Abstract
Key message
An ERF transcription factor OsERF101 is predominantly expressed in rice reproductive tissues and plays an important role in improving rice seed setting rate under drought stress.
Abstract
Drought reduces grain yield due to the cumulative damage effects to plant vegetative and reproductive developmental processes. However, the genes involved in these processes are still not completely understood. In this study, we identified a gene named OsERF101 as an important positive regulator in the adaptive responses to dehydration stress during the reproductive and vegetative stages. This gene encodes a member of APETALA2/Ethylene-Responsive Element Binding Protein (AP2/EREBP) family. OsERF101 was predominantly expressed in flowers, particularly in the tapetum and microspores under normal growth conditions. It was induced by drought, PEG6000 and abscisic acid (ABA) in leaves. During the vegetative stage, OsERF101-overexpression plants were more resistant to osmotic stress caused by PEG6000 compared to the control plants. They also had higher survival and seed setting rates than wild type when subjected to reproductive-stage drought stress. Further physiological analysis revealed that the pollen fertility was improved in the overexpression lines, while the knockout mutant and RNAi lines showed reduced pollen fertility and compromised drought tolerance during the reproductive stage. The increased proline content and peroxidase activity in OsERF101-overexpression plants might contribute to the improved drought-tolerance of plants. In addition, OsERF101-overexpression plants displayed ABA susceptible phenotype, in which the expression levels of ABA-responsive genes RD22, LEA3, and PODs were up-regulated. Taken together, our results indicate that OsERF101 is a gene that regulates dehydration responses during the vegetative and reproductive stages.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Drought limits plant growth, development and productivity (Bartels and Sunkar 2005). Due to the frequent occurrence of dry spell, it is important to develop crop cultivars that have better performance under drought conditions for maintaining the annual crop yield. To date, many genes have been identified to regulate water stress responses at the vegetative stage in rice and other cereals. However, whether they are also capable of improving the reproductive development and stabilizing grain yield under drought stress are often unknown. In our studies, we aim to find genes that can improve drought tolerance in terms of improving seed setting rate, and use these genes for developing drought-tolerant rice varieties.
Drought signaling pathways are usually classified into ABA-dependent and ABA-independent pathways (Shinozaki and Yamaguchi-Shinozaki 2007; Shinozaki et al. 2003), within which transcription factors play important roles in controlling gene expression. For example, DREB and NAC transcription factors regulate gene expression in ABA-independent pathway, while AREB, MYC and MYB function in ABA-dependent pathway (Shinozaki and Yamaguchi-Shinozaki 2007; Shinozaki et al. 2003). These transcription factors coordinate to regulate plant adaptation to various stresses, and disruption of them often cause altered stress response phenotypes. Their downstream target genes include a large array of drought-responsive genes, such as Responsive to Dehydration 22 (RD22) (Iwasaki et al. 1995; Yamaguchi-Shinozaki and Shinozaki 1993), Late Embryogenesis Abundant 3 (LEA3) (Hundertmark and Hincha 2008; Xiao et al. 2007), Stress-Induced type 2c Protein Phosphatase (SIPP2C1) (Li et al. 2013b), Drought-hypersensitive Mutant 2 (DSM2) (Du et al. 2010) and H-type Thioredoxins (OsTRXh1) (Zhang et al. 2011) in ABA-dependent pathway, and Early Responsive to Dehydration1 (ERD1) and Responsive to Dehydration 29A (RD29A) (Kiyosue et al. 1993; Yamaguchishinozaki and Shinozaki 1994) in ABA-independent pathway. These genes participate in regulating different physiological processes of drought response. For example, OsTRXh1 is involved in maintaining the balance between reactive oxygen species (ROS) production and scavenging (Du et al. 2010; Zhang et al. 2011). Overexpression of OsTRXh1 enhances abiotic stress tolerance by eliminating the excessive ROS (Mittler 2002).
Among the stress-related transcription factors, AP2/EREBP (APETALA2/Ethylene Responsive Element Binding Protein) genes constitute a large super-family in plants. They are classified into four subfamilies based on the number of AP2/ERF domains and the phylogenetic relationships: AP2, RAV (related to ABI3/VP1), dehydration-responsive element-binding protein (DREB) and ethylene-responsive factor (ERF) (Sakuma et al. 2002; Sharoni et al. 2011). Among them, DREB and ERF subfamilies contain a single AP2/ERF domain that can bind to DNA cis-regulatory elements directly (Nakano et al. 2006). Members from these two subfamilies are shown to be involved in diverse abiotic and biotic stress responses. ERF subfamily mainly regulate the expression of biotic stress-related genes through interaction with GCC box present in their promoters (Chakravarthy et al. 2003; Ohme-Takagi and Shinshi 1995; Wang et al. 2004), while the DREB subfamily modulate the expression of abiotic stress responsive genes by binding to C-repeat or dehydration response element (DRE) (Chen et al. 2008; Gilmour et al. 1998; Stockinger et al. 1997). In the last few years, the involvement of ERF subfamily in abiotic stress responses has also been reported. For example, tobacco stress-induced gene 1 (Tsi1) and tomato stress-responsive factor 1 (TSRF1), can bind to both GCC box and DRE sequences and promote salt or drought tolerance in transgenic plants (Park et al. 2001; Quan et al. 2010; Zhang et al. 2004). Rice ERF subfamily protein, submergence 1A (SUB1A), is important for plant survival from rapid dehydration following de-submergence, in addition to submergence and drought tolerance (Fukao et al. 2011). Another two ERF subfamily proteins OsERF922 and OsDERF1, negatively modulate rice salt and drought tolerance, respectively at the vegetative stage (Liu et al. 2012a; Wan et al. 2011).
There are 163 AP2/EREBP family genes in rice and 77 of them belong to ERF subfamily (Sharoni et al. 2011). Few of them have been characterized. In our previous studies, we analyzed the transcriptomic changes of rice flowers in response to drought to seek candidate genes that can promote the drought tolerance of reproductive tissues (Jin et al. 2013). Among the identified genes, OsERF101 gene was found to be induced by drought and expressed predominantly in flowers. Here we report the functional characterization of OsERF101 in rice. The results revealed that this gene plays an important role in regulating rice drought tolerance during both the vegetative and reproductive stages.
Materials and methods
Plant materials and growth conditions
Oryza sativa L. cv. Nipponbare (background for overexpression and RNAi lines) or O. sativa L. cv. Dongjin (background for T-DNA insertion mutant) were used for study in this work. Rice seeds were sterilized by 3% H2O2 for half an hour and then imbibed at 37 °C for 2 days in the dark. After imbibition, the germinating seeds were grown on floating plates in Yoshida nutrition solution (Yoshida et al. 1976) or grown in soil pots at 28 °C under a photoperiod of 12-h light/12-h dark cycle with the light intensity of approximately 375 µmol m−2 s−1 and the humidity around 50%.
Transgenic line construction and identification
In our previous studies on floral transcriptomic changes under drought stress, different sizes of rice florets (2–3, 3–4, 4–5 and 5–7 mm) were harvested to perform microarray analysis. The OsERF101 gene showed strong induced expression by drought stress in flowers. The full-length coding sequence of OsERF101 was amplified by reverse transcription (RT)-PCR using the RNA isolated from the flowers of drought-stressed rice plants. The OsERF101 overexpression construct was generated by inserting the open reading frame of OsERF101 into the pCAMBIA1301U vector, driven by the maize Ubiquitin gene promoter. The RNAi construct was generated by cloning an OsERF101 sequence-specific fragment (371 bp) into the pGEM-RNAi vector in inverted directions, then integrating the hairpin fragment into pCAMBIA1301U vector. The GUS reporter plasmid was constructed by fusing the OsERF101 promoter fragment (3080 bp upstream of the start codon) with the β-glucuronidase (GUS) reporter gene in the pCAMBIA1301 vector with ClonExpress™ One Step Cloning Kit (Vazyme Biotech, China).
The oserf101 knockout mutant and its parent wild type seeds (Dongjin) were ordered from Rice T-DNA Insertion Sequence Database (http://cbi.khu.ac.kr/RISD_DB.html) (Jeon et al. 2000; Jeong et al. 2006). The mutant was verified by genomic DNA PCR and RT-PCR.
All primers used for vector construction and mutant identification are listed in the Supplementary Table 1. All constructs were introduced into rice by Agrobacterium-mediated transformation method (Ozawa 2009).
Phylogenetic analysis
Similar sequences (E-value = 10−4) of OsERF101 were searched through BLAST in the phytozome database (http://www.phytozome.net/), and the results were inspected manually. Sequences were aligned with MUSCLE (MEGA 5.0), followed by manual alignment. Phylogenetic tree was constructed using an alignment of full-length protein sequences in neighbor-joining algorithm with MEGA 5.0. To test inferred phylogeny, we used bootstraps with 1,000 bootstrap replicates (Tamura et al. 2011).
PCR and quantitative RT-PCR (qRT-PCR) analyses
Genomic DNA was extracted based on the previously described method (Walbot 1988). The HPT (Hygromycin Phosphotransferase) gene was amplified for identifying the positive transgenic plants. Total RNA were isolated from flowers or leaves with TRIzol Reagent (TaKaRa, Japan), and then reverse-transcribed using PrimeScript™ RT reagent Kit with gDNA Eraser (TaKaRa, Japan). Gene-specific primers were designed using Primer Express 3.0 or Primer Premier 5.0 software. The rice Ubiquitin gene was used as an internal control. For analyzing the expression of OsERF101 and other marker genes in plants, qRT-PCR was performed with PrimeScript™ RT Master Mix (TaKaRa, Japan) using the CFX96™ Real-Time PCR Detection System (Bio-Rad, USA). Thermal cycling conditions were: 95 °C 1 min, 40 cycles of 95 °C 10 s, 60 °C 30 s, followed by dissociation curve analysis. Relative expression was normalized to the reference gene Ubiquitin. Mean and standard deviation values were calculated from three biological replicates.
GUS staining and RNA in situ hybridization
GUS staining was used to investigate the OsERF101 expression in T2 generation of OsERF101 promoter-GUS transgenic rice based on the methods described before (Jefferson et al. 1987). After staining and clearing, tissues were photographed using the stereo microscope S8APO (Leica).
For RNA in situ hybridization, wild-type flowers at various developmental stages were fixed in Histochoice MB Tissue Fixative reagent (Amresco, USA) for 16 h. After dehydration through an ethanol series and clearing through Histo-clear (Amresco, USA), tissues were embedded in Paraplast Plus (Sigma, USA) and sectioned at 3–6 mm thickness using an RM2245 Microtome (Leica Microsystems, Germany). For generating the RNA hybridization probe, a 389 bp OsERF101-specific cDNA fragment was amplified and cloned into pGEM-T vector (Promega, USA). The RNA probe was then transcribed in vitro using T7 or SP6 RNA polymerases provided in the DIG RNA labeling kit (Roche, Switzerland). RNA hybridization and immunological detection were performed following the protocol described previously (Yong et al. 2003). Tissues were photographed using a transmitted light microscopy (Carl Zeiss, Germany).
Trans-activation activity assay of OsERF101
The open reading frame of OsERF101 was cloned into the yeast vector pDEST-GBKT7, which was then transformed into AH109 host strain based on the described method (Gietz and Woods 2002). Yeasts were plated onto SD/-Trp dropout media for selection of the positive transformants. The positive colonies were then spotted onto SD/-Trp media with X-α-Gal for the reporter protein MEL1 activity assay. The blue colonies grown on SD/-Trp media indicated that OsERF101 activates the GAL4 reporter gene expression.
Abiotic stress treatments during vegetative and reproductive stages
The homozygous T3 generation of transgenic rice plants were used for phenotypic observation. For studying transcriptional responses to abiotic stresses, rice seedlings were grown in 1/8 Yoshida nutrition solution (Yoshida et al. 1976) for 14 days in a growth chamber under standard growth conditions described above, then NaCl, ABA or PEG6000 was added into the culture solution at a final concentration of 200 mM, 50 µM and 20%, respectively. Rice leaves were sampled at 0 h, 1 h, 3 h 7.5 h and 24 h after each treatment. OsERF101 expression level was then analyzed by qRT-PCR. For osmotic stress at the vegetative stage, rice seedlings were grown in 1/8 Yoshida nutrition media until five leaf-stage, then treated by 20% PEG6000 in the media. Three days later, the seedlings were analyzed and photographed.
For drought stress treatment at the reproductive stage, wild-type, overexpression, RNAi and knockout mutant were grown in soil pots in a greenhouse under standard growth conditions. To minimize experimental errors, each pot was filled with the same amount of soil and supplied with the same volume of water. After grown for about 2 months, rice plants started to switch into reproductive stage marked by the appearance of inflorescence primordia. Water was then drained off the pot until the soil water content dropped to 25%, and this level was maintained as described previously (Guo et al. 2016; Jin et al. 2013; Yao et al. 2018) for 1 week for Nipponbare genetic background plants, and for 10 days for Dongjin genetic background plants, due to its stronger drought tolerance. Control plants were irrigated normally all the time. After drought stress, watering was recovered till grain ripening. At least 40 plants for each line were analyzed, and the experiments were repeated for three times.
Physiological measurements of rice plants
After drought stress at the reproductive stage, the flag leaves were harvested for relative water content, chlorophyll content, proline content, superoxide dimutase (SOD) and peroxidase (POD) activity determination. Leaf relative water content as a percentage of (fresh weight − dry weight)/fresh weight was calculated (Kahn et al. 1993). Chlorophyll was extracted with 80% acetone; absorption of the extracts at 646.6 nm and 663.6 nm were measured and the total chlorophyll contents were calculated according to the following equation: Chltotal [µg ml−1] = 17.76 × A646.6 + 7.34 × A663.6 (Porra et al. 1989). Proline content was determined following the sulphosalicylic acid method (Troll and Lindsley 1955). SOD activity was measured based on its ability to inhibit photochemical reduction of nitro blue tetrazolium (NBT) chloride (Beauchamp and Fridovich 1971), and POD activity was decided by its H2O2-dependent oxidation activity for benzidine (Giannopolitis and Ries 1977). Rice pollen was stained by Alexander red to show the viability (Peterson et al. 2010). In addition, the survival rate and 1000-grain weight of the stressed plants were also surveyed for evaluating their drought tolerance trait.
Statistical analysis
All experiments were repeated three times unless otherwise stated. Significance of difference between samples and controls was performed using Student’s t-test.
Results
OsERF101 has trans-activation activity
Our previous comparative transcriptome analysis with rice flowers from normal and drought conditions has identified the gene of Os04g0398000 as a drought-inducible gene in flowers (Jin et al. 2013). Sequence analysis showed that this gene encodes an AP2/ERF transcription factor and was designated as OsERF101 in a genome-wide analysis of rice ERF family proteins (Nakano et al. 2006). A previous study has reported its function in regulating rice innate immunity and named it as OsRAP2.6 according to the sequence homology of its AP2/ERF domain to Arabidopsis RAP2.6 (Wamaitha et al. 2012). However, from the sequence analysis results, this gene has the highest similarity to AtRAP2.6L (RAP2.6-Like gene) (At5g13330) (identity 38.4%) instead of AtRAP2.6 (At1g43160) (identity 30.6%) in the full-length of protein sequence (Supplementary Fig. S1). We constructed the phylogenetic tree of OsERF101 homologous proteins using the DREB subfamily as the outgroup for rooting (Nakano et al. 2006). The phylogenetic analysis also showed that OsERF101 has closer relationship with AtRAP2.6L than AtRAP2.6. ERF family members from eudicots and monocots grouped respectively, indicating that the functional diversification within this branch predated the eudicot/monocot divergence (Fig. 1, Supplementary Fig. S1).
Phylogenetic analysis of the ERF subfamily members in various species. Bootstrap values were indicated on each branch point of the tree. Root was placed using an outgroup of the closest subfamily, DREB protein. Gm: Glycine max, Pt: Populus trichocarpa, At: Arabidopsis thaliana, Zm: Zea mays, Si: Setaria italica, Os: Oryza sativa, Bd: Brachypodium distachyon, Sb: Sorghum bicolor. Black diamonds show proteins from Arabidopsis; black triangles show proteins from rice
It has been reported that OsRAP2.6 is located in both nucleus and cytoplasm (Wamaitha et al. 2012). Our subcellular localization results using tobacco leaves also confirmed this result (Supplementary Fig. S2). So far, it is not known whether OsERF101 has transcription activation activity. To test this, we cloned OsERF101 into pDEST-GBKT7 vector, and transformed it into the yeast cell. Yeast cells carrying BD empty vector failed to turn blue on SD-/Trp supplemented with the chromogenic substrate X-α-Gal, while those containing BD-OsERF101 vector can activate the expression of reporter gene MEL1 and turned blue (Fig. 2), indicating that OsERF101 has transcriptional activation activity.
OsERF101 was preferentially expressed in the tapetum and microspores in flowers and induced by abiotic stress
To study the tissue-specific distribution of OsERF101 in rice, we used pOsERF101: GUS reporter lines to observe the GUS activity driven by OsERF101 promoter (Fig. 3a). GUS signal was not detected in root, callus, young leaf (leaves at five-leaf stage) and old leaf (leaves at three-tiller stage), but was slightly induced in young leaf and strongly induced in old leaf, especially after drought stress (Fig. 3a). GUS activity was strong in reproductive tissues including inflorescence primordia, lemma/palea of young flowers, anther and pistil.
Expressional studies of OsERF101 in rice. a GUS staining pattern of the pOsERF101:GUS transgenic lines grown under normal growth conditions. YL young leaf at five-leaf stage, YL-D drought stressed young leaf at five-leaf stage, OL old leaf at three-tiller stage, OL-D drought-stressed old leaf at three-tiller stage, IP inflorescence primordia, YF1 young flower at stage 4 or 5, YF2 young flower at stage 7 or 8, DS developing seeds on the fifth day post fertilization. Scale bars: 1 cm in root, callus, YL, YL-D, OL and OL-D; 0.2 cm in IP; 0.5 mm in YF1 and pistil; 1 mm in YF2, anther and DS. b RNA in situ hybridization analyses of the OsERF101 expression in the anthers under the normal conditions. IP inflorescence primordia; brown signal indicates the expression of OsERF101. Sense probe was used as the hybridization control. T tapetum, Msp microspore. Scale bars: 50 µm. c Time-dependent expression analyses of OsERF101 by qRT-PCR under different treatments. Two-week old seedlings were treated by PEG6000, ABA or NaCl in the culture media. PEG6000, 20%; ABA, 50 µM; NaCl, 200 mM. Values indicated means ± SD from three biological replicates. Significance of difference between 0 h and other time points was analyzed by Student’s t-test, *P < 0.05; **P < 0.01
To further examine the OsERF101 expression pattern in anther, we performed RNA in situ hybridization using the wild-type floral sections. The OsERF101 transcript was detected in the tapetum and microspores, especially at anther developmental stage 4, 5 and 8 (Fig. 3b). From the stage 10 on, the OsERF101 expression signal became weaker. Taken together, OsERF101 was expressed in developing microspores and tapetum, which suggests that it might play a role in the anther development.
qRT-PCR method was employed to examine the transcriptional responsiveness of OsERF101 to abiotic stresses. The result showed that OsERF101 was induced by 20% PEG6000 and ABA, but not by NaCl (Fig. 3c), indicating that OsERF101 is an osmotic stress- and ABA-responsive gene.
Overexpression of OsERF101 gene in rice plants enhanced dehydration tolerance at both vegetative and reproductive stages
To study OsERF101 function, we generated the overexpression and RNA interference (RNAi) transgenic plants of OsERF101 (Supplementary Fig. S3a). Meanwhile, we also ordered the oserf101 T-DNA insertion mutant from the seed stock at Kyung Hee University (RISD). PCR results showed that the T-DNA fragment was inserted in the second exon of OsERF101 and the genotypic identification showed that oserf101 is a null mutant (Supplementary Fig. S3b). Based on the RT-PCR and qRT-PCR results, representative overexpression lines and RNAi lines (Supplementary Fig. S3c, d) were chosen for further assessment for their tolerance to drought. Among them, OX6 has nearly the same OsERF101 expression level as the wild type and was used as a vector control (VC) (Supplementary Fig. S3d).
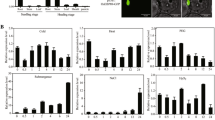
To assess the effects of altered OsERF101 expression level on dehydration/drought resistance, positive transgenic plants were grown in 1/8 Yoshida nutrition solution till five-leaf stage, then treated by 20% PEG6000. Three days after treatment, most of the leaves in control plants rolled severely while those of the OsERF101-overexpression lines only curled mildly (Fig. 4b, d). We also imposed drought stress on the plants grown in soil pots upon the onset of reproductive growth. The overexpression plants displayed stronger drought-resistance phenotype than control plants (Fig. 4f, h), while no difference between them was observed under normal growth conditions (Fig. 4a, c, e, g). After recovery for 3d, only 38.3% of the control plants survived, whereas up to 98.2% of the OsERF101-overexpression plants survived (Fig. 4i). The transgenic plants with higher OsERF101 expression level displayed a higher survival rate. In addition, the leaves of OsERF101-overexpression plants also lost water less quickly than WT control under drought stress (Fig. 4j). Photosynthetic capacity can be used as a reliable indicator to evaluate the energetic/metabolic imbalance of photosynthesis and yield performance under water deficit condition (Araus et al. 1998; Guo et al. 2008). Therefore, we examined the chlorophyll contents of the plants under normal and drought conditions, respectively. While drought stress reduced the chlorophyll contents of all plants, the chlorophyll content of plants with the highest level of OsERF101 (OX9) was still higher than that of the control plants (Fig. 4k). These results suggested that higher OsERF101 expression level could enhance plant tolerance to drought stress.
Phenotype of OsERF101 overexpression plants at the vegetative and reproductive stages under osmotic and drought stresses. a–d Phenotype of the OsERF101 overexpression seedlings under osmotic stress caused by 20% PEG6000. a, c Plants before stress; b, d plants after stress. A transgenic line with OsERF101 expression level similar to wild type was used as a vector control (VC). OX2, 8, 9, overexpression lines. Scale bars, 2 cm. e–h Phenotype of the OsERF101 overexpression plants at the reproductive stage under drought stress. e, g Before drought treatment; f, h after drought treatment. Scale bars, 10 cm. i The survival rates of different plants under drought stress at the reproductive stage. j The relative water contents (RWC) of leaves at the reproductive stage. k The chlorophyll contents of leaves at the reproductive stage. l The proline contents of leaves at the reproductive stage. m The POD activities of leaves at the reproductive stage. n The SOD activities of leaves at the reproductive stage. Drought stress experiments were repeated three times with similar results. Values were shown as means ± SD (n ≥ 40) in i–n. Significance of difference between WT control and the transgenic lines under the same conditions was analyzed by Student’s t-test, *P ≤ 0.05, **P ≤ 0.01
Proline accumulation is protective for plants to cope with water stress (Zhou et al. 2009). We measured the proline content in wild type and transgenic plants before and after drought stress. The proline content of OsERF101-overexpression lines (62.5–65.6 µg g−1) were higher than that of control plants (47.7 µg g−1) after drought stress (Fig. 4l). No consistent difference of proline content was detected among transgenic and control plants before drought stress (Fig. 4l). The results suggested that overexpression of OsERF101 enhanced the accumulation of free proline in rice to improve drought tolerance. Drought stress also activates the generation of reactive oxygen species (ROS), which causes multiple levels of damages to plants (Xia et al. 2009). We therefore examined the ROS-scavenging activities in both wild-type and transgenic plants. The results showed that under drought stress, overexpression of OsERF101 led to increased POD activity (207.4–216.5 U g−1 min−1 FW) compared to wild-type plants (173.6 U g−1 min−1 FW) (Fig. 4m), whereas no obvious difference was observed for SOD activity between transgenic and control plants under both normal and drought conditions (Fig. 4n). These results suggested that the increased drought tolerance of OsERF101-overexpression plants is at least partially due to increased proline accumulation and ROS clearing activities in plants.
Disruption of OsERF101 in rice reduced drought tolerance during the reproductive stage
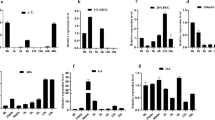
In addition to the overexpression lines, we also observed the phenotype of the OsERF101-RNAi plants and the oserf101 mutant (KO) under osmotic stress (Supplementary Fig. S4). The RNAi plants and KO mutant showed no growth phenotype distinct from the wild-type plants under normal conditions (Supplementary Fig. S4 a, c) and 20% PEG6000 treatment (Supplementary Fig. S4 b, d). However, when treated by drought at reproductive stage, OsERF101-RNAi lines showed more sensitive phenotype to drought stress than the control plants (Fig. 5a). Meanwhile, the KO mutant was also more susceptible to drought than its wild type control Dongjin (DJ) (Fig. 5a). Since DJ genetic background plants had stronger drought tolerance than Nipponbare plants, DJ plants (DJ wild type and KO mutant) were stressed three more days than Nipponbare rice (Nipponbare wild type and RNAi lines). About 32.3–35.7% of OsERF101-RNAi transgenic plants were recovered, compared with 58.1% of the wild type control plants after drought stress (Fig. 5b), and the KO mutant also had a survival rate at approximately 60% of the DJ control plants. The leaves of OsERF101 RNAi lines and KO mutant also lost water more quickly than their wild type siblings under drought (Fig. 5c). The chlorophyll content of RNAi and knockout plants were not significantly different from that of the control plants, although the drought stress seriously reduced the chlorophyll content of all plants (Fig. 5d).
Disruption of OsERF101 reduced rice drought tolerance. a Phenotype of OsERF101 RNAi (Ri) lines and the knockout (KO) mutant under normal and drought conditions. Scale bars: 10 cm. b The survival rate of different plants under drought stress at the reproductive stage. c The relative water contents (RWC) of leaves at the reproductive stage. d The chlorophyll content of leaves at the reproductive stage. e The proline content of different plants at the reproductive stage. f The POD activities of different plants at the reproductive stage. g The SOD activities of different plants at the reproductive stage. Drought stress experiments were done in triplicates. Values were shown as means ± SD (n ≥ 40) in b–g. Significance of difference between WT control and the transgenic lines or the knockout mutant under the same conditions was analyzed by Student’s t-test, *P ≤ 0.05, **P ≤ 0.01
We further examined the proline content, POD and SOD activities in the wild type, RNAi and KO plants during the reproductive stage. Results showed that after water stress, the proline content, POD and SOD activities were all reduced in the RNAi plants as well as in the KO mutant (Fig. 5e–g). However, before drought stress, most of them had no significant differences except POD. The POD activity in the oserf101 mutant was lower than that of wild-type plants even without stress, suggesting a possible ecotype influence on the gene function.
Overexpression of OsERF101 improved pollen fertility and seed setting rate under drought stress
OsERF101 is drought-responsive and mainly expressed in flowers (Fig. 3), suggesting that it could be involved in drought-adaptive response in reproductive tissues. Yield stability is an important crop trait for agricultural production and largely determined by the reproductive growth conditions of plants. Therefore, we analyzed the effects of OsERF101 expression on pollen development and gain yield under drought stress. The OsERF101-overexpression lines had significantly more viable pollen grains under drought stress (73.5%) than the control plants (62.3%) and OsERF101-RNAi lines (41.5%) (Fig. 6a, b). Meanwhile, the ERF101 overexpression plants also had higher seed setting rate (62.6%) (Fig. 6c). In the RNAi plants, the viable pollen grains are less than wild type both before and after drought stress, indicating that disruption of OsERF101 may affect pollen development. No significant difference in 1000-kernal weight was observed between transgenic plants and control plants, although the 1000-kernal weight of all plants decreased after drought stress (Fig. 6d). These results indicated that OsERF101 can improve pollen fertility and thus enhance seed setting rate under drought stress.
OsERF101 enhanced rice reproductive-stage drought tolerance. a Alexander red staining showing the fertility of pollen grains. The viable pollen grains were stained as dark purple and inviable blue. b Pollen fertilities of different lines under normal and drought conditions. c Seed setting rates of different lines under normal and drought conditions. d 1000-kernal weights of different lines under normal and drought conditions. The experiments were done in triplicates and values were shown as means ± SD (n ≥ 20) in b–d. Significance of difference between WT control and the transgenic lines under the same conditions was analyzed by Student’s t-test, *P ≤ 0.05, **P ≤ 0.01
Expression of ABA-responsive genes was increased in overexpression lines
To seek the molecular cues underlying the phenotype of OsERF101 overexpression and mutant plants, we compared the expression levels of stress-responsive genes in flowers from the overexpression, KO mutant and the wild-type control plants, under normal and drought conditions, respectively. The results showed that many drought-responsive genes such as ABA8OX3, ABA2, DREB2, LEA3, P5CS1, OAT and PODs genes were strikingly up-regulated in the overexpression lines (OX8 and OX9) after drought stress, but down-regulated in the KO mutant for most of them. ABA degradation gene ABA8OX3 and ABA synthesis gene ABA2 were both up-regulated in the overexpression plants compared to wild type after drought stress (Fig. 7a), suggesting a possible feedback regulation mechanism of ABA on ABA homeostasis. In order to track the time-dependent expression level changes of ABA-responsive genes under osmotic stress, we selected three marker genes, POD2, LEA3 and RD22, for further analysis. 20% PEG6000 was used to simulate the drought stress. Results showed that POD2 expression level increased all the time whereas LEA3 and RD22 firstly increased and then dropped under osmotic stress, and for most of the time, these genes were expressed at a higher level in overexpression lines than that in the wild-type plants (Fig. 7b). OsERF101 probably improves rice drought tolerance by indirectly or directly up-regulating the expressions of these drought-responsive genes. Correspondingly, LEA3 and RD22 genes were expressed at relatively lower levels in the knockout mutant than wild type. Since LEA3 and RD22 are the typical ABA-responsive genes (Hundertmark and Hincha 2008; Iwasaki et al. 1995; Xiao et al. 2007; Yamaguchi-Shinozaki and Shinozaki 1993), to further explore the possibility that OsERF101 enhanced drought tolerance through ABA signaling pathway, we treated the three-leaf-stage rice with 50 µM ABA. Results showed that OsERF101 overexpression plants grew more slowly than the control plants (Fig. 7c, d), indicating that overexpression of OsERF101 confers sensitivity to ABA treatment and OsERF101 is involved in ABA signaling pathway.
ABA signaling pathway was affected in the transgenic plants. a Expression level changes of stress-responsive genes in the flowers of the overexpression lines and the knockout mutant compared with wild type control. The flowers of stage 5–8 after drought treatment for 1 week were harvested for qRT-PCR. Values indicate means ± SD from three biological replicates. b Time-dependent expression curves of ABA-responsive genes after 20% PEG6000 treatment at the vegetative stage. Leaves from 2-week seedlings were analyzed. Values indicate means ± SD from three biological replicates. c Phenotype of three-leaf-stage rice seedlings treated by 50 µM ABA for 2 days. d Seedling growth measurements after ABA treatment. VC has similar ERF101 expression level to wild type and was used as a vector control. Values were shown as means ± SD (n ≥ 40). Significance of difference between WT or VC control and the transgenic lines under the same conditions was analyzed by Student’s t-test, *P ≤ 0.05, **P ≤ 0.01
Discussion
Sessile plants have evolved distinct mechanisms to respond and adapt to drought stress. We found that OsERF101 plays a role in this process. Previous work reported that in Arabidopsis, the ERF transcription factor AtRAP2.6 is involved in JA-regulated plant disease resistance (Ali et al. 2013; He et al. 2004). Overexpression of AtRAP2.6 resulted in a dwarf phenotype with extensive secondary branching and small siliques, indicating a development role of AtRAP2.6 as well (Krishnaswamy et al. 2011). OsERF101 was named as OsRAP2.6 since it has an AP2/ERF domain with 94% identity to that of AtRAP2.6 and also functions in suppressing blast fungus growth in rice (Wamaitha et al. 2012). However, we found that this gene is more similar to AtRAP2.6L in the full-length sequence and phylogenetic relationship. Arabidopsis RAP2.6L is reported to play a role in abiotic stress tolerance such as salt, drought and water logging stress (Krishnaswamy et al. 2011; Liu et al. 2012b), but not in biotic stress response. Our results also showed that rice OsERF101 was induced by ABA and osmotic stress, and overexpression of OsERF101 improves plant survival and pollen fertility under the drought stress, supporting a role of this gene in abiotic stress response. Therefore, OsERF101 might encode an intermediate phylogenetic type of ERF protein in evolution and functions in both biotic and abiotic stress responses.
Drought stress can lead to excessive ROS generation, and cause damages and even cell death in plants (Li et al. 2013a). ROS scavenging enzymes, such as SOD and POD, are important for plants to deal with oxidative stress induced by environmental stresses. Under drought stress, the activity of PODs in OsERF101 overexpression plants was increased while it was reduced in the RNAi lines and knockout mutant (Figs. 4, 5). Consistent with this, two genes encoding PODs (POD1 and POD2) were up-regulated in the OsERF101 overexpression lines shown by qRT-PCR. SOD activity displayed no significant changes in OsERF101 overexpression plants but was slightly decreased in the RNAi and mutant plants (Figs. 4, 5). On the other hand, overproduction of proline in plants may lead to increased tolerance against abiotic stresses through osmotic adjustment or membrane stabilization (Verbruggen and Hermans 2008; Zhou et al. 2009). We also found that the proline content was increased in the overexpression lines but decreased in the RNAi and mutant plants compared to wild type control. In support of this, the proline biosynthetic genes OsP5CS1 and OsOAT were transcriptionally up-regulated and the catabolic gene OsP5CDH was down-regulated in the overexpression lines, and as a contrast, OsP5CS1 and OsP5CDH genes had the opposite changes in KO mutant. Taken together, OsERF101 could protect plant cells by promoting ROS clearing and osmo-protectant accumulation, and OsERF101 is a positive regulator of drought response.
Improving grain yield is an important goal for breeding rice elite cultivar, and one of the important processes affecting crop yield is the reproductive process. The stress tolerance properties at this stage largely determine the final output of crop growth. Therefore, seeking for genes which can improve reproductive development under stresses is important for molecular breeding. OsERF101 is predominantly expressed in flowers under normal growth conditions, and the RNA in situ hybridization results also showed that it is expressed in tapetum and microspores, indicating a possible role of this gene in male development. This speculation is supported by the evidence that RNA interference and knock out mutation of OsERF101 reduced the pollen fertility under normal growth conditions. OsERF101 is induced by drought stress in flowers (Jin et al. 2013) and in leaves, and overexpression of this gene improves the pollen fertility and seed setting rate under drought stress, suggesting a dual role of OsERF101 in reproductive development and in drought responses. Our results demonstrated that OsERF101 regulates male development process under drought stress.
ABA regulates the expression level of a large number of genes under abiotic stresses (Manavalan et al. 2012; Seiler et al. 2011; Wilkinson and Davies 2002; Yoshida et al. 2010). In ABA signaling pathway, ABA2 is involved in ABA biosynthesis (Cheng et al. 2002; Gonzalez-Guzman et al. 2002); whereas ABA8OX1 and ABA8OX3 are responsible for ABA degradation (Kushiro et al. 2004). We found that the expression levels of both ABA synthesis and degradation genes were increased in the flowers of OsERF101-overexpression lines. It is likely that drought stress increased ABA level in the overexpression lines and it conversely activated the expression of ABA degradation genes such as ABA8OX3 for protecting the anthers from the destructive impact of excess ABA on reproductive development (Ji et al. 2011). The OsERF101-overexpression plants also showed ABA-sensitive phenotype (Fig. 7b), suggesting that the ABA metabolism or ABA signaling in these plants were influenced. qRT-PCR results revealed that drought stress induced the expression of five ABA-responsive genes more strongly in the overexpression plants than in wild-type plants (Fig. 7), further supporting a role of OsERF101 in ABA signaling pathway. Considering that OsERF101 is an ABA-inducible gene, the function of OsERF101 in ABA signaling pathway remains further investigation.
Interestingly, OsERF101, as a transcription factor, is localized both in cytosol and in nucleus based on our results (Supplementary Fig. S2) and another report (Wamaitha et al. 2012). It remains unknown why it is in cytosol. Wamaitha et al. proposed that OsERF101 was phosphorylated by OsMAPK3/6 in both nucleus and cytoplasm in rice innate immunity (Wamaitha et al. 2012). It is worthy of further study whether this modification is related to its localization status. Other dually-localized protein with transcriptional activity have also been reported, such as Arabidopsis bZIP28, NAC089 and PIF7 (Leivar et al. 2008; Liu et al. 2007; Yang et al. 2014). There are various mechanisms underlying the translocation behavior of transcription factors. Studying whether OsERF101 changes its localization under drought stress will provide new insights on its action of mode.
In summary, our results revealed that OsERF101 positively regulates drought responses. OsERF101 increases the expression levels of drought-responsive genes for synthesizing proline and for eliminating harmful ROS to protect vegetative and reproductive tissues against water deficit stress.
References
Ali MA, Abbas A, Kreil DP, Bohlmann H (2013) Overexpression of the transcription factor RAP2.6 leads to enhanced callose deposition in syncytia and enhanced resistance against the beet cyst nematode Heterodera schachtii in Arabidopsis roots BMC Plant Biol. https://doi.org/10.1186/1471-2229-13-47
Araus JLAT, Voltas J, Nakkoul H, Nachit MM (1998) Chlorophyll fluorescence as a selection criterion for grain yield in durum wheat under Mediterranean conditions. Field Crop Res 55:209–223
Bartels D, Sunkar R (2005) Drought and salt tolerance in plants. Crit Rev Plant Sci 24:23–58
Beauchamp C, Fridovich I (1971) Superoxide dismutase: improved assays and an assay applicable to acrylamide gels. Anal Biochem 44:276–287
Chakravarthy S, Tuori RP, D’Ascenzo MD, Fobert PR, Despres C, Martin GB (2003) The tomato transcription factor Pti4 regulates defense-related gene expression via GCC box and non-GCC box cis elements. Plant Cell 15:3033–3050. https://doi.org/10.1105/tpc.017574
Chen JQ, Meng XP, Zhang Y, Xia M, Wang XP (2008) Over-expression of OsDREB genes lead to enhanced drought tolerance in rice. Biotechnol Lett 30:2191–2198. https://doi.org/10.1007/s10529-008-9811-5
Cheng WH et al (2002) A unique short-chain dehydrogenase/reductase in Arabidopsis glucose signaling and abscisic acid biosynthesis and functions. Plant Cell 14:2723–2743
Du H, Wang N, Cui F, Li X, Xiao J, Xiong L (2010) Characterization of the beta-carotene hydroxylase gene DSM2 conferring drought and oxidative stress resistance by increasing xanthophylls and abscisic acid synthesis in rice. Plant Physiol 154:1304–1318
Fukao T, Yeung E, Bailey-Serres J (2011) The submergence tolerance regulator SUB1A mediates crosstalk between submergence and drought tolerance in rice. Plant Cell 23:412–427
Giannopolitis CN, Ries SK (1977) Superoxide dismutases: II. Purification and quantitative relationship with water-soluble protein in seedlings. Plant Physiol 59:315–318
Gietz RD, Woods RA (2002) Transformation of yeast by lithium acetate/single-stranded carrier DNA/polyethylene glycol method. Methods Enzymol 350:87–96
Gilmour SJ, Zarka DG, Stockinger EJ, Salazar MP, Houghton JM, Thomashow MF (1998) Low temperature regulation of the Arabidopsis CBF family of AP2 transcriptional activators as an early step in cold-induced COR gene expression. Plant J 16:433–442
Gonzalez-Guzman M et al (2002) The short-chain alcohol dehydrogenase ABA2 catalyzes the conversion of xanthoxin to abscisic aldehyde. Plant Cell 14:1833–1846
Guo PG, Baum M, Varshney RK, Graner A, Grando S, Ceccarelli S (2008) QTLs for chlorophyll and chlorophyll fluorescence parameters in barley under post-flowering drought. Euphytica 163:203–214. https://doi.org/10.1007/s10681-007-9629-6
Guo C, Yao L, You C, Wang S, Cui J, Ge X, Ma H (2016) MID1 plays an important role in response to drought stress during reproductive development. Plant J 88:280–293. https://doi.org/10.1111/tpj.13250
He P et al (2004) Activation of a COI1-dependent pathway in Arabidopsis by Pseudomonas syringae type III effectors and coronatine. Plant J 37:589–602. https://doi.org/10.1111/j.1365-313X.2003.01986.x
Hundertmark M, Hincha DK (2008) LEA (late embryogenesis abundant) proteins and their encoding genes in Arabidopsis thaliana. BMC Genomics 9:118–130. https://doi.org/10.1186/1471-2164-9-118
Iwasaki T, Yamaguchishinozaki K, Shinozaki K (1995) Identification of a cis-regulatory region of a gene in Arabidopsis thaliana whose induction by dehydration is mediated by abscisic-acid and requires protein-synthesis. Mol Gen Genet 247:391–398. https://doi.org/10.1007/Bf00293139
Jefferson RA, Kavanagh TA, Bevan MW (1987) GUS fusions: beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J 6:3901–3907
Jeon JS et al (2000) T-DNA insertional mutagenesis for functional genomics in rice. Plant J 22:561–570
Jeong DH et al (2006) Generation of a flanking sequence-tag database for activation-tagging lines in japonica rice. Plant J 45:123–132. https://doi.org/10.1111/j.1365-313X.2005.02610.x
Ji XM et al (2011) Control of abscisic acid catabolism and abscisic acid homeostasis is important for reproductive stage stress tolerance in cereals. Plant Physiol 156:647–662. https://doi.org/10.1104/pp.111.176164
Jin Y, Yang H, Wei Z, Ma H, Ge X (2013) Rice male development under drought stress: phenotypic changes and stage-dependent transcriptomic reprogramming. Mol Plant 6:1630–1645. https://doi.org/10.1093/mp/sst067
Kahn TL, Fender SE, Bray EA, O’Connell MA (1993) Characterization of expression of drought- and abscisic acid-regulated tomato genes in the drought-resistant species Lycopersicon pennellii. Plant Physiol 103:597–605
Kiyosue T, Yamaguchi-Shinozaki K, Shinozaki K (1993) Characterization of cDNA for a dehydration-inducible gene that encodes a CLP A, B-like protein in Arabidopsis thaliana L. Biochem Biophys Res Commun 196:1214–1220
Krishnaswamy S, Verma S, Rahman MH, Kav NN (2011) Functional characterization of four APETALA2-family genes (RAP2.6. RAP2.6L, DREB19 and DREB26) in Arabidopsis. Plant Mol Biol 75:107–127. https://doi.org/10.1007/s11103-010-9711-7
Kushiro T et al (2004) The Arabidopsis cytochrome P450 CYP707A encodes ABA 8′-hydroxylases: key enzymes in ABA catabolism. EMBO J 23:1647–1656. https://doi.org/10.1038/sj.emboj.7600121
Leivar P et al (2008) The Arabidopsis phytochrome-interacting factor PIF7, together with PIF3 and PIF4, regulates responses to prolonged red light by modulating phyB levels. Plant Cell 20:337–352. https://doi.org/10.1105/tpc.107.052142
Li XY et al (2013a) Overexpression of Arachis hypogaea AREB1 gene enhances drought tolerance by modulating ROS scavenging and maintaining endogenous ABA content. Int J Mol Sci 14:12827–12842. https://doi.org/10.3390/ijms140612827
Li YS et al (2013b) A novel nuclear protein phosphatase 2C negatively regulated by ABL1 is involved in abiotic stress and panicle development in rice. Mol Biotechnol 54:703–710. https://doi.org/10.1007/s12033-012-9614-8
Liu JX, Srivastava R, Che P, Howell SH (2007) An endoplasmic reticulum stress response in Arabidopsis is mediated by proteolytic processing and nuclear relocation of a membrane-associated transcription factor, bZIP28. Plant Cell 19:4111–4119. https://doi.org/10.1105/tpc.106.050021
Liu D, Chen X, Liu J, Ye J, Guo Z (2012a) The rice ERF transcription factor OsERF922 negatively regulates resistance to Magnaporthe oryzae and salt tolerance. J Exp Bot 63:3899–3911. https://doi.org/10.1093/jxb/ers079
Liu P, Sun F, Gao R, Dong H (2012b) RAP2.6L overexpression delays waterlogging induced premature senescence by increasing stomatal closure more than antioxidant enzyme activity. Plant Mol Biol 79:609–622. https://doi.org/10.1007/s11103-012-9936-8
Manavalan LP, Chen X, Clarke J, Salmeron J, Nguyen HT (2012) RNAi-mediated disruption of squalene synthase improves drought tolerance and yield in rice. J Exp Bot 63:163–175. https://doi.org/10.1093/jxb/err258
Mittler R (2002) Oxidative stress, antioxidants and stress tolerance. Trends Plant Sci 7:405–410
Nakano T, Suzuki K, Fujimura T, Shinshi H (2006) Genome-wide analysis of the ERF gene family in Arabidopsis rice. Plant Physiol 140:411–432. https://doi.org/10.1104/pp.105.073783
Ohme-Takagi M, Shinshi H (1995) Ethylene-inducible DNA binding proteins that interact with an ethylene-responsive element. Plant Cell 7:173–182. https://doi.org/10.1105/tpc.7.2.173
Ozawa K (2009) Establishment of a high efficiency Agrobacterium-mediated transformation system of rice (Oryza sativa L.). Plant Sci 176:522–527
Park JM, Park CJ, Lee SB, Ham BK, Shin R, Paek KH (2001) Overexpression of the tobacco Tsi1 gene encoding an EREBP/AP2-type transcription factor enhances resistance against pathogen attack and osmotic stress in tobacco. Plant Cell 13:1035–1046
Peterson R, Slovin JP, Chen JQ (2010) A simplified method for differential staining of aborted and non-aborted pollen grains. Int J Plant Biol 1:66–69
Porra RJ, Thompson WA, Kriedemann PE (1989) Determination of accurate extinction coefficients and simultaneous equations for assaying chlorophyll’s a and b extracted with four different solvents: verification of the concentration of chlorophyll standards by atomic spectroscopy. Biochim Biophys Acta 1051:15–28
Quan R, Hu S, Zhang Z, Zhang H, Huang R (2010) Overexpression of an ERF transcription factor TSRF1 improves rice drought tolerance. Plant Biotechnol J 8:476–488. https://doi.org/10.1111/j.1467-7652.2009.00492.x
Sakuma Y, Liu Q, Dubouzet JG, Abe H, Shinozaki K, Yamaguchi-Shinozaki K (2002) DNA-binding specificity of the ERF/AP2 domain of Arabidopsis DREBs, transcription factors involved in dehydration- and cold-inducible gene expression. Biochem Biophys Res Commun 290:998–1009. https://doi.org/10.1006/bbrc.2001.6299
Seiler C et al (2011) ABA biosynthesis and degradation contributing to ABA homeostasis during barley seed development under control and terminal drought-stress conditions. J Exp Bot 62:2615–2632. https://doi.org/10.1093/jxb/erq446
Sharoni AM et al (2011) Gene structures, classification and expression models of the AP2/EREBP transcription factor family in rice. Plant Cell Physiol 52:344–360. https://doi.org/10.1093/pcp/pcq196
Shinozaki K, Yamaguchi-Shinozaki K (2007) Gene networks involved in drought stress response and tolerance. J Exp Bot 58:221–227. https://doi.org/10.1093/jxb/erl164
Shinozaki K, Yamaguchi-Shinozaki K, Seki M (2003) Regulatory network of gene expression in the drought and cold stress responses. Curr Opin Plant Biol 6:410–417
Stockinger EJ, Gilmour SJ, Thomashow MF (1997) Arabidopsis thaliana CBF1 encodes an AP2 domain-containing transcriptional activator that binds to the C-repeat/DRE, a cis-acting DNA regulatory element that stimulates transcription in response to low temperature and water deficit. Proc Natl Acad Sci USA 94:1035–1040
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28:2731–2739. https://doi.org/10.1093/molbev/msr121
Troll W, Lindsley J (1955) A photometric method for the determination of proline. J Biol Chem 215:655–660
Verbruggen N, Hermans C (2008) Proline accumulation in plants: a review. Amino Acids 35:753–759. https://doi.org/10.1007/s00726-008-0061-6
Walbot (1988) Preparation of DNA from single rice seedlings. Rice Genet Newsl 5:149–151
Wamaitha MJ, Yamamoto R, Wong HL, Kawasaki T, Kawano Y, Shimamoto K (2012) OsRap2.6 transcription factor contributes to rice innate immunity through its interaction with receptor for activated kinase-C 1 (RACK1). Rice (N Y) 5:35. https://doi.org/10.1186/1939-8433-5-35
Wan L, Zhang J, Zhang H, Zhang Z, Quan R, Zhou S, Huang R (2011) Transcriptional activation of OsDERF1 in OsERF3 and OsAP2-39 negatively modulates ethylene synthesis and drought tolerance in rice. PLoS ONE 6:e25216. https://doi.org/10.1371/journal.pone.0025216
Wang H et al (2004) Ectopic overexpression of tomato JERF3 in tobacco activates downstream gene expression and enhances salt tolerance. Plant Mol Biol 55:183–192. https://doi.org/10.1007/s11103-004-0113-6
Wilkinson S, Davies WJ (2002) ABA-based chemical signalling: the co-ordination of responses to stress in plants. Plant Cell Environ 25:195–210
Xia XJ et al (2009) Reactive oxygen species are involved in brassinosteroid-induced stress tolerance in cucumber. Plant Physiol 150:801–814. https://doi.org/10.1104/pp.109.138230
Xiao B, Huang Y, Tang N, Xiong L (2007) Over-expression of a LEA gene in rice improves drought resistance under the field conditions. Theor Appl Genet 115:35–46. https://doi.org/10.1007/s00122-007-0538-9
Yamaguchishinozaki K, Shinozaki K (1994) A novel cis-acting element in an Arabidopsis gene is involved in responsiveness to drought, low-temperature or high-salt stress. Plant Cell 6:251–264. https://doi.org/10.1105/Tpc.6.2.251
Yamaguchi-Shinozaki K, Shinozaki K (1993) The plant hormone abscisic acid mediates the drought-induced expression but not the seed-specific expression of rd22, a gene responsive to dehydration stress in Arabidopsis thaliana. Mol Gen Genet 238:17–25
Yang ZT et al (2014) The membrane-associated transcription factor NAC089 controls ER-stress-induced programmed cell death in plants. PLoS Genet 10:e1004243. https://doi.org/10.1371/journal.pgen.1004243
Yao L et al (2018) The AWPM-19 family protein OsPM1 mediates abscisic acid influx and drought response in rice. Plant Cell. https://doi.org/10.1105/tpc.17.00770
Yong WD et al (2003) Vernalization-induced flowering in wheat is mediated by a lectin-like gene VER2. Planta 217:261–270. https://doi.org/10.1007/s00425-003-0994-7
Yoshida S, Forno D, Cock JH, Gomez KA (1976) Laboratory manual for physiological studies of rice. International Rice Research Institute, Manila, Philippines, pp 61–66
Yoshida T et al (2010) AREB1, AREB2, and ABF3 are master transcription factors that cooperatively regulate ABRE-dependent ABA signaling involved in drought stress tolerance and require ABA for full activation. Plant J 61:672–685. https://doi.org/10.1111/j.1365-313X.2009.04092.x
Zhang H et al (2004) Tomato stress-responsive factor TSRF1 interacts with ethylene responsive element GCC box and regulates pathogen resistance to Ralstonia solanacearum. Plant Mol Biol 55:825–834. https://doi.org/10.1007/s11103-004-2140-8
Zhang CJ, Zhao BC, Ge WN, Zhang YF, Song Y, Sun DY, Guo Y (2011) An apoplastic h-type thioredoxin is involved in the stress response through regulation of the apoplastic reactive oxygen species in rice. Plant Physiol 157:1884–1899. https://doi.org/10.1104/pp.111.182808
Zhou W, Li Y, Zhao BC, Ge RC, Shen YZ, Wang G, Huang ZJ (2009) Overexpression of TaSTRG gene improves salt and drought tolerance in rice. J Plant Physiol 166:1660–1671. https://doi.org/10.1016/j.jplph.2009.04.015
Acknowledgements
This research work was supported by the projects from National Nature Science Foundation of China No. 31770274 to XG and No. 31700208 to YJ, Shanghai Science and Technology Innovation Action Plan No. 18JC1411800 to XG, the Open Research Funds of the State Key Laboratory of Genetic Engineering, Fudan University No. SKLGE-1617 to XZ, and Shanghai Engineering Research Center of Plant Germplasm Resources No. 17DZ2252700 to YJ.
Author information
Authors and Affiliations
Contributions
XG and HM designed the experiments. XG and YJ wrote the article. YJ, WP, XZ and XC did the experiments. ML provided assistance in manuscript preparation.
Corresponding authors
Additional information
Accession numbers Sequence data of genes reported in this paper can be found in RAP-DB (http://rapdb.dna.affrc.go.jp/) in the following numbers: OsERF101 and OsRAP2.6 (Os04g0398000), ABA8OX3 (Os09g0457100), ABA2 (Os04g0448900), GAMYB (Os01g0812000), DREB2 (Os01g0165000), LEA3 (Os05g0542500), ERF34 (Os04g0550200), POD1 (Os03g0369000), POD2 (Os01g0326300), RD22 (Os01g0733500), OsP5CS1 (Os05g0455500), OsP5CDH (Os05g0536400), OsOAT (Os03g643300), and in TAIR (http://www.arabidopsis.org): AtRAP2.6 (At1g43160) and AtRAP2.6L (At5g13330).
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Jin, Y., Pan, W., Zheng, X. et al. OsERF101, an ERF family transcription factor, regulates drought stress response in reproductive tissues. Plant Mol Biol 98, 51–65 (2018). https://doi.org/10.1007/s11103-018-0762-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11103-018-0762-5