Abstract
Flavin monooxygenases (FMO) play a key role in tryptophan (Trp)-dependent indole-acetic acid (IAA) biosynthesis in plants and regulate plant growth and development. In this study, the full-length genomic DNA and cDNA of OsFMO (t), a FMO gene that was originally identified from a rolled-leaf mutant in rice, was isolated and cloned from wild type of the rolled-leaf mutant. OsFMO (t) was found to have four exons and three introns, and encode a protein with 422 amino acid residues that contains two basic conserved motifs, with a ‘G×G××G’ characteristic structure. OsFMO(t) showed high amino acid sequence identity with FMO proteins from other plants, in particular with YUCCA from Arabidopsis, FLOOZY from Petunia, and OsYUCCA1 from rice. Our phylogenetic analysis showed that OsFMO(t) and the homologous FMO proteins belong to the same clade in the evolutionary tree. Overexpression of OsFMO (t) in transformed rice calli produced IAA-excessive phenotypes that showed browning and lethal effects when exogenous auxins such as naphthylacetic acid (NAA) were added to the medium. These results suggested that the OsFMO(t) protein is involved in IAA biosynthesis in rice and its overexpression could lead to the malformation of calli. Spatio-temporal expression analysis using RT-PCR and histochemical analysis for GUS activity revealed that expression of OsFMO (t) was totally absent in the rolled-leaf mutant. However, in the wild type variety, this gene was expressed at different levels temporally and spatially, with the highest expression observed in tissues with fast growth and cell division such as shoot apexes, tender leaves and root tips. Our results demonstrated that IAA biosynthesis regulated by OsFMO (t) is likely localized and might play an essential role in shaping local IAA concentrations which, in turn, is critical for regulating normal growth and development in rice.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
As the endogenous auxin in plants, indole-acetic acid (IAA) is known to regulate diverse processes in plant growth and development (Palme et al., 1991). Recent findings have shown that local auxin biosynthesis plays a key role in shaping the local auxin gradient and is essential for regulating many developmental processes, such as embryogenesis, seedling growth, vascular patterning, leaf formation, etc. (Cheng et al., 2006, 2007; Stepanova et al., 2008; Tao et al., 2008). Due to the complex pathways involved in endogenous IAA biosynthesis, such as several tryptophan (Trp)- dependent and Trp-independent pathways (see figure 1A), the molecular mechanisms and the physiological roles of de novo auxin biosynthesis in plants are still poorly understood (Zhao, 2010).
The endogenous IAA biosynthetic pathways. Adapted and revised from Won et al. (2011), Zhao et al. (2002) and Zhao (2012). (A) Trp-dependent pathways of IAA biosynthesis. (B) A simple two-step pathway for IAA biosynthesis. TAM, tryptamine; IAOx, indole-3-acetaldoxime; IAM, indole-3-acetamide; IAN, indole-3-acetonitrile; IAAld, indole-3-acetaldehyde. TAA, tryptophan aminotransferases of Arabidopsis; YUC, flavin monooxygenases of Arabidopsis.
It has been clearly demonstrated that Trp-dependent auxin biosynthesis is essential for plant developmental processes (Cheng et al., 2006, 2007; Gallavotti et al., 2008; Zhao, 2010). Genetic studies so far have undoubtedly revealed that YUC (flavin monooxygenases of Arabidopsis) and TAA (tryptophan aminotransferases of Arabidopsis) are major players in Trp-dependent IAA biosynthetic pathway (Cheng et al., 2006; Cheng et al., 2007; Zhao et al., 2001; Zhao, 2010, 2012). Other enzymes, such as CYP79B2/B3, nitrilases, aldehyde oxidases and pyruvate decarboxylases, are probably not the main contributors to IAA biosynthesis (Normanly et al., 1997; Zhao et al., 2002; Vande Broek et al., 2005; Sugawara et al., 2009; Mashiguchi et al., 2011).
YUC flavin monooxygenase was previously proposed to catalyze the conversion of tryptamine to N-hydroxyl tryptamine, which may be utilized for IAA production (see figure 1B) (Zhao et al., 2001; Ljung et al., 2002) and TAA tryptophan aminotransferase participated in the step from Trp to indole-3-pyruvate (IPA) (Stepanova et al., 2008; Tao et al., 2008). The enzymes, YUC and TAA, which were previously considered to participate in separate pathways in IAA biosynthesis, are now thought to function in the same auxin biosynthesis pathway (reviewed by Zhao 2010, 2012). Trp is converted into IPA by TAA and IPA is then used as a substrate by YUC to produce IAA in Arabidopsis (Mashiguchi et al., 2011; Won et al., 2011). Genetic studies have shown that this simple two-step pathway is the main mechanism for de novo IAA biosynthesis in plants, and the rate-limiting step for IAA biosynthesis is the second step that is catalyzed by YUC (see figure 1B; Zhao 2010, 2012). The mutation of YUC-like FMO genes in A. thaliana and Petunia, resulted in varied levels of local endogenous IAA (Zhao et al., 2001; Santamaria et al., 2002) and abnormal phenotypes, such as leaf-rolling in these plants (Cheng et al., 2006, 2007).
Rice ‘rolled-leaf’ related genes are important for high photosynthetic efficiency breeding (Lu et al., 2005). Some of these genes encode MYB transcription factor-like proteins (Luo et al., 2007), GARP-like proteins of the KANADI family in A. thaliana (Yan et al., 2006; Zhang et al., 2009) and AGO family proteins (Shao et al., 2005; Shi et al., 2007). In the genetic analysis of a rolled-leaf mutant induced by γ rays from Qinghuazhan (QHZ), a variety of indica rice (Oryza sativa L.), Yi et al. (2007) identified an open reading frame (ORF) encoding flavin monooxygenase (FMO), temporarily named OsFMO (t), as the candidate for the rolled-leaf mutation. In the present study, we isolated OsFMO (t) from QHZ, the corresponding wild type variety, and conducted a sequence analysis based on the sequence of the FMO gene in Nipponbare (O. sativa L. japonica) in the public rice sequence database (http://www.ncbi.nih.gov). We also overexpressed the gene OsFMO (t) to study the role of this gene in auxin biosynthesis. These studies coupled with our spatio-temporal studies suggest an essential role of OsFMO (t) in local de novo IAA biosynthesis in rice.
Materials and methods
Plant materials
Qinghuazhan (QHZ, O. sativa L. indica), the wild type variety (provided by College of Agronomy, South China Agricultural University, Guangzhou, China) corresponding to γ-rl, a rolled-leaf mutant (Yi et al., 2007), was used as the plant material for gene cloning and characterization of OsFMO (t). DNA was extracted from the tender leaves of QHZ’s seedlings by CTAB method (Murray and Thompson 1980). Total RNA was extracted from the tender leaves of seedlings 10 days after germination using Trizol reagent according to the manufacturer’s instructions (Invitrogen, Carlsbad, USA). The methods of extraction and measurement of IAA were adapted from Wang et al. (2002). Rice calli, 100 mg, were weighed accurately and grounded in liquid nitrogen. To these samples, 1 mL of 80% (v/v) precooled (4°C) methanol containing 0.01% butylated hydroxytoluene (BHT) was immediately added, and the samples were then incubated for 4 h at 4°C before centrifuging for 10 min at 10,000 g. The pellets were extracted again in 0.5 mL of 80% (v/v) precooled (4°C) methanol containing 0.01% BHT and recentrifuged in the same manner. Supernatants of these first and second extractions were pooled together and then cleaned-up by the Sep-Pak C18 Cartridge (Waters, Milford, USA). The Sep-Pak C18 Catridge was preconditioned with 5 mL of methanol and 5 mL of 70% (v/v) methanol. The sample solution was passed through the preconditioned C18 column. The column was then rinsed with 5 mL methanol. The elution was dealt with a nitrogen drying step and redissolved in 100 μL of 0.05 mol/L phosphate buffer (pH 7.0). The extract was filtered through a 0.22-μm filter and a portion of 20 μL was injected into a Waters 2695 Series HPLC system equipped with fluorescence detector. Separation was performed on a C18 column (150 mm × 2.1 mm ID, 5 μm) from Elite (Dalian, China). A 5.0 mm-C18 security guard column from Phenomenex (Torrance, USA) was attached to the analytical column. A solvent system consisting of acetonitrile and formic acid-water was used. The gradient profile of the mobile phase was from 5 to 40% (v/v) of acetonitrile and 95 to 60% (v/v) of formic acid over 20 min. The flow rate was 0.4 mL/min.
cDNA cloning and sequence analysis of \(\emph{OsFMO}_{\emph{(t)}}\)
Primers (table 1) were designed using the public rice sequence database (http://www.ncbi.nih.gov) for Nipponbare (O. sativa L. japonica) to amplify full-length genomic DNA and cDNA from OsFMO (t). Using a reverse-transcription kit (Invitrogen, Carlsbad, USA), single-stranded cDNA of the OsFMO (t) gene was first synthesized and used as a template for amplification of the full-length cDNA. PCR was conducted in 50 μL reaction volumes that included 2.0 μL templates, 5.0 μL 10× buffer, 3.0 μL 25 mM MgSO4, 5.0 μL 2 mM dNTPs, 0.5 μL of each primer (5 μM) and 1.0 μL KOD-plus Taq DNA polymerase; distilled water was added to a total volume of 50 μL. In the amplification of cDNA, actin primers (0.5 μL each) were also added as internal standard. The PCR programme was as follows: 94°C for 2 min; 30 cycles of 94°C for 30 s, 55°C for 60 s, 68°C for 2 min; and 68°C for 10 min. The PCR products were examined by electrophoresis using a 1% agarose gel.
The generated nucleotide sequences were verified using BLAST tools (http://blast.ncbi.nlm.nih.gov/). The MEGA 4.0 program (http://www.megasoftware.net/mega4/mega.html) was used for phylogenetic analysis and a phylogenetic tree was constructed using the neighbour-joining (NJ) method (bootstrapping with 1000 replicates). The resulting nucleotide sequences were deposited in GenBank under accession numbers HQ443270 and HQ443271.
Construction of the \(\emph{OsFMO}_{\emph{(t)}}\) overexpression vector
To directly investigate the differential effect of overexpression, we constructed a transgene driven by the maize pUbi promoter. First, the full-length cDNA of OsFMO (t) was amplified from QHZ plants by RT-PCR and digested with BamHI and SpeI, then inserted into a pUbi promoter-driven-pCAMBIA1380, which was used as the binary vector for Agrobacterium-mediated transformation of Zhonghua 11 (O. sativa L. japonica). The pUbi::OsFMOt-Ov, the overexpression fusion vector with the promoter Ubi and OsFMO (t) gene, was verified by sequencing.
Rice transformation and growth conditions
Calli derived from seeds of Zhonghua 11 (O. sativa L. japonica) were used for genetic transformation. GUS-fusion constructs and OsFMO (t) overexpression recombinant vectors were stably transferred into rice using the Agrobacterium-mediated transformation according to the procedures of Hiei et al. (1994). All transformants were confirmed for the presence of the foreign GUS or hygromycin-resistant gene (Hpt) by PCR and Southern blotting. Transgenic plants were transplanted into fields and planted under routine growth conditions.
Expression analysis of \(\emph{OsFMO}_{\emph{(t)}}\)
To understand the expression pattern of OsFMO (t) in QHZ, appropriate primers (table 1) were designed according to the sequences in the region close to OsFMO (t), so as to amplify the promoter region. The amplified fragment was then double digested with BamHI and HindIII and inserted into pCAMBIA1300G to construct pOsFMOt::GUS, the fusion vector with an OsFMO (t) promoter and a GUS reporter gene. The fusion vector was transferred into Zhonghua 11 (O. sativa L. japonica) by Agrobacterium-mediated transformation, and the empty vector pCAMBIA1300G was used as a negative control. Histochemical staining of transformants and observations were conducted according to the methods described by Jefferson et al. (1987). The histochemical GUS assay was performed in a staining solution containing 1.0 mg/mL 5-bromo-4-chloro-3-indoxyl-β-D-glucuronide in 0.1 M sodium phosphate buffer (pH 7.0), 10 mM Na2 EDTA, 0.5 mM potassium ferricyanide/ferrocyanide and 0.1% Triton X-100. Samples were infiltrated under vacuum for 10 min and then incubated overnight at 37°C. The staining solution was removed, and the samples were cleared in 70% ethanol.
RT-PCR analysis was performed similar to the procedure described above in the ‘cDNA cloning and sequence analysis of OsFMO (t)’ section, except that the number of cycles was increased to 35.
Results and discussion
Isolation of \(\emph{OsFMO}_{\emph{(t)}}\) and gene sequence analysis
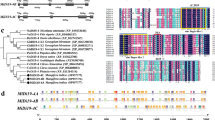
The full-length genomic DNA of OsFMO (t) was found to be 2504 bp (GenBank accession no. HQ443270). Its alignment with the FMO gene (Os03g0162000) from Nipponbare (O. sativa L. japonica) from the rice database (http://www.ncbi.nih.gov) showed that they were completely identical in base sequence. The full-length cDNA sequence of OsFMO (t) (1266 bp) (GenBank accession no. HQ443271) alignment with the cDNA clone (accession no. AK072466) of the FMO gene in Nipponbare showed that these two sequences were also identical. Comparison of the genomic and cDNA sequences of the OsFMO (t) gene, revealed that the gene consisted of four exons and three introns (figure 2A).
(A) Exons and introns of the OsFMO (t) gene. The black and white regions indicate exons and introns, respectively. (B) Comparison of amino acid sequences among OsFMO(t) and the homologous FMO proteins in plants. Two basic conserved motifs are indicated by boxes. YUCCA, YUCCA2 and YUCCA3, FMO-like proteins in A. thaliana; FLOOZY, FMO-like protein in Petunia; OsYUCCA1, FMO-like protein in O. sativa.
Previous studies (Schlenk, 1998; Krueger and Williams, 2005) have indicated that almost all FMO proteins are highly conserved in different kinds of organisms and share a highly conserved domain. Homologic search of FMO genes (http://www.ncbi.nlm.nih.gov/) showed that they are widely distributed in nematodes, bacteria, mammals (including humans) and higher plants. The deduced amino acid sequence of OsFMO(t) containing 422 residues was found to be similar to OsYUCCA1 in rice (Yamamoto et al., 2007), YUCCA in A. thaliana (Zhao et al., 2001) and FLOOZY in Petunia (Santamaria et al., 2002) with a sequence identity of 71, 71 and 70%, respectively. Kubo et al. (1997) and Hou et al. (2011) demonstrated that the conserved domain of FMO contained two basic conserved motifs i.e., a FAD-binding motif and a NADP-binding motif, both of which have the same characteristic structure of ‘G×G××G’ in their amino acid sequences which is essential for binding with flavin-adenine dinucleotide (FAD) and nicotinamide adenine dinucleotide phosphate (NADP), respectively, and hence plays a key role in FMO-catalyzed processes. The results of multiple sequence alignment (see figure 2B) among OsFMO(t) and its homologous proteins YUCCA (Zhao et al., 2001), FLOOZY (Santamaria et al., 2002) and OsYUCCA1 (Yamamoto et al., 2007) showed that these two basic motifs are also highly conserved among these proteins, possessing the same characteristics in base sequence as reported (Kubo et al., 1997; Hou et al., 2011).
By searching the rice genome database (http://www.ncbi.) with the amino acid sequence of OsFMO(t) as query, 32 other putative FMO genes located on different rice chromosomes were found. Results (figure 3) from phylogenetic analysis of these FMO proteins in rice and other homologous FMO proteins identified in A. thaliana and Petunia indicated that the FMO family members in the rice genome were divided into two clades designated as clade I and clade II. In clade I, 19 members of the rice FMO family, excluding OsFMO(t), were clustered with some FMO-like proteins from A. thaliana indicating that clade I was more distant to OsFMO(t) in evolution and probably had different functions relative to OsFMO(t) in rice. In clade II, OsFMO(t) which was investigated in the present study and OsYUCCA1, identified by Yamamoto et al. (2007), were found to be clustered together with YUCCA, YUCCA2 and YUCCA3 from A. thaliana and FLOOZY from Petunia, suggesting that these proteins are closer in evolutionary distance.
Phylogenetic tree of the homologous FMO-like proteins from A. thaliana, Petunia and O. sativa. At, A. thaliana; Os, O. sativa; YUCCA, YUCCA2 and YUCCA3, FMO-like proteins in A. thaliana; FLOOZY, FMO-like protein in Petunia; OsYUCCA1~7, FMO-like proteins in O. sativa. Bootstrap values (1000 replicates) are displayed near the nodes. The scale indicates the number of amino acid residue substitutions per site.
FMO usually participates in xenobiotic metabolism in animals (Krueger and Williams, 2005; Hou et al., 2011). In plants, FMO proteins play an important role in biosynthesis of endogenous IAA, as demonstrated for YUCCA (Zhao et al., 2001). FMO catalyzes the biochemical reaction that converts IPA to IAA, which is the limiting step of the major pathways for Trp-dependent IAA biosynthesis in plants (Zhao, 2012). In our study, OsFMO (t), a FMO gene encoding flavin monooxygenase, was cloned and characterized from the wild type variety (QHZ) of γ-rl, a rice rolled-leaf mutant. The sequence analysis and phylogenetic tree demonstrated that OsFMO (t) has high sequence identity with FMO genes such as YUCCA (Zhao et al., 2001), FLOOZY (Santamaria et al., 2002) and OsYUCCA1 (Yamamoto et al., 2007); and also showed a closer evolutionary relationship with these genes compared to other rice FMO proteins. Therefore, OsFMO (t) may exert functions similar to those of FMO genes in different plants and might play critical roles in auxin biosynthesis and regulating growth and development of rice.
Phenotypes and gene expression analysis of rice transformants with pUbi::OsFMOt-Ov
To investigate the possible role(s) of OsFMO (t), we transformed pUbi::OsFMOt-Ov into calli of rice (supplementary data in electronic supplementary material at http://www.ias.ac.in/jgenet/). As shown in figure 4C, all hygromycin-resistant (Hpt + ) calli (45 total) that were transformed with the construct would not proliferate and differentiate normally in the auxin-supplemented differentiation medium (NAA added at 1.0 mg/L), and large numbers of hairy roots were observed. These calli ultimately browned and died (figure 4C). On the other hand, when vector pCAMBIA1380 without OsFMO (t) (control) was transformed, the calli (Hpt + , 33 total) regenerated normal shoots in the same medium (figure 4A). Further, when calli overexpressing OsFMO (t) were grown in the medium without NAA (figure 4D), around 42% of them sustained for around two months, though eventually still unable to regenerate into shoots. Additionally, this type of Hpt + calli showed significant phenotypes, such as large numbers of adventitious roots growing (figure 4a, red solid arrow), abundant root hairs (figure 4, e&f), malformed leaves and stems (figure 4, a,b&c, red solid arrow) and adventitious roots on the malformed stems (figure 4, c&d), which were associated with IAA overproduction phenotypes reported previously (Zhao et al., 2001; Yamamoto et al., 2007).
Phenotypes of hygromycin-resistant (Hpt + ) calli transformed by pUbi::OsFMOt-Ov. (A–D) Differentiation cultivation of the Hpt + calli (A & B, transformed by pCAMBIA1380, the negative control; (C & D), transformed by pUbi::OsFMOt-Ov); a–f, IAA-excessive phenotypes from OsFMO (t) overexpression (a, red solid arrow shows adventitious roots; b, abnormal leaf; c, red solid arrow shows abnormal stem; d, adventitious roots on abnormal stem; e & f, adventitious roots with abundant root hairs); (A & C), medium with exogenous naphthylacetic acid (NAA) at a final concentration of 1.0 mg/L; (B & D), medium without exogenous NAA; (E), RT-PCR analysis (35 cycles) of OsFMO (t); (F), IAA analysis of rice calli. CK, the Hpt + callus transformed by pCAMBIA1380; I & II, two Hpt + calli transformed by pUbi::OsFMOt-Ov.
Using RT-PCR (35 cycles), expression of OsFMO (t) was analysed within two of these survived calli (Hpt + ) transformed by pUbi::OsFMOt-Ov. The expression level of OsFMO (t) was different between them (shown in figure 4E), suggesting an expression variation caused by different transformation events. Expression product was not detected in the Hpt + calli transformed with the control vector, indicating that the expression level of this gene is low in normal tissues. In addition, the IAA contents in the two survived calli increased with different levels as compared to the control (figure 4F). These results indicated that transformation of pUbi::OsFMOt-Ov did increase OsFMO (t) transcription level in the calli, and the excessive auxin produced in these transformants due to the overexpression of OsFMO (t) (which encodes FMO associated with IAA biosynthesis in vivo), could seriously affect shoot regeneration which, in turn, might inhibit normal growth of the calli that ultimately led to the death of the transformants.
Genetic analyses with overexpression and loss of function mutations revealed that FMO plays an important role in de novo IAA synthesis, both in dicots (A. thaliana) and monocots (rice) (Cheng et al., 2006, 2007; Zhao, 2010). Overexpression of FMO genes is usually accompanied by IAA-excessive phenotypes in plants. For example, overexpression of YUCCA in A. thaliana (Zhao et al., 2001) led to vigorous growth of explants and emergence of large numbers of root hairs in MS medium without IAA. OsYUCCA1-overexpression and NAL7-overexpression in rice also led to the emergence of identical auxin-excessive phenotypes, such as the appearance of large numbers of hairy roots and difficulty in regenerating transgenic seedlings (Yamamoto et al., 2007; Fujino et al., 2008). Our study reiterates this fact, as seen from figure 4C, where calli from transformants overexpressing OsFMO (t) showed browning and eventually died. These calli showed the IAA overproduction phenotypes even in the absence of NAA from the medium (figure 4D, a–f), suggesting that OsFMO (t) plays a key role in biosynthesis of endogenous IAA in rice.
Spatio-temporal study using histochemical staining and RT-PCR
By using rice plants developed from calli transformed with pOsFMOt::GUS (supplementary data in electronic supplementary material), expression of OsFMO (t) in various tissues was observed. As shown in figure 5A, GUS activity was detected in roots and tender leaves of seedlings five days after germination, and was high in root tips and the prophyll leaf tip (figure 5a′). The intensity of GUS staining was also strongly enhanced in shoot apex (figure 5b′) and lateral root tips (figure 5c′) at 15-day-old seedling stage. Intense staining was also observed in some parts of the adult plant, e.g. tender auricles (figure 5e′), filaments (figure 5f′), paleae and lemmas (figure 5g′), as well as episperm (figure 5h′) of seeds during the grain filling stage and lateral roots on the matured roots (figure 5d′); but not in other matured parts such as the matured leaves. In summary, GUS staining was mainly observed in the tender parts of plants undergoing active growth and cell division and became undetectable in the old tissues. The results indicated that OsFMO (t) expression was closely associated with the development of tender organs during rice growth and development.
(A) The results of histochemical staining in rice plants transformed with pOsFMOt::GUS. (a–i) Negative control; (a′–i′) various tissues of transgenic rice plants harbouring pOsFMOt::GUS; (a & a′) seedling at five days after germination; (b & b′) stem apex at 15-day-old seedling stage; (c & c′) lateral roots at 15-day-old seedling stage; (d & d′) lateral roots on older root at tillering stage; (e & e′) auricles at 30-day-old seedling stage; (f & f′) spikelet and filaments at heading stage; (g & g′) spikelets at grain filling stage; (h & h′) seed coat at grain filling stage; (i & i′) rice calli at preliminary differentiation stage. Scale bar = 2 mm. (B) Reverse transcription pattern of OsFMO (t) in different organs (35 cycles). γ-rl, the rolled-leaf mutant; QHZ, the wild type variety; DAG: day after germination.
RT-PCR (35 cycles) was also performed to further analyse the OsFMO (t) gene expression in tender organs at different stages, including roots and aerial parts at the seedling stage (five days after germination, DAG); stem apices and tender leaves at the active tillering stage (60 DAG); and tender roots, leaves and panicles at the booting stage (80 DAG). The rice housekeeping gene Actin was amplified at the same time as an internal standard control as shown in figure 5B. OsFMO (t) gene expression products were amplified in tender roots, leaves and panicles of QHZ (wild type variety), indicating that OsFMO (t) was expressed in the young parts of the seedlings and plants, with a higher expression in tender leaves and panicles at the booting stage. However, amplifications of the specific products failed in more mature tissues with slow cell growth such as in matured leaves and old roots (data not shown). This might be due to the fact that the expression of this gene in these mature tissues was too low to be detected by RT-PCR. Our previous study (Yi et al., 2007) showed that a large deletion might occur around the OsFMO (t) locus in the genome of γ-rl, the rolled-leaf mutant. Consistent with our previous findings, we did not observe expression of this gene in any of the organs of γ-rl. (figure 5B).
For a long time, the predominant view has been that the shoots were the only source of auxin biosynthesis. It has been postulated that polar auxin transport is responsible for auxin distribution to other parts of the plant and auxin gradient maintenance (Tanaka et al., 2006; Grieneisen et al., 2007). However, recent findings clearly demonstrate that de novo auxin biosynthesis is regulated both temporally and spatially and contributes to local auxin gradient generation and maintenance (Cheng et al., 2007; Stepanova et al., 2008; Tao et al., 2008; Zhao, 2010, 2012). FMO genes such as TAA and YUC expressed with temporal and spatial precision in all organs including shoots, roots and flowers and leaves in Arabidopsis and each organ appears to be selfsufficient in terms of controlling auxin gradients for growth and development (Zhao, 2010, 2012). Our results from histochemical staining and RT-PCR indicate that the expression of OsFMO (t) varies in different organs as well as different developing stages. Generally, its expression level is higher in tender tissues with faster growth, especially in root tips and shoot apices with vigorous cell division; which are the major sources of endogenous IAA in plants. Since FMOs are involved in catalyzing the limiting step of Trp-dependent de novo IAA biosynthesis, the higher expression of the OsFMO (t) gene restricted to these small parts suggests that de novo auxin biosynthesis regulated by this gene is possibly highly localized and local auxin biosynthesis appears to be an important aspect of shaping local auxin concentrations for regulating organ growth and development. Mutations in this gene might affect normal development of organs such as leaves.
OsYUCCA1 was the first reported FMO gene in rice and is located on chromosome 1 (Yamamoto et al., 2007). OsFMO (t), located on chromosome 3 (Yi et al., 2007), may play a role that is different from OsYUCCA1. OsYUCCA1 expressed highly in all the investigated tissues; while OsFMO (t) expression were detected only in young and fast growth organs, as demonstrated in this study. Transformed calli with OsYUCCA1 overexpression could successfully regenerate into seedlings despite of abnormal phenotypes. The calli overexpressing OsFMO (t), however, survived only on medium without NAA and failed to regenerate, indicated that OsFMO (t) and OsYUCCA1 had similarity as well as differences in tissue expression specificity and functions.
Fujino et al. (2008) reported the spontaneous rice narrow-leaf mutant, nal7, was resulted from a single base mutation in NAL7 also located on chromosome 3. Overexpression of both OsFMO (t) and NAL7 gene could produce same IAA-excessive phenotypes. Preliminary studies indicated that a large deletion in the vicinity of OsFMO (t) occurred in the genome of γ-rl mutant (Yi et al., 2007). RT-PCR anaylsis could not detect expression of OsFMO (t) in different organs of this mutant. On the other hand, expression of NAL7 was detected in various organs in the nal7 mutant. In some organs such as in panicles, its expression in the mutant was even higher than that in the wild type (Fujino et al., 2008). In addition, the phenotypes of γ-rl and nal7 mutants were also different, γ-rl displayed as rolled leaf in the whole plant, reduced flag-leaf area, increased tiller number, enlarged midrib, reduced lateral veins number, reduced total root length, root number (data not shown), etc.; while nal7 only observed with narrowed leaf, abnormal bulliform cells, but no changes in its root development and tiller number (Fujino et al., 2008). The results indicated that OsFMO (t) and NAL7 had similarity as well as differences in mutation phenotypes and functions.
In summary, the OsFMO (t) gene was cloned and characterized in the present study. The results from sequence identity and phylogenetics analyses suggested that this gene possessed similar functions to that of other homologous FMO genes in plants. The results from overexpression transformation and gene expression analysis indicated that OsFMO (t) is involved in endogenous synthesis of IAA in rice and expressed in young plant tissues with vigorous growth and cell division. Our study suggests a possible essential role for OsFMO (t) in local biosynthesis of IAA and maintenance of local IAA concentrations, which are undoubtedly critical for regulating growth and development of rice.
References
Cheng Y., Dai X. and Zhao Y. 2006 Auxin biosynthesis by the YUCCA flavin monooxygenases controls the formation of floral organs and vascular tissues in Arabidopsis. Genes Dev. 20, 1790–1799.
Cheng Y., Dai X. and Zhao Y. 2007 Auxin synthesized by the YUCCA flavin monooxygenases is essential for embryogenesis and leaf formation in Arabidopsis. Plant Cell 19, 2430–2439.
Fujino K., Matsuda Y., Ozawa K., Nishimura T., Koshiba T., Fraaije M. W. and Sekiguchi H. 2008 NARROW LEAF 7 controls leaf shape mediated by auxin in rice. Mol. Genet. Genomics 279, 499–507.
Gallavotti A., Barazesh S., Malcomber S., Hall D., Jackson D., Schmidt R. J. and McSteen P. 2008 Sparse inflorescence 1 encodes a monocot specific YUCCA-like gene required for vegetative and reproductive development in maize. Proc. Natl. Acad. Sci. USA 105, 15196–15201.
Grieneisen V. A., Xu J., Marée A. F., Hogeweg P. and Scheres B. 2007 Auxin transport is sufficient to generate a maximum and gradient guiding root growth. Nature 449, 1008–1013.
Hiei Y., Ohta S., Komari T. and Kumashiro T. 1994 Efficient transformation of rice (Oryza sativa L.) mediated by Agrobacterium and sequence analysis of the boundaries of the T-DNA. Plant J. 6, 271–282.
Hou X., Liu S., Pierri F., Dai X., Qu L. and Zhao Y. 2011 Allelic analyses of the Arabidopsis YUC1 locus reveal residues and domains essential for the functions of YUC family of flavin monooxygenases. J. Integr. Plant Biol. 53, 54–62.
Jefferson R., Kavanagh T. and Bevan W. 1987 GUS fusions: β-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 6, 3901–3907.
Krueger S. K. and Williams D. E. 2005 Mammalian flavin-containing monooxygenases: structure/function, genetic polymorphisms and role in drug metabolism. Pharmacol. Ther. 106, 357–387.
Kubo A., Itoh S. and Itoh K. 1997 Determination of FAD-binding domain in flavin-containing monooxygenase 1 (FMO1). Arch. Biochem. Biophys. 345, 271–277.
Ljung K., Hul A. K., Kowalczyk M., Marchant A., Celenza J., Cohen J. D. and Sandberg G. 2002 Biosynthesis, conjugation, catabolism and homeostasis of indole-3-acetic acid in Arabidopsis thaliana. Plant Mol. Biol. 50, 309–332.
Lu J., Lang Y., Zhang Z. and Zhu Q. 2005 Comparison of plant morphology and population structure and photosynthetic characteristics among a series of near-isogenic lines with different leaf rolling index in rice. J. Yangzhou Univ. (Agri. Life Sci. Edition) 26, 56–60.
Luo Z., Yang Z., Zhong B., Li Y., Xie R., Zhao F. et al. 2007 Genetic analysis and fine mapping of a dynamic rolled leaf gene, RL10(t), in rice (Oryza sativa L.). Genome 50, 811–817.
Mashiguchi K., Tanaka K., Sakai T., Sugawara S., Kawaide H., Natsume M. et al. 2011 The main auxin biosynthesis pathway in Arabidopsis. Proc. Natl. Acad. Sci. USA 108, 18512–18517.
Murray M. G. and Thompson W. F. 1980 Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res. 8, 4321–4325.
Normanly J., Grisafi P., Fink G. R. and Bartel B. 1997 Arabidopsis mutants resistant to the auxin effects of indole-3-acetonitrile are defective in the nitrilase encoded by the NIT1 gene. Plant Cell 9, 1781–1790.
Palme K., Hesse T., Moore I., Campos N., Feldwisch J., Garbers C. et al. 1991 Hormonal modulation of plant growth: the role of auxin perception. Mech. Dev. 33, 97–106.
Santamaria R. T., Bliek M., Ljung K., Sandberg G., Mol J. N., Souer E. and Koes R. 2002 FLOOZY of petunia is a flavin-mono-oxygenase-like protein required for the specification of leaf and flower architecture. Genes Dev. 16, 753–763.
Schlenk D. 1998 Occurrence of flavin-containing monooxygenases in non-mammalian eukaryotic organisms. Comp. Biochem. Physiol. C Pharmacol. Toxicol. Endocrinol. 121, 185–195.
Shao Y., Chen Z., Zhang Y., Chen E., Qi D., Miao J. and Pan X. 2005 One major QTL mapping and physical map construction for rolled leaf in rice. Acta Genet. Sin. 32, 501–506.
Shi Z., Wang J., Wan X., Shen G., Wang X. and Zhang J. 2007 Over-expression of rice OsAGO7 gene induces upward curling of the leaf blade that enhanced erect-leaf habit. Planta 226, 99– 108.
Stepanova A. N., Robertson-Hoyt J., Yun J., Benavente L. M., Xie D., Dolezal K. et al. 2008 TAA1-mediated auxin biosynthesis is essential for hormone crosstalk and plant development. Cell 133, 177–191.
Sugawara S., Hishiyama S., Jikumaru Y., Hanada A., Nishimura T., Koshiba T. et al. 2009 Biochemical analyses of indole-3-acetaldoxime-dependent auxin biosynthesis in Arabidopsis. Proc. Natl. Acad. Sci. USA 106, 5430–5435.
Tanaka H., Dhonukshe P., Brewer P. B. and Friml J. 2006 Spatiotemporal asymmetric auxin distribution: a means to coordinate plant development. Cell Mol. Life Sci. 63, 2738–2754.
Tao Y., Ferrer J. L., Ljung K., Pojer F., Hong F., Long J. A. et al. 2008 Rapid synthesis of auxin via a new tryptophan dependent pathway is required for shade avoidance in plants. Cell 133, 164–176.
Vande Broek A., Gysegom P., Ona O., Hendrickx N., Prinsen E., Van Impe J. and Vanderleyden J. 2005 Transcriptional analysis of the Azospirillum brasilense indole-3-pyruvate decarboxylase gene and identification of a cis-acting sequence involved in auxin responsive expression. Mol. Plant Microbe Interact. 18, 311–323.
Wang R., Xiao L., Lin W., Cao Y. and Bo X. 2002 High performance liquid chromatographic determination of internal hormones in inter-subspecific hybrid rice. Chin. J. Chromatography 20, 148–150.
Won C., Shen X., Mashiguchi K., Zheng Z., Dai X., Cheng Y. et al. 2011 Conversion of tryptophan to indole-3-acetic acid by TRYPTOPHAN AMINOTRANSFERASES OF ARABIDOPSIS and YUCCAS in Arabidopsis. Proc. Natl. Acad. Sci. USA 108, 18518–18523.
Yamamoto Y., Kamiya N., Morinaka Y., Matsuoka M. and Sazuka T. 2007 Auxin biosynthesis by the YUCCA genes in rice. Plant Physiol. 143, 1362–1371.
Yan C., Yan S., Zhang Z., Liang G., Lu J. and Gu M. 2006 Genetic analysis and gene fine mapping for a rice novel mutant (rl9(t)) with rolling leaf character. Chin. Sci. Bull. 51, 63–69.
Yi J., Zhuang C., Wang X., Cao Y., Liu Y. and Mei M. 2007 Genetic analysis and molecular mapping of a rolling leaf mutation gene in rice. J. Integr. Plant Biol. 49, 1746–1753.
Zhang G., Xu Q., Zhu X., Qian Q. and Xue H. 2009 SHALLOT-LIKE1 is a KANADI transcription factor that modulates rice leaf rolling by regulating leaf abaxial cell development. Plant Cell 21, 719–735.
Zhao Y. 2010 Auxin biosynthesis and its role in plant development. Annu. Rev. Plant Biol. 61, 49–64.
Zhao Y. 2012 Auxin biosynthesis: a simple two-step pathway converts tryptophan to indole-3-acetic acid in plants. Mol. Plant 5, 334–338.
Zhao Y., Christensen S. K., Fankhauser C., Cashman J. R., Cohen J. D., Weige D. and Chory J. 2001 A role for flavin monooxygenase-like enzymes in auxin biosynthesis. Science 291, 306–309.
Zhao Y., Hull A. K., Gupta N. R., Goss K. A., Alonso J., Ecker J. R., Normanly J., Chory J. and Celenza J. L. 2002 Trp-dependent auxin biosynthesis in Arabidopsis: involvement of cytochrome P450s CYP79B2 and CYP79B3. Genes Dev. 16, 3100–3112.
Acknowledgements
This work was supported by grants from the National Natural Sciences Foundation of China (nos. 30671279 and 31071070).
Author information
Authors and Affiliations
Additional information
[Yi J., Liu L., Cao Y., Li J. and Mei M. 2013 Cloning, characterization and expression of \(\emph{OsFMO}_{(t)}\) in rice encoding a flavin monooxygenase. J. Genet. 92, xx–xx]
Jicai Yi and Lanna Liu contributed equally to this work
Electronic Supplementary Material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
YI, J., LIU, L., CAO, Y. et al. Cloning, characterization and expression of \(\emph{OsFMO}_{{\mathbf{(}\emph{t}\mathbf{)}}}\) in rice encoding a flavin monooxygenase . J Genet 92, 471–480 (2013). https://doi.org/10.1007/s12041-013-0297-0
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12041-013-0297-0