Abstract
Protein phosphatase 2Cs (PP2Cs) have been demonstrated to play critical roles in regulation of plant growth/development, abscisic acid signaling pathway and adaptation to environmental stresses. Here we report the cloning and molecular characterization of a novel rice protein phosphatase 2C gene, OsBIPP2C2 (Oryza sativa L. BTH-induced protein phosphatase 2C 2). OsBIPP2C2 has three alternatively spliced transcripts and the largest transcript OsBIPP2C2a encodes a 380 aa protein containing all 11 conserved catalytic subdomains of PP2Cs. Expression of OsBIPP2C2a was significantly induced by benzothiadiazole (BTH), one of defense-related signal molecules in plants. Expression of OsBIP2C2a was induced by infection with the blast fungus, Magnaporthe grisea, and the pathogen-induced expression of OsBIPP2C2a in BTH-treated rice seedlings was much earlier and stronger than those in water-treated seedlings. Overexpression of OsBIPP2C2a in transgenic tobacco plants resulted in increased disease resistance against tobacco mosaic virus and Phytophthora parasitica var. nicotianae. Importantly, the OsBIPP2C2a-overexpressing transgenic tobacco plants showed constitutive expression of defense-related genes. These results suggest that OsBIPP2C2a may play an important role in disease resistance through activation of defense response.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Protein phosphatases play critical roles in balance of protein dephosphorylation by removing the phosphor-groups from their phosphorylated targets. Among them, protein phosphatase 2Cs (PP2Cs) have been studied extensively for their biochemical functions in plant growth, development and responses to hormones and abiotic stresses (Schweighofer et al. 2004). In Arabidopsis, 76 genes were identified to encode putative PP2C-type phosphatases (Kerk et al. 2002). Implication of PP2Cs in regulation of abscisic acid (ABA) response is well established in Arabidopsis. Biochemical and genetic analyses have resulted in the identification of at least four PP2Cs, including ABI1, ABI2, PP2CA, and HAB1 (formerly named AtP2C-HA), as negative regulators of ABA signal transduction (Meyer et al. 1994; Stone et al. 1994; Leung et al. 1997; Gosti et al. 1999; Merlot et al. 2001; Tahtiharju and Palva 2001; Gonzalez-Garcia et al. 2003; Saez et al. 2004; Kuhn et al. 2006; Yoshida et al. 2006; Nishimura et al. 2007). Functions of ABI1 and ABI2 appear to be highly overlapped, including ABA-regulated seeds dormancy, guard cell closure, and acclimation to various stresses (Pei et al. 1997; Leung et al. 1997; Gosti et al. 1999; Chak et al. 2000; Merlot et al. 2001; Saez et al. 2006). Other identified Arabidopsis PP2Cs include kinase-associated protein phosphatase (KAPP) and POL, which regulate the normal developmental process of tissues and organs (Stone et al. 1998; Shah et al. 2002; Yu et al. 2003).
A number of PP2Cs identified from other plant species have also been shown to play roles in growth and response to various abiotic stresses. FsPP2C1 and FsPP2C2, two PP2C genes from beech (Fagus sylvatica) seeds, were induced by ABA in dormant seeds (Lorenzo et al. 2001, 2002). Results from transgenic Arabidopsis overexpressing FsPP2C1 gene indicate that negative regulation of ABA signaling by FsPP2C1 is a factor contributing to promote the transition from seed dormancy to germination during early stage of stratification (Gonzalez-Garcia et al. 2003). Interestingly, constitutive expression of FsPP2C2 in transgenic Arabidopsis under the cauliflower mosaic virus 35S promoter resulted in enhanced sensitivity to ABA and osmotic stress in seeds and vegetative tissues as well as dwarf and delayed flowering, suggesting that FsPP2C2 is a positive regulator of ABA (Reyes et al. 2006). Thus, FsPP2C2 is different from other identified plant PP2Cs that act as negative regulators of ABA signaling. MP2C, a wound-induced alfalfa PP2C gene, was found to be activated within 10 min and gradually decreased to non-detectable levels after 20 min wounding treatment (Meskiene et al. 1998). NtPP2C1, a tobacco PP2C gene, was strongly activated by drought, but repressed by oxidative stress and heat shock, suggesting that NtPP2C1 may operate at the junction of drought, heat shock and oxidative stress (Vranova et al. 2000). Moreover, a lotus PP2C gene, LiNPP2C1, was shown to be expressed specifically during establishment of root nodules with Mesorhizobium, but not in the early stage (Kapranov et al. 1999), indicating the involvement of PP2Cs in plant-microbe interactions.
Compared with the extensive studies on the functions of MAP kinases, little attention has been paid on the involvement of PP2Cs in plant biotic stress responses. Recently, several lines of evidence suggest a role for PP2Cs in regulation of plant defense responses. Tobacco DBP1 contains a typical catalytic domain of PP2C at its C terminus and a transcription factor domain at the N terminus and was found to modulate transcription of a defense gene, CEVI1, whose expression was up-regulated only in compatible interaction between citrus exocortis viroid and tobacco, but not in incompatible interaction (Carrasco et al. 2003). 14-3-3G is directly involved in regulating DBP1 function by promoting nuclear export and subsequent cytoplasmic retention of DBP1 under conditions that in turn alleviate DBP1-mediated repression of target gene expression (Carrasco et al. 2006). Arabidopsis AP2C1 is a novel stress signal regulator that inactivates the stress-responsive MAPKs. Mutant ap2c1 plants produce higher amounts of jasmonic acid and are more resistant to phytophagous mites. Plants overexpressing AP2C1 gene display reduced ethylene production and defense-related gene expression, and compromised innate immunity against the necrotrophic pathogen Botrytis cinerea (Schweighofer et al. 2007).
We are interested in the function of protein phosphorylation/dephosphorylation, which is modulated by protein kinases and phosphatases, respectively, in regulation of plant defense response. We previously found that two MAP kinase genes, OsBIMK1 and OsBIMK2, and a PP2C gene, OsBIPP2C1, play roles in rice disease resistance responses (Song and Goodman 2002a; Song et al. 2006; Hu et al. 2006). Here we report the isolation and identification of a new rice PP2C gene, OsBIPP2C2. Among the predicted three alternatively spliced transcripts of OsBIPP2C2 gene, accumulation of OsBIPP2C2a transcript was increased in rice seedlings after treatment with benzothiadiazole, a disease resistance signal molecule and infection by Magnaporthe grisea, a fungal pathogen causing rice blast disease. Overexpression of OsBIPP2C2a in transgenic tobacco plants resulted in enhanced disease resistance and constitutive expression of defense-related genes. Our results suggest that OsBIPP2C2a may play an important role in disease resistance through activation of defense response.
Materials and methods
Plant growth and treatments
Rice (Oryza sativa L. subsp. indica cv. Yuanfengzao) seedlings were grown under greenhouse conditions at 22/27°C (night/day) and 3-week-old seedlings were used for all experiments. Rice seedlings were treated by foliar spraying with 0.3 mM BTH solution (Novartis Crop Protection Inc., Research Triangle Park, NC, USA) or with sterilized distilled water as a control. At 3 days after BTH treatment, the seedlings were inoculated with spore suspension (5 × 105 spores per ml in 0.05% Tween-20) of Magnaporthe grisea (strain 85-14B1 of race ZB1) and then the inoculated seedlings were kept at 100% relative humidity in darkness for 36 h (Luo et al. 2005a). Samples were collected at different time points as indicated after treatment and stored at −80°C until further use.
Cloning of OsBIPP2C2
Two differentially expressed clones (BNHN-n22 and BNHN-n23), obtained previously from screenings of suppression subtractive hybridization libraries (Song and Goodman 2002a), show high level of similarity to plant PP2C genes. To obtain the full-length cDNA of this putative PP2C gene, the 5′- and 3′-missing ends were amplified by PCR using phage DNA prepared from a rice cDNA library (Song and Goodman 2002a) as template. To amplify the 3′-end, a gene-specific primer, BNHN-n22-1F (5′-GGA CGG GTA TAG CAA ACA GA-3′) and a vector anchored universal reverse primer, T7 (5′-GTA ATA CGA CTC ACT ATA GGG C-3′) were used for amplification of the 3′-end; a gene-specific primer, BNHN-n22-1R (5′-CTT CCT CGT GGC TTC CTT TA-3′), and another vector anchored forward universal primer, T3 (5′-AAT TAA CCC TCA CTA AAG GGA-3′) were used for amplification of the 5′-end. The full-length cDNAs were amplified using a gene-specific primer, BNHN-n22-2F (5′-GCT TGT GTT GGA TCG GAG T-3′), located in the 5′-UTR, and the vector universal primer T7. A pair of primers, BNHN-n22-3F (5′-CAT ATG TTG AGG GCG GTG GC-3′, underlined is an NdeI site) and BNHN-n22-3R (5′-GAA TTC TAC CAA CCG GAG CC-3′, underlined is an EcoRI site) was used for cloning of the ORF sequences. All PCR products were cloned into pUCm T-vector (Sangon, Shanghai, China) by T/A cloning and confirmed by sequencing. The correct clones for the full-length cDNAs were designated as pUCm-OsBIPP2C2.
DNA sequencing and analysis
DNA sequencing was performed on both strands on the MegaBACE 1000 DNA Analysis System (Amersham Biosciences, UK) at the Center of Analysis and Measurement in Zhejiang University. Sequence edits and ORF prediction were carried out using DNASTAR software (LaserGene, Madison, WI, USA). Similarity searches were done using BLAST at the NCBI GenBank database (http://www.ncbi.nlm.nih.gov/BLAST/). Sequence alignments were conducted using ClustalW (http://www.ebi.ac.uk/clustalw/).
Total RNA extraction and Northern blotting
Total RNA was extracted from rice and tobacco leaf samples using a standard acid-guanine method (Sambrook and Russell 2001). Twenty (rice) or ten (tobacco) micrograms of total RNA were separated on 1% agarose-formaldehyde gel, and transferred onto Hybond N+ (Amersham Biosciences, UK) nylon membrane in 20 × SSC by capillary action overnight. A 3′-end 700 bp fragment of OsBIPP2C2 was prepared from plasmid pUCm-OsBIPP2C2 by digesting with PstI. Fragments for tobacco PR-1a and Sar8.2b, two defense-related genes were prepared as previously described (Song and Goodman 2002b; Luo et al. 2005b). cDNA fragments were labeled with [α-32P]-dCTP by the random priming method using a Random Primed DNA Labeling Kit (Takara, Dalian, China). Pre-hybridization was performed at 42°C for 30 min in ULTRAhyb buffer (Ambion Inc., Austin, USA) and hybridization was carried out overnight at 42°C in the same hybridization buffer with the [α-32P]-labeled probe. After hybridization, the blots were washed twice with 2 × SSC, 0.1%SDS and 1 × SSC, 0.1%SDS for 5 min each at 42°C, and autoradiographed by exposure to X-ray film (Lucky Film Corporation, Baoding, China) for 2 days at −80°C.
Construction of binary vector and generation of transgenic tobacco
The ORF sequence of OsBIPP2C2 was released by digestion with XbaI/PstI from plasmid pUCm-OsBIPP2C2 and ligated into XbaI/PstI sites of plant binary vector CHF3, in which the OsBIPP2C2 gene was placed under the control of the CaMV 35S promoter in the sense orientation. The resulting plasmid, CHF3-OsBIPP2C2, was introduced into Agrobacterium tumefaciens EHA105 by electroporation. Agrobacteria were grown at 28°C with shaking (200 rpm) in YEP broth containing 100 μg/ml streptomycin and 100 μg/ml spectinomycin and bacterial suspension of OD600 = 0.80 was used for transformation. Transformation of tobacco (cv. Xanthi nc) was performed using Agrobacterium-mediated leaf disc transformation method. Transgenic lines with single copy of OsBIPP2C2a gene were selected and T3 homozygous seeds were used for study. The seeds of the transgenic tobacco lines were germinated on 1/2 MS medium containing 200 μg/ml kanamycin in a growth chamber under 16 h day/8 h night at temperature of 22–25°C for 3–4 weeks and the surviving plants were transferred into natural soil and grown in the growth room for another 3–4 weeks.
Genomic DNA of transgenic plants was extracted by CTAB protocol (Luo et al. 2005b). Approximately 100 ng of genomic DNA was used for detection of the transgene in transgenic plants by PCR with primers BNHN-n22-3F and BNHN-n22-3R. Genomic DNA was digested with HindIII, separated on agarose gel, and transferred onto Hybond N+ (Amersham Biosciences, UK) nylon membrane by capillary action. Southern blotting analysis was performed with the same protocol as Northern blotting analysis.
Disease resistance assays
Inoculation of the tobacco plants with TMV was performed using standard mechanical rubbing method. For preparation of the inoculums, TMV-infected tobacco leaves were ground in a mortar with a pestle in 5 ml of potassium phosphate buffer (50 mM, pH 7.0). Fully expanded leaves were dusted with dry carborundum and inoculated by gently rubbing the upper leaf surface with 100 μl of the viral suspension inoculums, followed by rinsing with deionized water immediately. Control plants were also dusted with carborundum and mock inoculated with sample volume of potassium phosphate buffer. Following inoculation, plants were maintained at 23–26°C under continuous illumination provided by cool-white fluorescent lamps. The lesion numbers on the inoculated leave of at least six plants were counted 5 days after inoculation.
Phytophthora parasitica var. nicotianae was cultivated on PDA medium (potato, 200 g/L; dextrose 20 g/L; agar 15 g/L; pH 6.5) at 25°C for 4–6 days. The mycelia were collected and suspended in water to OD600 ≈ 0.5 for inoculation. Six two-month-old tobacco plants were inoculated by pouring of mycelium suspension at the base of the stem (20 mL/plant) and were then incubated in growth room under the conditions described above. Disease severity was evaluated three weeks after inoculation on a four-scale standard as the follows: 0, the inoculated plants have no significant symptom; 1, lesions on stem base of the inoculated plants do not encircle the stem and the plants have no wilted leaves; 2, lesions on stem base encircle the stem and 2–3 lower leaves of the inoculated plants become yellowish and/or wilted; 3, lesions on stem base encircle the stem and the inoculated plants become wilted. Disease index was calculated by dividing sum of plants scored in each rating × corresponding rating value with total number of plants × 3.
All experiments were performed independently three times, and the data from these experiments were subjected to statistical analysis using DPS software (Beijing China-agri BRAINS Science & Technology Co. Ltd, http://www.nbs.net.cn/yzjaa.htm).
Results
Cloning of OsBIPP2C2 and identification of two alternatively spliced transcripts
In our previous studies aimed at elucidation of molecular biology of rice disease resistance response, we isolated and characterized hundreds of differentially expressed cDNA clones (Song and Goodman 2002a). Among them, two differentially expressed clones, BNHN-n22 (GenBank Accession No. BI118738) and BNHN-n23 (GenBank Accession No. BI118739), contain cDNA inserts of 344 bp and 399 bp, respectively, which show high level of similarity to genes encoding plant PP2Cs. A rice PP2C gene, OsBIPP2C1 (Oryza sativa L. BTH induced protein phosphatase 2C 1) was previously identified (Hu et al. 2006), we thus designated this new PP2C gene as OsBIPP2C2. Rapid amplification of cDNA ends approach was used to amplify the missing 5′- and 3′-end sequences and a full-length cDNA was assembled. Database searches revealed that OsBIPP2C2 corresponds to locus Os03g55320, which is annotated to have three alternatively spliced transcripts, Os03g55320.1, Os03g55320.2 and Os03g55320.3 (Fig. 1a). In amplification of OsBIPP2C2 ORF, two fragments of ~1,140 and ~1,050 bp in size were amplified. Cloning and sequencing revealed that these two fragments were 1,143 and 1,040 bp in size, respectively, and had identical sequences of 696 bp at 5′-end. Alignments of these two transcripts with predicted Os03g55320 transcripts, indicate that the large (1,143 bp) and small (1,040 bp) transcripts correspond to Os03g55320.1 and Os03g55320.3, respectively. We designated the large transcript as OsBIPP2C2a and the short one as OsBIPP2C2c. OsBIPP2C2a is supported by two full-length cDNAs (GenBank Accession Nos. AK102620 and AK105764) isolated from rice subsp. Japonica by Knowledge-based Oryza Molecular biological Encyclopedia (http://cdna01.dna.affrc.go.jp/cDNA/); while OsBIPP2C2c lacks support of full-length cDNA available in public database. Another full-length cDNA, AK066083, confirms the predicted transcript of Os03g55320.2. The OsBIPP2C2c transcript seems to be transcribed from Os03g55320 locus with an advanced transcription stop due to an alternatively splicing event. This was the reason that the OsBIPP2C2c sequence was amplified using a pair of primers that were designated for amplification of the OsBIPP2C2 ORF. Database searching and alignment analyses revealed that there are 3 nucleotide differences between the OsBIPP2C2a sequence and its corresponding rice genomic sequence; but these differences do not result in any changes in protein sequence. Because OsBIPP2C2c does not encode a typical PP2C, OsBIPP2C2a was chosen for further study.
Alternatively spliced transcripts of OsBIPP2C2 gene and alignment of OsBIPP2C2a with other PP2Cs. a Exon/intron organizations, splicing patterns of the OsBIPP2C2 gene. Exons are indicated by black boxes and the introns are indicated by thick inverted V-shape lines. For OsBIPP2C2c transcript, 3′ end sequence after stop codon is also presented as bold line. Numbers of nucleotides are indicated above for introns and below for exon in bp. b Alignment analysis of and conserved domains in OsBIPP2C2a protein. The putative subdomains of PP2C are indicated by numbers above the sequences and residues putatively involved in binding metal ions are marked by filled stars. Plant PP2Cs used for alignment are Arabidopsis thaliana ABI1 (CAA54383) and Fagus sylvatica FsPP2C2 (CAB90634)
Characterization of OSBIPP2C2 protein
The full-length cDNAs of OsBIPP2C2a contains a predicted ORF of 1,143 bp, encoding a 380 aa protein with calculated molecular weight of 41.8 kD and isoelectric point of 9.4. OsBIPP2C2a contains all 11 conserved catalytic subdomains present in all PP2Cs (Rodriguez 1998) with six residues putatively involved in binding of the phosphate and metal ions (Das et al. 1996) (Fig. 1b). However, OsBIPP2C2b and OsBIPP2C2c lack domains 10–11 and domains 7–11, respectively. OsBIPP2C2a has a relatively shorter N-terminal extensions (44 aa) than those of other plant PP2Cs (110–140 aa).
Because PP2Cs family comprises of a large number of members in plants, we thus searched for possible function of OsBIPP2C2a by comparing phylogenetic relationship between OsBIPP2C2a and other previously studied plant PP2Cs with defined biological functions. Phylogenetic tree analysis revealed that OsBIPP2C2a shows a relatively low level of identity (<45%) to other reported plant PP2Cs (Fig. 2) at amino acid sequence level. OsBIPP2C2a is more close (45% identity) to a biochemically identified Fagus sylvatica FsPP2C2 (Lorenzo et al. 2002), which has been shown to be involved in osmotic stress tolerance in seeds and vegetative tissues through regulating ABA sensitivity (Reyes et al. 2006). However, OsBIPP2C2a has less than 25% of identity to other well-identified plant PP2Cs such as Arabidopsis ABI1 (Leung et al. 1994; Meyer et al. 1994), ABI2 (Leung et al. 1997; Rodriguez et al. 1998) and AtPP2CA (Kuromori and Yamamoto 1994), tobacco NtPP2C1 and NtPP2C2 (Vranova et al. 2000), and Medicago sativa MP2C (Meskiene et al. 1998). Furthermore, OsBIPP2C2a has only 21% of identity to OsBIPP2C1, previously identified rice PP2C that plays a role in disease resistance (Hu et al. 2006). Thus, OsBIPP2C2a may be a new stress-related PP2C.
Phylogenetic tree analysis of OsBIPP2C2a. Alignment was done using ClustalW program in DNAstar software and the PP2C proteins used are: Arabidopsis thaliana ABI1 (CAA54383), ABI2 (CAA70162), AtP2CA (BAA07287), KAPP (AAB38148) and HAB1 (At1g72770); Nicotiana tabacum NtPP2C1 (CAC10358), NtPP2C2 (CAC10359) and DBP1 (AAM75346); Fagus sylvatica FsPP2C1 (CAB90633) and FsPP2C2 (CAB90634); Lotus japonicus LiNPP2C1 (AAD17804); Medicago sativa MP2C (CAA72341); Mesembryanthemum crystallinum, Mpc2 (AAC36697), Mpc5 (AAC36698), Mpc6 (AAC36699) and Mpc8 (AAC36700); Zea mays ZMPP2 (AAG43835) and ZmKAPP (AAB93832); Oryza sativa OsKAPP (AAC26828)
Expression of OsBIPP2C2a in rice defense response
The differentially expressed clones BNHN-n22 and BNHN-n23, which contain cDNA inserts derived from OsBIPP2C2a, were obtained from suppression subtractive hybridization library constructed by subtraction of cDNAs from BTH-induced rice tissues with those from water control. This implied that OsBIPP2C2 gene might be induced by BTH treatment in rice. We first analyzed whether expression of OsBIPP2C2a was induced by BTH treatment. Because OsBIPP2C2 has multiple alternatively spliced transcripts, a 700 bp fragment at 3′-end of OsBIPP2C2a that covers the different sequence among the three alternatively spliced transcripts was used as probe. The specificity of the probe used was further confirmed by RT-PCR, in which only the OsBIPP2C2a transcript could be amplified as confirmed by sequencing the RT-PCR product. Northern blot analysis revealed that OsBIPP2C2a has a very low level of basic expression in healthy leaf tissues of rice seedlings grown under normal conditions (Fig. 3a). However, treatment of 3-week-old seedlings with BTH induced markedly the expression of OsBIPP2C2a (Fig. 3b). OsBIPP2C2a transcript accumulated significantly at 12 h, peaked at 24 h and maintained at high level until 48 h after treatments (Fig. 3b). The OsBIPP2C2a transcript returned to background level at 72 h after BTH treatment (Fig. 3b).
Induction of OsBIPP2C2a expression in defense responses. a and b OsBIPP2C2a expression in 3-week-old rice seedlings after treatment by foliar spraying with 0.3 mM of BTH or water as control. c and d Expression of OsBIPP2C2a in response to infection by Magnaporthe grisea. Three-week-old seedlings were treated by foliar spraying 0.3 mM BTH solution and inoculated with M. grisea 3 days after treatment. Leaf samples were collected at various time points (h) as indicated after BTH treatment (a and b) or after inoculation (c and d). Twenty micrograms of total RNA were fractionated on 1.5% agarose formaldehyde gel and hybridized with the 32P-labeled 700 bp fragment of OsBIPP2C2a cDNA as probe. The corresponding ethidium bromide gel image confirms equal loading of RNA samples
The expression pattern of OsBIPP2C2a in disease resistance response against the blast fungus, M. grisea was further analyzed. In this experiment, we treated the rice seedlings by spraying BTH solution and inoculated with spores of M. grisea 3 days after BTH treatment. The BTH-treated seedlings showed an induced defense response against infection by the blast fungus. In the BTH-treated rice seedlings, OsBIPP2C2a transcript accumulated to a high level as early as 12 h and peaked at 24 h after inoculation with the fungus. High level of the OsBIPP2C2a transcript was maintained within first 36 h and declined to background level after 36 h of inoculation (Fig. 3c). In contrast, no significant accumulation of OsBIPP2C2a transcript was detected in water-treated control seedlings within the first 24 h after inoculation. However, a relatively low level of expression was also observed in water-treated control seedlings during 36–48 h after inoculation (Fig. 3d). These results suggest that OsBIPP2C2a transcript could be induced by infection with the blast fungus and the expression level in the BTH-induced rice plants was much earlier and stronger than those in the uninduced plants, especially during the early stage of infection.
Generation and characterization of OsBIPP2C2a transgenic tobacco plants
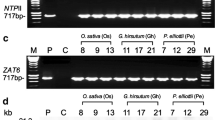
To gain further insights into the biological function of the OsBIPP2C2 gene, we performed a functional analysis in transgenic tobacco plants by overexpression of OsBIPP2C2a under control of the cauliflower mosaic virus (CaMV) 35S promoter. Sixteen independent transgenic lines were obtained and all of them contained the OsBIPP2C2a gene as confirmed by PCR detection of its cDNA sequence with genomic DNA as templates and sequencing of the amplified fragments (Fig. 4a). Copy number of the OsBIPP2C2a gene in transgenic lines was analyzed by Southern blotting and five transgenic lines with single copy of the transgene were identified (Fig. 4b). The transgene OsBIPP2C2a expressed in the transgenic lines, although the expression level varied to some extent (Fig. 4c). No significant abnormal morphological and growth/development phenotypes were observed in OsBIPP2C2a transgenic plants. Independent transgenic lines #03, #07, #33 and #36 were chosen for further studies as these lines contained a single copy of the transgene and showed high level of OsBIPP2C2a expression in transgenic plants.
Generation and characterization of OsBIPP2C2a-overexpressing transgenic tobacco plants. a Detection of OsBIPP2C2a in transgenic plants by PCR. b Southern blot analysis of copy number of the OsBIPP2C2a gene in the transgenic lines. Genomic DNA was digested with HindIII, separated on agarose gel, and transferred onto nylon membrane. The blot was hybridized with 32P-labeled fragment as a probe. c Expression of OsBIPP2C2a in transgenic plants. Ten micrograms of total RNA extracted from leaf samples of 6-week-old plants were fractionated and hybridized with 32P-labeled OsBIPP2C2a fragment as a probe. The corresponding ethidium bromide gel image shows the relative levels of RNA loaded for each sample. P Plasmid, WT untransformed wild-type plants; #03, #07, #33, #36 and #41, independent transgenic lines
OsBIPP2C2a-overexpressing plants enhanced disease resistance
To explore the possible role of OsBIPP2C2a in defense responses, we analyzed the disease resistance levels of OsBIPP2C2a-overexpressing tobacco plants against tobacco mosaic virus (TMV) and P. parasitica var. nicotianae. Under our experimental conditions, typical necrotic lesions were observed on the leaves of the wild-type and the OsBIPP2C2a-overexpressing transgenic plants 3–5 days after inoculation with TMV (Fig. 5). However, the lesion numbers in the leaves of the OsBIPP2C2a-overexpressing transgenic lines #03, #07, #33 and #36 were significantly reduced (P ≤ 0.05) by 39–55% as compared with that of the wild-type plants (Fig. 5). Likewise, disease symptoms, typical wilting of the lower old leaves and rotting of the stem bases were seen on the transgenic and the wild-type plants 10 days after drench-inoculation with P. parasitica var. nicotianae (Fig. 6). Disease indexes at 10 and 20 days after inoculation in the transgenic lines #03, #07 and #36 were significantly reduced (P ≤ 0.05) by 43–90 and 33–84%, respectively, as compared with those of wild-type plants (Fig. 6). These results suggest that overexpression of the rice OsBIPP2C2a in tobacco plants enhances disease resistance against TMV and P. parasitica var. nicotianae.
Enhanced disease resistance against tobacco mosaic virus in OsBIPP2C2a-overexpressing tobacco plants. a Representative disease symptom on transgenic and wild-type plants. b Disease severity as measured by lesion numbers. Data presented are the means (standard deviations from three independent experiments. The bars with the same letter are not significantly different at P ≤ 0.05. Photographs were taken 7 days after inoculation. WT untransformed wild-type plants; #03, #07, #33 and #36, independent transgenic lines
Enhanced disease resistance against Phytophthora parasitica in OsBIPP2C2a-overexpressing tobacco plants. a Representative disease symptom on transgenic and wild-type plants. b Disease index on the transgenic and wild type plants 10 and 20 days after inoculation with P. parasitica. Open and solid columns represent the disease indexes at 10 and 20 days after inoculation, respectively. Data presented are the means (standard deviations from three independent experiments. The bars with the same letter are not significantly different at P ≤ 0.05. Photographs were taken 20 days after inoculation. WT untransformed wild-type plants; #03, #07 and #36, independent transgenic lines
Constitutive expression of defense genes in OsBIPP2C2a transgenic plants
To further confirm the disease resistance phenotype observed in OsBIPP2C2a-overexpressing tobacco plants, we analyzed and compared expression levels of PR-1a and Sar8.2b, two defense-related genes, in OsBIPP2C2a transgenic plants with those in wild-type plants. As shown in Fig. 7, under the experimental conditions, no significant expression of PR-1a and Sar8.2b was detected in the wild-type plants. However, elevated expressions of these two PR genes were observed in OsBIPP2C2a-overexpressing plants in absence of pathogens or other chemical inducers (Fig. 7). The increased levels of PR-1a and Sar8.2b expression varied among the transgenic lines tested. These results indicate that overexpression of the rice OsBIPP2C2a in transgenic tobacco plants results in constitutive activation of defense responses, which in turn confers enhanced disease resistance against TMV and P. parasitica var. nicotianae.
Expression of PR-1a and Sar8.2b in OsBIPP2C2a-overexpressing tobacco plants. Leaf samples were collected from six-week-old tobacco plants of different transgenic lines. Ten micrograms of total RNA were fractionated on agarose formaldehyde gel and hybridized with the 32P-labeled fragments of PR-1a and Sar8.2b cDNAs. The corresponding ethidium bromide gel image shows the relative levels of RNA loaded for each sample. WT untransformed wild-type plants; #02 and #07, independent transgenic lines
Discussion
Protein phosphatase 2Cs have been extensively studied for their involvement in regulation of plant growth and development as well as in the ABA-responsive signaling pathway and in adaptation to environmental stresses (Rodriguez et al. 1998; Schweighofer et al. 2004). However, our knowledge on the involvement and function of PP2Cs in regulation of defense responses in plants is very limited. Recently, it was demonstrated that the Arabidopsis AP2C1 plays a key role in regulation of innate immunity against Botrytis cinerea (Schweighofer et al. 2007). We previously identified a rice PP2C gene, OsBIPP2C1 and found that overexpression of OsBIPP2C1 in tobacco led to enhanced disease resistance (Hu et al. 2006). In the present study, we identified a new PP2C gene, OsBIPP2C2, whose expression was induced in rice defense response. Our functional analysis by ectopic expression of OsBIPP2C2a, one of three alternatively spliced transcripts of OsBIPP2C2 gene, in transgenic tobacco plants suggest that OsBIPP2C2 may function in regulation of defense responses through activation of defense-related gene expression.
The OsBIPP2C2 gene (Os03g55320) is predicted to have three alternatively spliced transcripts. Our cloning of OsBIPP2C2a and OsBIPP2C2c, and the available full-length cDNAs in GenBank database, confirm the existence of these three alternatively spliced transcripts in rice. The OsBIPP2C2a transcript encodes a typical PP2C containing all 11 conserved subdomains of the PP2C catalytic domain, while the other two transcripts encode C-terminal truncated putative PP2C proteins lacking subdomains 8–11. This is similar to the situation for rice OsMAPK5, whose large transcript OsMAPK5a encodes a functional MAPK with typical kinase activity, but the small transcript OsMAPK5b encodes a truncated MAPK with no kinase activity (Xiong and Yang 2003). The OsBIPP2C2a transcript was accumulated in rice tissues in response to BTH treatment and pathogen attack. We tried to detect the OsBIPP2C2c transcript in rice by RT-PCR using a pair of specific primers located in the flanking sequences of the alternative splicing sites. However, we were unable to detect the OsBIPP2C2c transcript in healthy rice seedlings and its accumulation, like the OsBIPP2C2a transcript, in response to BTH treatment and pathogen infection (data not shown). Therefore, it is likely that OsBIPP2C2a transcript is the prevalent transcript of the OsBIPP2C2 gene. However, the biological role of the other two alternatively spliced transcripts of the OsBIPP2C2 gene or the biochemical activity of their products, if any, remain to be explored further.
Benzothiadiazole has been previously shown to be an effective inducer of induces defense response in rice against blast, bacterial leaf blight and sheath blight diseases (Gorlach et al. 1996; Song et al. 2001; Ge et al. 1999; Zhang et al. 2003). In this study, we observed that treatment of rice seedlings with BTH induced expression of OsBIPP2C2 gene, resulting in accumulation of the OsBIPP2C2a transcript. This is supported from the other side by the fact that the two differentially expressed clones that contain partial cDNA of OsBIPP2C2 gene were isolated from BTH-treated rice seedlings. Moreover, accumulation of OsBIPP2C2a transcript was also induced by M. grisea. Thus, it is likely that OsBIPP2C2 gene is responsive and inducible to chemical inducers of defense response, including BTH, and pathogen infection. OsBIPP2C1, a rice PP2C gene that we identified previously, and the Arabidopsis AP2C1 were also shown to be up-regulated by different biotic and abiotic stress factors, including pathogen infection (Hu et al. 2006; Schweighofer et al. 2007). Another feature of OsBIPP2C2 expression is that the accumulation kinetics of OsBIPP2C2a transcript showed different patterns in the BTH-induced rice plants and the uninduced plants after infection with M. grisea, and this pathogen-induced expression of OsBIPP2C2a was much earlier and stronger in the BTH-induced rice plants than those in the uninduced plants. It is likely that expression of OsBIPP2C2 gene, especially the OsBIPP2C2a transcript, can be induced during early stage of the disease resistance response in rice against M. grisea and thus OsBIPP2C2a might be involved in regulation of defense response.
Direct involvement of OsBIPP2C2a in disease resistance response was further studied through functional analysis by ectopic overexpression in transgenic tobacco plants. The transgenic tobacco plants overexpressing the OsBIPP2C2a transcript of OsBIPP2C2 gene showed enhanced disease resistance against TMV and P. parasitica var. nicotianae, leading to a significant reduction in disease severity. This is similar to our previous observation that overexpression of OsBIPP2C1 in transgenic tobacco resulted in enhanced disease resistance (Hu et al. 2006). Importantly, coincided with the enhanced disease resistance in OsBIPP2C2a-overexpressing plants is the constitutive expression of defense-related genes including PR-1 and Sar8.2b. Such relationship between enhanced disease resistance and elevated levels of defense gene expression has been already documented previously for some defense-related transcriptional factors, e.g. constitutive expression of genes encoding for rice OsBIHD1 or OsBIERF3 in tobacco and AtWRKY70 or AtWRKY18 in Arabidopsis resulted in increased disease resistance and elevated levels of PR gene expression (Luo et al. 2005b; Cao et al. 2006; Chen and Chen 2002; Li et al. 2004). The coincidence of the enhanced disease resistance with elevated PR gene expression suggests that OsBIPP2C2a in transgenic tobacco plants acts as a positive regulator of defense response by modulating PR gene expression. This is different from the Arabidopsis AP2C1, which is a negative modulator of innate immunity because plants with increased AP2C1 levels compromised disease resistance against B. cinerea and expression of PDF1.2 gene (Schweighofer et al. 2007). Taken together, it is likely that plant PP2Cs have much diverse biological functions and relatively narrower substrate specificity than those in other organisms, probably due to the large number of PP2Cs in plants as compared with the numbers in yeast and human (Kerk et al. 2002; Stark 1996; Cheng et al. 2000).
In conclusion, our studies present evidence supporting that OsBIPP2C2a plays an important role in disease resistance; however, the mechanism by which OsBIPP2C2a regulates defense response remains to be explored. Further functional genomics studies with knockout/knockdown mutants and/or overexpression in transgenic rice are underway and results from these studies will provide new insights into the biological function and the mechanism of OsBIPP2C2 in rice.
Abbreviations
- ABA:
-
Abscisic acid
- BTH:
-
Benzothiadiazole
- CaMV:
-
Cauliflower mosaic virus
- PP2C:
-
Protein phosphatase 2C
- TMV:
-
Tobacco mosaic virus
References
Cao Y, Wu Y, Zheng Z, Song F (2006) Overexpression of the rice EREBP-like gene OsBIERF3 enhances disease resistance and salt tolerance in transgenic tobacco. Physiol Mol Plant Pathol 67:202–211
Carrasco JL, Ancillo G, Mayda E, Vera P (2003) A novel transcription factor involved in plant defense endowed with protein phosphatase activity. EMBO J 22:3376–3384
Carrasco JL, Castello MJ, Vera P (2006) 14-3-3 mediates transcriptional regulation by modulating nucleocytoplasmic shuttling of tobacco DNA-binding protein phosphatase-1. J Biol Chem 281:22875–22881
Chak RK, Thomas TL, Quatrano RS, Rock CD (2000) The genes ABI1 and ABI2 are involved in abscisic acid- and drought-inducible expression of the Daucus carota L. Dc3 promoter in guard cells of transgenic Arabidopsis thaliana (L.) Heynh. Planta 210:875–883
Chen C, Chen Z (2002) Potentiation of developmentally regulated plant defense response by AtWRKY18, a pathogen-induced Arabidopsis transcription factor. Plant Physiol 129:706–716
Cheng A, Kaldis P, Solomon MJ (2000) Dephosphorylation of human cyclin-dependent kinases by protein phosphatase type 2C alpha and beta isoforms. J Biol Chem 275:34744–34749
Das AK, Helps NR, Cohen PT, Barford D (1996) Crystal structure of the protein serine/threonine phosphatase 2C at 2.0 A resolution. EMBO J 15:6798–6809
Ge X, Song F, Zheng Z (1999) Systemic acquired resistance to Magnaporthe grisea in rice induced by BTH. Acta Agri Zhejiangensis 11:311–314
Gonzalez-Garcia MP, Rodriguez D, Nicolas C, Rodriguez PL, Nicolas G, Lorenzo O (2003) Negative regulation of abscisic acid signaling by the Fagus sylvatica FsPP2C1 plays a role in seed dormancy regulation and promotion of seed germination. Plant Physiol 133:135–144
Gorlach J, Volrath S, Knauf-Beiter G, Hengy G, Beckhove U, Kogel K-H, Oostendorp M, Staub T, Ward E, Kessmann H, Ryals J (1996) Benzothiadiazole, a novel class of inducers of systemic acquired resistance, activates gene expression and disease resistance in wheat. Plant Cell 8:629–643
Gosti F, Beaudoin N, Serizet C, Webb AA, Vartanian N, Giraudat J (1999) ABI1 protein phosphatase 2C is a negative regulator of abscisic acid signaling. Plant Cell 11:1897–1910
Hu X, Song F, Zheng Z (2006) Molecular characterization and expression analysis of a rice protein phosphatase 2C gene, OsBIPP2C1, and overexpression in transgenic tobacco conferred enhanced disease resistance and abiotic tolerance. Physiol Plant 127:225–236
Kapranov P, Jensen TJ, Poulsen C, de Bruijn FJ, Szczyglowski K (1999) A protein phosphatase 2C gene, LjNPP2C1, from Lotus japonicus induced during root nodule development. Proc Natl Acad Sci USA 96:1738–1743
Kerk D, Bulgrien J, Smith DW, Barsam B, Veretnik S, Gribskov M (2002) The complement of protein phosphatase catalytic subunits encoded in the genome of Arabidopsis. Plant Physiol 129:908–925
Kuhn JM, Boisson-Dernier A, Dizon MB, Maktabi MH, Schroeder JI (2006) The protein phosphatase AtPP2CA negatively regulates abscisic acid signal transduction in Arabidopsis, and effects of abh1 on AtPP2CA mRNA. Plant Physiol 140:127–139
Kuromori T, Yamamoto M (1994) Cloning of cDNAs from Arabidopsis thaliana that encode putative protein phosphatase 2C and a human Dr1-like protein by transformation of a fission yeast mutant. Nucleic Acids Res 22:5296–5301
Leung J, Bouvier-Durand M, Morris PC, Guerrier D, Chefdor F, Giraudat J (1994) Arabidopsis ABA response gene ABI1: features of a calcium-modulated protein phosphatase. Science 264:1448–1452
Leung J, Merlot S, Giraudat J (1997) The Arabidopsis ABSCISIC ACID- INSENSITIVE2 (ABI2) and ABI1 genes encode homologous protein phosphatases 2C involved in abscisic acid signal transduction. Plant Cell 9:759–771
Li J, Brader G, Palva ET (2004) The WRKY70 transcription factor: a node of convergence for jasmonate-mediated and salicylate-mediated signals in plant defense. Plant Cell 16:319–331
Lorenzo O, Rodriguez D, Nicolas G, Rodriguez PL, Nicolas C (2001) A new protein phosphatase 2C (FsPP2C1) induced by abscisic acid is specifically expressed in dormant beechnut seeds. Plant Physiol 125:1949–1956
Lorenzo O, Nicolas C, Nicolas G, Rodriguez D (2002) Molecular cloning of a functional protein phosphatase 2C (FsPP2C2) with unusual features and synergistically up-regulated by ABA and calcium in dormant seeds of Fagus sylvatica. Physiol Plant 114:482–490
Luo H, Song F, Goodman RM, Zheng Z (2005a) Up-regulation of OsBIHD1, a rice gene encoding BELL homeodomain transcriptional factor, in disease resistance responses. Plant Biol 7:459–468
Luo H, Song F, Zheng Z (2005b) Overexpression in transgenic tobacco reveals different roles for the rice homeodomain gene OsBIHD1 in biotic and abiotic stress responses. J Exp Bot 56:2673–2682
Merlot S, Gosti F, Guerrier D, Vavasseur A, Giraudat J (2001) The ABI1 and ABI2 protein phosphatases 2C act in a negative feedback regulatory loop of the abscisic acid signalling pathway. Plant J 25:295–303
Meskiene I, Bogre L, Glaser W, Balog J, Brandstotter M, Zwerger K, Ammerer G, Hirt H (1998) MP2C, a plant protein phosphatase 2C, functions as a negative regulator of mitogen-activated protein kinase pathways in yeast and plants. Proc Natl Acad Sci USA 95:1938–1943
Meyer K, Leube MP, Grill E (1994) A protein phosphatase 2C involved in ABA signal transduction in Arabidopsis thaliana. Science 264:1452–1455
Nishimura N, Yoshida T, Kitahata N, Asami T, Shinozaki K, Hirayama T (2007) ABA-Hypersensitive Germination1 encodes a protein phosphatase 2C, an essential component of abscisic acid signaling in Arabidopsis seed. Plant J 50:935–949
Pei ZM, Kuchitsu K, Ward JM, Schwarz M, Schroeder JI (1997) Differential abscisic acid regulation of guard cell slow anion channels in Arabidopsis wild-type and abi1 and abi2 mutants. Plant Cell 9:409–423
Reyes D, Rodriguez D, Gonzalez-Garcia MP, Lorenzo O, Nicolas G, Garcia-Martinez JL, Nicolas C (2006) Overexpression of a protein phosphatase 2C from beech seeds in Arabidopsis shows phenotypes related to abscisic acid responses and gibberellin biosynthesis. Plant Physiol 141:1414–1424
Rodriguez PL (1998) Protein phosphatase 2C (PP2C) function in higher plants. Plant Mol Biol 38:919–927
Rodriguez PL, Leube MP, Grill E (1998) Molecular cloning in Arabidopsis thaliana of a new protein phosphatase 2C (PP2C) with homology to ABI1 and ABI2. Plant Mol Biol 38:879–883
Saez A, Apostolova N, Gonzalez-Guzman M, Gonzalez-Garcia MP, Nicolas C, Lorenzo O, Rodriguez PL (2004) Gain-of-function and loss-of-function phenotypes of the protein phosphatase 2C HAB1 reveal its role as a negative regulator of abscisic acid signalling. Plant J 37:354–369
Saez A, Robert N, Maktabi MH, Schroeder JI, Serrano R, Rodriguez PL (2006) Enhancement of abscisic acid sensitivity and reduction of water consumption in Arabidopsis by combined inactivation of the protein phosphatases type 2C ABI1 and HAB1. Plant Physiol 141:1389–1399
Sambrook J, Russell DW (2001) Molecular cloning: a laboratory manual, 3rd edn. Cold Spring Harbor Laboratory Press, Cold Spring Harbor
Schweighofer A, Hirt H, Meskiene I (2004) Plant PP2C phosphatases: emerging functions in stress signaling. Trends Plant Sci 9:236–243
Schweighofer A, Kazanaviciute V, Scheikl E, Teige M, Doczi R, Hirt H, Schwanninger M, Kant M, Schuurink R, Mauch F, Buchala A, Cardinale F, Meskiene I (2007) The PP2C-type phosphatase AP2C1, which negatively regulates MPK4 and MPK6, modulates innate immunity, jasmonic acid, and ethylene levels in Arabidopsis. Plant Cell 19:2213–2224
Shah K, Russinova E, Gadella TW Jr, Willemse J, De Vries SC (2002) The Arabidopsis kinase-associated protein phosphatase controls internalization of the somatic embryogenesis receptor kinase 1. Genes Dev 16:1707–1720
Song FM, Goodman RM (2002a) OsBIMK1, a rice MAP kinase gene involved in disease resistance responses. Planta 215:997–1005
Song F, Goodman RM (2002b) Cloning and identification of the promoter of the tobacco Sar8.2b gene, a gene involved in systemic acquired resistance. Gene 290:115–124
Song F, Ge X, Zheng Z, Xie Y (2001) Benzothiadiazole induces systemic acquired resistance in rice against bacterial leaf blight. Chin J Rice Sci 15:323–326
Song D, Chen J, Song F, Zheng Z (2006) A novel rice MAPK gene, OsBIMK2, is involved in disease-resistance responses. Plant Biol 8:587–596
Stark MJ (1996) Yeast protein serine/threonine phosphatases: multiple roles and diverse regulation. Yeast 12:1647–1675
Stone JM, Collinge MA, Smith RD, Horn MA, Walker JC (1994) Interaction of a protein phosphatase with an Arabidopsis serine/threonine receptor kinase. Science 266:793–795
Stone JM, Trotochaud AE, Walker JC, Clark SE (1998) Control of meristem development by CLAVATA1 receptor kinase and kinase-associated protein phosphatase interactions. Plant Physiol 117:1217–1225
Tahtiharju S, Palva T (2001) Antisense inhibition of protein phosphatase 2C accelerates cold acclimation in Arabidopsis thaliana. Plant J 26:461–470
Vranova E, Langebartels C, van Montagu M, Inze D, van Camp W (2000) Oxidative stress, heat shock and drought differentially affect expression of a tobacco protein phosphatase 2C. J Exp Bot 51:1763–1764
Xiong L, Yang Y (2003) Disease resistance and abiotic stress tolerance in rice are inversely modulated by an abscisic acid-inducible mitogen-activated protein kinase. Plant Cell 15:745–759
Yoshida T, Nishimura N, Kitahata N, Kuromori T, Ito T, Asami T, Shinozaki K, Hirayama T (2006) ABA-hypersensitive germination3 encodes a protein phosphatase 2C (AtPP2CA) that strongly regulates abscisic acid signaling during germination among Arabidopsis protein phosphatase 2Cs. Plant Physiol 140:115–126
Yu LP, Miller AK, Clark SE (2003) POLTERGEIST encodes a protein phosphatase 2C that regulates CLAVATA pathways controlling stem cell identity at Arabidopsis shoot and flower meristems. Curr Biol 13:179–188
Zhang W, Ge X, Song F, Zheng Z (2003) Systemic acquired resistance of rice against sheath blight disease induced by benzothiadiazole. Acta Phytophylacica Sinica 30:171–176
Acknowledgments
We are grateful to Dr. Xueping Zhou, Institute of Biotechnology, Zhejiang University for TMV samples and Mr. Rongyao Chai (Zhejiang Academy of Agricultural Science) for the Magnaporthe grisea isolate 85-14B1. This study was supported by projects from National Natural Science Foundation of China (grants no. 30771399), National 863 Project (2007AA10Z140) and the Ph.D. Programs Foundation of Ministry of Education of China (No. 20070335111).
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by A. Atanassov.
X. Hu and H. Zhang contributed equally to this study.
The nucleotide sequences of OsBIPP2C2a and OsBIPP2C2c have been deposited in GenBank under accession numbers of AY8313942 and AY831395, respectively.
Rights and permissions
About this article
Cite this article
Hu, X., Zhang, H., Li, G. et al. Ectopic expression of a rice protein phosphatase 2C gene OsBIPP2C2 in tobacco improves disease resistance. Plant Cell Rep 28, 985–995 (2009). https://doi.org/10.1007/s00299-009-0701-7
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00299-009-0701-7