaf rolling exhibited different or even contradictory expression patterns in RL-D. These inconsistencies, in conjunction with the fact that RL-D is a dominant mutant, implied that a novel molecular mechanism underlies the leaf-rolling phenotype of RL-D.
The tentative candidate PP2C gene (herein named OsPP2C) was identified in the mapped region (Fig. 4). Re-sequencing identified base variations in the first exon of the OsPP2C (Os03g0395100) gene in the RL-D mutant that may lead to premature translation termination or the production of a truncated form of the OsPP2C protein (Fig. S1). This mutation may alter the subcellular location of the OsPP2C protein in the RL-D mutant (Fig. 7). Moreover, processes associated with protein phosphorylation or modification were significantly enriched in the genes differentially expressed in RL-D (Table 1). The OsPP2C gene encodes a protein phosphatase, which may participate in the regulation of multiple signaling transduction pathways via the dephosphorylation of cyclin-dependent kinase (CDK) (Tougane et al. 2010; Umbrasaite et al. 2010; Fan et al. 2013). In rice, PP2C phosphatases may participate in pollen germination, disease resistance, and responses to drought and oxidative stress (Fujii and Toriyama 2008; Hu et al. 2009; You et al. 2014). Although PP2C phosphatases have not previously been implicated in rolled-leaf formation, our results indicated that the leaf-rolling phenotype may be associated with the truncation of PP2C phosphatase or the alteration of its subcellular location in RL-D, although this possibility requires further study. Interestingly, qRT-PCR analysis showed OsPP2C was significantly upregulated just before the appearance of leaf rolling in the mutant plants (that is, at the five-leaf stage) (Fig. 6b). However, no base variations were observed in the promoter region of the OsPP2C gene in RL-D. We speculated that the alteration of subcellular location or the inactivation of the OsPP2C protein in RL-D may stimulate the production of additional OsPP2C mRNA due to a feedback loop. In addition, because RL-D is a “gain-of-function” dominant mutant, the genetic complementation of the RL-D PP2C gene into the WT and experiments knocking-out the WT PP2C gene are needed to confirm OsPP2C as the candidate gene for RL-D.
In the present study, the mapped interval (15,402,194–16,145,546 bp) of the RL-D locus is adjacent to the centromere region (17,882,716–20,185,397) of chromosome 3 (http://plants.ensembl.org/Oryza_sativa/Location/Overview?db=core;r=3:15402194-16145546), which may be one of coldspots on Chr. 3, thus leading to low recombination rate within the mapped interval. The previous study showed that the average physical distance per centimorgan is observed to be 244 kb for the rice genome, which varies with position along the chromosome, with centromere regions exhibiting > 1 Mb/cM (Chen et al. 2002). Generally, the genetic recombination in centromere regions and other heterochromatic regions is severely suppressed (Chen et al. 2002). The previous study also indicated that many low (coldspot)-recombinaniton regions outside centromeric regions were scattered along chromosomes, which exhibited low recombination rate (Wu et al. 2003). However, the repetitive content and gene density in these coldspots are similar to those of regions with normal recombination rates, and the factors that suppress genetic recombination in these regions remain unknown (Wu et al. 2003). Another study indicated that the locus of the rice dwarf1 gene was adjacent to the centromere of chromosome 5, and the mapped region of the locus exhibited low recombination frequency. Finally, the authors used an F2 mapping population with a size as large as 13,000 individuals, including 3,185 recessive (dwarf) plants, to fine-map the dwarf1 locus in rice (Ashikari et al. 1999). In fact, none of the markers we developed in the current mapped interval was recombined with the RL-D locus. As a result, the RL-D locus was only roughly mapped in a 743-kb region (Fig. 4). We supposed that the size of mapping population used in the present study was not large enough for fine mapping.
Conclusions
In summary, our results indicated that the dominant rolled-leaf phenotype of the RL-D mutant represents a novel rolled-leaf germplasm with beneficial agronomic traits, which can be used to develop desired phenotypes. Genetic analyses and re-sequencing assays suggested that OsPP2C (Os03g0395100) may be the candidate gene for the RL-D locus. We propose that the “gain of function” caused by the mutation of the OsPP2C gene in RL-D may regulate the rolled-leaf phenotype by affecting the development of the secondary cell wall, including sclerenchyma tissues and leaf polarity via dephosphorylation in a complex manner; this process may present a novel mechanism underlying leaf rolling in rice. However, the details of OsPP2C function, as well as the molecular mechanisms underlying the association of this gene with the development of sclerenchymatous cells, remain to be investigated. Our results help to clarify the leaf-rolling mechanism in rice, and also lay a foundation for further studies of RL-D and its application in rice breeding.
Availability of Data and Materials
All data supporting the conclusions of this article are provided with the article and its supplementary information files.
References
Abdel-Latif A, Osman G (2017) Comparison of three genomic DNA extraction methods to obtain high DNA quality from maize. Plant Methods 13:1
Anders S, Huber W (2010) Differential expression analysis for sequence count data. Genome Biol 11(10):1–12
Anders S, Huber W (2013) Differential expression of RNA-Seq data at the gene level-the DESeq package. European Molecular Biology Laboratory (EMBL) Heidelberg Germany
Ashikari M, Jianzhong WU, Yano M, Sasaki T, Yoshimura A (1999) Rice gibberrellin-insensitive dwarf mutant gene Dwarf1 encodes the α-subunit of GTP-binding protein. Proc Natl Acad Sci USA 96(18):10284–10289
Bowman JL, Eshed Y, Baum SF (2002) Establishment of polarity in angiosperm lateral organs. Trends Genet 18(3):134–141
Chen M, Presting G, Barbazuk WB, Goicoechea JL, Blackmon B, Fang G, Kim H, Frisch D, Yu Y, Sun S, Higingbottom S, Phimphilai J, Phimphilai D, Thurmond S, Gaudette B, Li P, Liu J, Hatfield J, Main D, Farrar K, Henderson C, Barnett L, Costa R, Williams B, Walser S, Atkins M, Hall C, Budiman MA, Tomkins JP, Luo M, Bancroft I, Salse J, Regad F, Mohapatra T, Singh NK, Tyagi AK, Soderlund C, Dean RA, Wing RA (2002) An integrated physical and genetic map of the rice genome. Plant Cell 14(3):537–545
Chen QL, Xie QJ, Gao J, Wang WY, Sun B, Liu BH, Zhu HT, Peng HF, Zhao HB, Liu CH, Wang J, Zhang JL, Zhang GQ, Zhang ZM (2015) Characterization of Rolled and Erect Leaf 1 in regulating leave morphology in rice. J Exp Bot 66(19):6047
Chen SB, Tao LZ, Zeng LR, Vega-Sanchez ME, Umemura K, Wang GL (2006) A highly efficient transient protoplast system for analyzing defence gene expression and protein-protein interactions in rice. Mol Plant Pathol 7(5):417–427
Chen YL, Liang HL, Ma XL, Lou SL, Xie YY, Liu ZL, Chen LT, Liu YG (2013) An efficient rRice mutagenesis system based on suspension-cultured cells. J Integr Plant Biol 55(2):122–130
Dai MQ, Hu YF, Zhao Y, Liu HF, Zhou DX (2007) A WUSCHEL-LIKE HOMEOBOX gene represses a YABBY gene expression required for rice leaf development. Plant Physiol 144(1):380–390
Davin LB, Lewis NG (2005) Lignin primary structures and dirigent sites. Curr Opin Biotechnol 16(4):407–415
Eshed Y, Izhaki A, Baum SF, Floyd SK, Bowman J (2004) Asymmetric leaf development and blade expansion in Arabidopsis are mediated by KANADI and YABBY activities. Development 131(12):2997–3006
Fahlgren N, Montgomery TA, Howell MD, Allen E, Dvorak SK, Alexander AL, Carrington JC (2006) Regulation of AUXIN RESPONSE FACTOR3 by TAS3 ta-siRNA affects developmental timing and patterning in Arabidopsis. Curr Biol 16(9):939–944
Fan LS, Hao HQ, Xue YQ, Zhang L, Song K, Ding ZJ, Botella MA, Wang HY, Lin JX (2013) Dynamic analysis of Arabidopsis AP2 σ subunit reveals a key role in clathrin-mediated endocytosis and plant development. Development 140(18):3826–3837
Fang JJ, Guo TT, Xie ZW, Chun Y, Zhao JF, Peng LX, Zafar SA, Yuan SJ, Xiao LT, Li XY (2021) The URL1-ROC5-TPL2 transcriptional repressor complex represses the ACL1 gene to modulate leaf rolling in rice. Plant Physiol 185(4):1722–1744
Fang KL, Zhao FM, Cong YF, Sang XC, Du Q, Wang DZ, Li YF, Ling YH, Yang ZL, He GH (2012) Rolling-leaf14 is a 2OG-Fe (II) oxygenase family protein that modulates rice leaf rolling by affecting secondary cell wall formation in leaves. Plant Biotechnol J 10(5):524–532
Fujii S, Toriyama K (2008) DCW11, down-regulated gene 11 in CW-type cytoplasmic male sterile rice, encoding mitochondrial protein phosphatase 2C is related to cytoplasmic male sterility. Plant Cell Physiol (japan) 9(4):633–640
Fujino K, Matsuda Y, Ozawa K, Nishimura T, Koshiba T, Fraaije MW, Sekiguchi H (2008) NARROW LEAF 7 controls leaf shape mediated by auxin in rice. Mol Genet Genom MGG 279(5):499–507
Gui JS, Shen JH, Li LG (2011) Functional characterization of evolutionarily divergent 4-coumarate: coenzyme a Ligases in rice. Plant Physiol 157(2):574–586
Guo TT, Wang DF, Fang JJ, Zhao JF, Yuan SJ, Xiao LT, Li XY (2019) Mutations in the rice OsCHR4 gene, encoding a CHD3 family chromatin remodeler, induce narrow and rolled leaves with increased cuticular wax. Int J Mol Sci 20(10):2567
Hibara KI, Obara M, Hayashida E, Abe M, Ishimaru T, Satoh H, Itoh JI, Nagato Y (2009) The ADAXIALIZED LEAF1 gene functions in leaf and embryonic pattern formation in rice. Dev Biol 334:345–354
Hu J, Zhu L, Zeng DL, Gao ZY, Guo LB, Fang YX, Zhang GH, Dong GJ, Yan MX, Liu J, Qian Q (2010) Identification and characterization of NARROW AND ROLLED LEAF 1, a novel gene regulating leaf morphology and plant architecture in rice. Plant Mol Biol 73(3):283–292
Hu XB, Zhang HJ, Li GJ, Yang YX, Zheng Z, Song FM (2009) Ectopic expression of a rice protein phosphatase 2C gene OsBIPP2C2 in tobacco improves disease resistance. Plant Cell Rep 28(6):985–995
Hunter C, Willmann MR, Wu G, Yoshikawa M, Poethig G-N, S, (2006) Trans-acting siRNA-mediated repression of ETTIN and ARF4 regulates heteroblasty in Arabidopsis. Development 133:2973–2981
Itoh J, Hibara K, Sato Y, Nagato Y (2008) Developmental role and auxin responsiveness of class III homeodomain leucine zipper gene family members in rice. Plant Physiol 147(4):1960–1975
Juarez MT, Kui JS, Thomas J, Heller BA, Timmermans MCP (2004) MicroRNA-mediated repression of rolled leaf1 specifies maize leaf polarity. Nature 428(6978):84–88
Kerstetter RA, Bollman K, Taylor RA, Bomblies K, Poethig RS (2001) KANADI regulates organ polarity in Arabidopsis. Nature 411(6838):706–709
Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL (2013) TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol 14(4):295–311
Lander ES, Green P, Abrahamson J, Barlow A, Daly MJ, Lincoln SE, Newberg LA (1987) MAPMAKER: an interactive computer package for constructing primary genetic linkage maps of experimental and natural populations. Genomics 1(2):174–181
Li C, Zou XH, Zhang CY, Shao QH, Liu J, Liu B, Li HY, Zhao T (2016) OsLBD3–7 overexpression induced adaxially rolled leaves in rice. PLoS One 11(6):e0156413
Li L, Shi ZY, Li L, Shen GZ, Wang XQ, An LS, Zhang JL (2010) Overexpression of ACL 1 (abaxially curled leaf 1) increased bulliform cells and induced abaxial curling of leaf blades in rice. Mol Plant 3(5):807–817
Li WQ, Zhang MJ, Gan PF, Qiao L, Yang SQ, Miao H, Wang GF, Zhang MM, Liu WT, Li HF, Shi CH, Chen KM (2017) CLD1/SRL1 modulates leaf rolling by affecting cell wall formation, epidermis integrity and water homeostasis in rice. Plant J 92(5):904–923
Li XJ, Yang YY, Yao JL, Chen GX, Li XH, Zhang QF, Wu CY (2009) FLEXIBLE CULM 1 encoding a cinnamyl-alcohol dehydrogenase controls culm mechanical strength in rice. Plant Mol Biol 69(6):685–697
Li YY, Shen A, Xiong W, Sun QL, Luo Q, Song T, Li ZL, Luan WJ (2016b) Overexpression of OsHox32 results in pleiotropic effects on plant type architecture and leaf development in rice. Rice (n y) 9(1):46
Li ZY, Mo WP, Jia LQ, Xu YC, Tang WJ, Yang WQ, Guo YL, Lin RC (2019) Rice FLUORESCENT1 is involved in the regulation of chlorophyll. Plant Cell Physiol 60(10):2307–2318
Liu HL, Xu YY, Xu ZH, Chong K (2007a) A rice YABBY gene, OsYABBY4, preferentially expresses in developing vascular tissue. Dev Genes Evol 217(9):629–637
Liu PP, Montgomery TA, Fahlgren N, Kasschau KD, Nonogaki H, Carrington JC (2007b) Repression of AUXIN RESPONSE FACTOR10 by microRNA160 is critical for seed germination and post-germination stages. Plant J 52:133–146
Liu XF, Li M, Liu K, Tang D, Sun MF, Li YF, Shen Y, Du GJ, Cheng ZK (2016) Semi-Rolled Leaf2 modulates rice leaf rolling by regulating abaxial side cell differentiation. J Exp Bot 67(8):2139–2150
Liang JY, Guo SY, Sun B, Liu Q, Chen XH, Peng HF, Zhang ZM, Xie QJ (2018) Constitutive expression of REL1 confers the rice response to drought stress and abscisic acid. Rice (n y) 11(1):59
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCt Method. Methods 25(4):402–408
Mallory AC, Bartel DP, Bartel B (2005) MicroRNA-directed regulation of Arabidopsis AUXIN RESPONSE FACTOR17 is essential for proper development and modulates expression of early auxin response genes. Plant Cell 17:1360–1375
Mallory AC, Reinhart BJ, Jones-Rhoades MW, Tang GL, Zamore PD, Barton MK, Bartel DP (2004) MicroRNA control of PHABULOSA in leaf development: importance of pairing to the microRNA 5′ region. EMBO J 23(16):3356–3364
Mao XZ, Cai T, Olyarchuk JG, Wei LP (2005) Automated genome annotation and pathway identification using the KEGG Orthology (KO) as a controlled vocabulary. Bioinformatics 21(19):3787–3793
Matsumoto H, Yasui Y, Kumamaru T, Hirano HY (2018) Characterization of a half-pipe-like leaf1 mutant that exhibits a curled leaf phenotype. Genes Genet Syst 92(6):287–291
Muthayya S, Sugimoto JD, Montgomery S, Maberly GF (2014) An overview of global rice production, supply, trade, and consumption. Ann N Y Acad Sci 1324:7–14
Shi ZY, Wang J, Wan XS, Shen GZ, Wang XQ, Zhang JL (2007) Over-expression of rice OsAGO7 gene induces upward curling of the leaf blade that enhanced erect-leaf habit. Planta 226(1):99–108
Tanaka K, Murata K, Yamazaki M, Onosato K, Miyao A, Hirochika H (2003) Three distinct rice cellulose synthase catalytic subunit genes required for cellulose synthesis in the secondary wall. Plant Physiol 133(1):73–83
Tonnessen BW, Manosalva P, Lang JM, Baraoidan M, Bordeos A, Mauleon R, Oard J, Hulbert S, Leung H, Leach JE (2015) Rice phenylalanine ammonia-lyase gene OsPAL4 is associated with broad spectrum disease resistance. Plant Mol Biol 87(3):273–286
Toriba T, Harada K, Takamura A, Nakamura H, Ichikawa H, Suzaki T, Hirano HY (2007) Molecular characterization the YABBY gene family in Oryza sativa and expression analysis of OsYABBY1. Mol Genet Genomics 277(5):457–468
Tougane K, Komatsu K, Bhyan SB, Sakata Y, Ishizaki K, Yamato KT, Kohchi T, Takezawa D (2010) Evolutionarily conserved regulatory mechanisms of abscisic acid signaling in land plants:characterization of ABSCISIC ACID INSENSITIVE1-like type 2C protein phosphatase in the liverwort Marchantia polymorpha. Plant Physiol 152(3):1529–1543
Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, Baren MJ, Salzberg SL, Wold BJ, Pachter L (2010) Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol 28(28):511–515
Umbrasaite J, Schweighofer A, Kazanaviciute V, Magyar Z, Ayatollahi Z, Unterwurzacher V, Choopayak C, Boniecka J, Murray JA, Bogre L, Meskiene I (2010) MAPK Phosphatase AP2C3 Induces Ectopic Proliferation of Epidermal Cells Leading to Stomata Development in Arabidopsis. PLoS One 5(12):e15357
Waites R, Selvadurai HR, Oliver IR, Hudson A (1998) The PHANTASTICA gene encodes a MYB transcription factor involved in growth and dorsoventrality of lateral organs in Antirrhinum. Cell 93(5):779–789
Wang XL, Wang FH, Chen HQ, Liang XY, Huang YM, Yi JC (2017) Comparative genomic hybridization and transcriptome sequencing reveal that two genes, OsI_14279 (LOC_Os03g62620) and OsI_10794 (LOC_Os03g14950) regulate the mutation in the γ-rl rice mutant. Physiol Mol Biol Plants 23(4):745–754
Wang YH, Xue YB, Li JY (2005) Towards molecular breeding and improvement of rice in China. Trends Plant Sci 10(12):610–614
Wu J, Mizuno H, Hayashi-Tsugane M, Ito Y, Chiden Y, Fujisawa M, Katagiri S, Saji S, Yoshiki S, Karasawa W, Yoshihara R, Hayashi A, Kobayashi H, Ito K, Hamada M, Okamoto M, Ikeno M, Ichikawa Y, Katayose Y, Yano M, Matsumoto T, Sasaki T (2003) Physical maps and recombination frequency of six rice chromosomes. Plant J 36(5):720–730
Wu RH, Li BS, He S, Wassmann F, Yu CH, Qin GJ, Schreiber L, Qu LJ, Gu HY (2011) CFL1, a WW domain protein, regulates cuticle development by modulating the function of HDG1, a class IV homeodomain transcription factor, in rice and Arabidopsis. Plant Cell 23(9):3392–3411
Xiang JJ, Zhang GH, Qian Q, Xue HW (2012) Semi-rolled leaf1 encodes a putative glycosylphosphatidylinositol- anchored protein and modulates rice leaf rolling by regulating the formation of bulliform cells. Plant Physiol 159(4):1488–1500
Xu PZ, Ali A, Han BL, Wu XJ (2018) Current advances in molecular basis and mechanisms regulating leaf morphology in rice. Front Plant Sci 9:1528
Xu SB, Tao YF, Yang ZQ, Chu JY (2002) A simple and rapid methods used for silver staining and gel preservation. Hereditas 24:335–336
Xu Y, Wang YH, Long QZ, Huang JX, Wang YL, Zhou KN, Zheng M, Sun J, Chen H, Chen SH, Jiang L, Wang CM, Wan JM (2014) Overexpression of OsZHD1, a zinc finger homeodomain class homeobox transcription factor, induces abaxially curled and drooping leaf in rice. Planta 239(4):803–816
Yang JW, Fu JX, Li J, Cheng XL, Li F, Dong JF, Liu ZL, Zhuang CX (2014) A novel co-immunoprecipitation protocol based on protoplast transient gene expression for studying protein-protein interactions in rice. Plant Mol Biol Rep 32:153–161
Yang SQ, Li WQ, Miao H, Gan PF, Qiao L, Chang YL, Shi CH, Chen KM (2016) REL2, a gene encoding an unknown function protein which contains DUF630 and DUF632 domains controls leaf rolling in rice. Rice (n y) 9(1):37
Yi JC, Liu LN, Cao YP, Li JZ, Mei MT (2013) Cloning, characterization and expression of OsFMO(t) in rice encoding a flavin monooxygenase. J Genet 92(3):471–480
You J, Zong W, Hu HH, Li XH, Xiao JH, Xiong LZ (2014) A STRESS-RESPONSIVE NAC1-regulated protein phosphatase gene rice Protein Phosphatase18 modulates drought and oxidative stress tolerance through abscisic acid-independent reactive oxygen species scavenging in rice. Plant Physiol 166(4):2100–2114
Young MD, Wakefield MJ, Smyth GK, Oshlack A (2010) Gene ontology analysis for RNA-seq: accounting for selection bias. Genome Biol 11(2):R14
Zhang GH, Hou X, Wang L, Xu J, Chen J, Fu X, Shen NW, Nian JQ, Jiang ZZ, Hu J, Zhu L, Rao YC, Shi YF, Ren DY, Dong GJ, Gao ZY, Guo LB, Qian Q, Luan S (2021) PHOTO-SENSITIVE LEAF ROLLING 1 encodes a polygalacturonase that modifies cell wall structure and drought tolerance in rice. New Phytol 229:890–901
Zhang GH, Xu Q, Zhu XD, Qian Q, Xue HW (2009) SHALLOT-LIKE1 is a KANADI transcription factor that modulates rice leaf rolling by regulating leaf abaxial cell development. Plant Cell 21:719–735
Zhao SS, Zhao L, Liu FX, Wu YZ, Zhu ZF, Sun CQ, Tan LB (2016) NARROW AND ROLLED LEAF 2 regulates leaf shape, male fertility, and seed size in rice. J Integr Plant Biol 58(12):983–996
Zou LP, Sun XH, Zhang ZG, Liu P, Wu JX, Tian CJ, Qiu JL, Lu TG (2011) Leaf rolling controlled by the homeodomain Leucine Zipper Class IV gene Roc5 in rice. Plant Physiol 156(3):1589–1602
Acknowledgements
We thank Dr. Yuanling Chen at South China Agricultural University for providing rice seeds of Rolled Leaf-Dominant (RL-D) mutant.
Funding
This work was supported by the National Natural Science Foundation of China (Nos. 30671279 and 31071070), the Guangdong Basic and Applied Basic Research Foundation (No. 2020A1515010906), and the Special Fund for Scientific Innovation Strategy-construction of High Level Academy of Agriculture Science (No. R2019PY-JX001).
Author information
Authors and Affiliations
Contributions
XMG and FHW carried out the experiment and prepared the figures and tables. JCY designed the research, analyzed the data, and wrote the manuscript. HMC, XLL, and SCZ contributed to plant materials management and data evaluation. JLZ contributed for critically analyzing the data and reading this manuscript. All authors approved the final version of manuscript.
Corresponding authors
Ethics declarations
Ethics Approval and Consent to Participate
Not applicable.
Consent for Publication
Not applicable.
Competing Interests
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Key Message
• This study investigated the dominant rolled-leaf (RL-D) rice mutant and presented a novel dominant rolled-leaf germplasm that may help to improve rice leaf morphology. The results also suggested that the RL-D phenotype might result from abnormal sclerenchyma tissues, possibly regulated by a protein phosphatase encoding gene OsPP2C via the dephosphorylation pathway.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Gong, X., Wang, F., Chen, H. et al. Genetic Mapping and Transcriptomic Analysis Revealed the Molecular Mechanism Underlying Leaf-Rolling and a Candidate Protein Phosphatase Gene for the Rolled Leaf-Dominant (RL-D) Mutant in Rice. Plant Mol Biol Rep 40, 256–270 (2022). https://doi.org/10.1007/s11105-021-01318-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11105-021-01318-2