Abstract
Type 2C protein phosphatase plays an important role in the signal transduction of stress response in plants. In this paper, we identified a novel stress-induced type 2C protein phosphatase gene OsSIPP2C1 from rice. OsSIPP2C1 contains a complete open reading frame of 1,074 bp, encoding a protein with 357 amino acids. OsSIPP2C1 expression was up-regulated by high salt, PEG6000 and exogenous ABA, and enhanced in the abl1 mutant under normal, salt, or drought condition. Interestingly, OsSIPP2C1 expression was increased during the early panicle development. Subcellular localization assay using rice protoplast cells indicated that OsSIPP2C1 was predominantly located in the nucleus. Together, it is suggested that a nuclear PP2C protein OsSIPP2C1 negatively regulated by ABL1 is involved in abiotic stress and panicle development in rice.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Environmental stresses such as cold, drought, and salinity adversely affect plant growth and crop production. Plants can initiate a number of molecular, cellular, and physiological changes to respond and adapt to various stresses. It has been established that two distinct pathways (ABA dependent and ABA independent) are involved in stress-responsive gene expression [1, 2]. The phytohormone ABA plays a central role in abiotic stress response, especially in plant response to drought or salt stress, as well as in the regulation of plant development. The protein phosphorylation and dephosphorylation mediated by protein kinases and protein phosphatases play important roles in ABA signal transduction. The protein phosphatases can be divided into two major classes: protein tyrosine phosphatases and protein serine/threonine phosphatases [3, 4]. The protein serine/threonine phosphatases are classified into protein phosphatase P (PPP) and protein phosphatase M (PPM) families. The PPP family includes type I (PP1), type 2A (PP2A), and type 2B (PP2B), whereas the PPM family includes type 2C (PP2C) and pyruvate dehydrogenase phosphatase [5].
The roles for PP2C in the ABA-dependent signaling pathway have been extensively studied. In Arabidopsis, it was estimated that seventy-six genes encode PP2C-type phosphatases [6]. The group A of PP2C family contains most of the identified genes involved in ABA signaling, and they act as negative regulators in the pathway. ABI1 and ABI2 are the best studied PP2Cs [7–10]. Through genetic screening, it was obtained two mutants abi1-1 and abi2-1 with ABA-insensitive phenotypes. Both ABI1 and ABI2 encode PP2C proteins and their expression is up-regulated by ABA. Further studies have suggested that ABI1 and ABI2 negatively regulate ABA signaling with overlapping functions, and that other PP2Cs are also involved in ABA signaling as ABI1 and ABI2 contribute nearly 50 % of the ABA-induced PP2C activity [8].
In recent years, it has been made a significant progress in the research on ABA receptors. The pyrabactin resistance 1 (PYR1)/PYR1-LIKE (PYL)/regulatory components of ABA receptors (RCAR) family proteins were recognized as the ABA receptors that inhibit PP2C activity, such as ABI1 and ABI2, in an ABA-dependent manner [11–14]. ABI1, ABI2 and some other group A PP2Cs can interact with and directly dephosphorylate the subclass III SNF1-related protein kinase 2 (SnRK2) [15, 16]. Recently, a SnRK2-PP2C complex structure was revealed where the kinase activation loop docks into the active site of PP2C, while the conserved ABA-sensing tryptophan of PP2C inserts into the kinase catalytic cleft, thus mimicking receptor-PP2C interactions [17]. In the absence of ABA, PP2Cs repress ABA signaling by dephosphorylation and inactivation of SnRK2s. However, under ABA conditions induced by environmental or other stimuli, the ABA receptors PYR/PYL/RCAR proteins bind to PP2C and release SnRK2s [15, 16]. After that, SnRK2s can phosphorylate the downstream substrates to positively activate ABA response [15, 16]. Recently, it was found that Arabidopsis HAI2/AtAIP1 could interact with ABA receptors while aip1 null mutant plants exhibited reduced sensitivity to ABA and glucose during seed germination, suggesting that AIP1 is associated with ABA-mediated cell signaling and functions as a positive regulator of ABA [18]. Therefore, plant PP2Cs may play both positive and negative roles in ABA signaling.
The genes coding for PP2C proteins have been identified in different plant species, mostly in Arabidopsis [3]. Although a bioinformatics survey has identified 78 PP2C genes in rice [19], few rice PP2C genes have been functionally investigated [20, 21]. OsBIPP2C1 and OsBIPP2C2a are two PP2C genes induced by disease resistance inducers and pathogen infections in rice [20, 21]. Overexpression of OsBIPP2C1 or OsBIPP2C2a could enhance disease resistance. A rice PP2C gene Xb15 was reported to negatively regulate XA21-mediated innate immune response [22]. Based on a microarray analysis of salt responsive genes in rice, we identified and cloned a new PP2C gene OsSIPP2C1 from rice. Our data indicated that OsSIPP2C1 was involved in both reproductive development and abiotic stress response, and negatively regulated by ABL1 transcription factor.
Materials and Methods
Plant Materials
The rice (Oryza sativa L. sub. japonica) cultivars Jiucaiqing, Zhonghua11, and Zhonghua11 mutant abl1 [23] were used in this study. The seeds were sterilized in 0.1 % HgCl2 and germinated at 30 °C. The seedlings were cultured with Yoshida’s culture solution in growth chamber as previously described [24]. For tissue-specific gene expression analysis, the roots and young culms from two-week-old seedlings, the leaves, culms, nodes, anthers, and panicles at different lengths from adult plants were harvested and immediately frozen in liquid nitrogen, respectively.
Stress Treatments
The two-week-old seedlings were used for stress treatments as previously described [25]. For salt, osmotic and ABA treatments, the seedlings were cultured in Yoshida’s culture solution supplemented with 150 mM NaCl, 20 % (w/v) PEG6000, and 0.1 mM ABA, respectively. For cold treatment, the seedlings were transferred to the growth chamber with the temperature of 4 °C.
RNA Isolation and Reverse Transcription
The total RNA was extracted using Trizol reagent (Invitrogen, USA) according to the manufacturer’s protocol. The RNA was subsequently treated with DNase I (Promega, USA) to remove the remaining genomic DNA. The first strand cDNA was synthesized with 2 μg total RNA using Reverse Transcription system (Promega, USA).
Semi-quantitative RT-PCR
The RT-PCR primers for OsSIPP2C1 were 5′-GGCGAGGTGTGACTTCTA-3′ and 5′-GCTTGTGGTCGGAGGATA-3′. The PCR program included an initial denaturation at 94 °C for 5 min, 32 cycles at 94 °C for 30 s, 58 °C for 50 s, 72 °C for 1 min, and a final extension at 72 °C for 10 min. In addition, an 800-bp PCR fragment of rice actin gene Rac1 was amplified as an internal control to insure equal amount of cDNA used in each sqRT-PCR reaction. The primers for Rac1 were 5′-GGAACTGGTATGGTCAAGG-3′ and 5′-AGTCTCATGGATAACCGCAG-3′.
Subcellular Localization
A green fluorescent protein (GFP) fusion protein was constructed using the full-length OsSIPP2C1 cDNA with a C-terminal fusion of the GFP clone under control of CaMV 35S promoter. The rice protoplast preparation and transformation were conducted as previously described [25]. Subcellular distribution of the GFP fusion protein was examined by the confocal laser scanning microscopy (Leica, TCS SP2). Cells were labeled with the DNA dye 4, 6-diamidino-2-phenylindole (DAPI) to visualize the nucleus.
In Silico Sequence Analysis
Multiple sequences alignments were produced by the ClustalX and GeneDOC programs. For promoter sequence analysis, 1,200-bp upstream sequence of OsSIPP2C1 was obtained from Rice Genome Annotation Project (http://rice.plantbiology.msu.edu/) and analyzed with MatInspector program (http://www.genomatix.de/). In silico gene expression, analysis was performed with Genevestigator platform (https://www.genevestigator.com/).
Results
Cloning and Sequence Analysis of OsSIPP2C1
Through a microarray-based investigation on salt-induced genes in rice seedlings, 1,834 genes were found to be up-regulated (>2 fold) by salt stress (data not shown). Among these salt-induced genes, an EST (probe ID: Os.9022.1.S1_at) showed a 23.3-fold induction after salt treatment. Database search indicated that this EST sequence encodes a previously unknown type 2C protein phosphatase. As this gene is largely induced by salt stress, we named this gene Oryza sativa salt-induced PP2C Protein 1 (OsSIPP2C1) under GenBank accession number AK063334. The full-length OsSIPP2C1 gene containing a complete ORF of 1,074 bp was cloned from rice seedlings treated with salt by RT-PCR. The predicted protein product of OsSIPP2C1 comprises 357 amino acids with calculated molecular mass of 37.6 kDa.
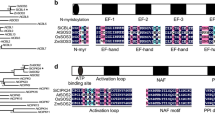
OsSIPP2C1 showed homologies to other plant PP2C proteins including Arabidopsis ABI1 (45 % similarity), AtPP2CA (55 % similarity) [26, 27], and maize ZmPP2C1 (79 % similarity) (Fig. 1). In particular, the catalytic domain is well conserved among these PP2C proteins, whereas the N-terminal extension of OsSIPP2C1 diverges with ABI1 and ABI2 (Fig. 1). Eleven typical motifs [28] are conserved among these PP2Cs including OsSIPP2C1 (Fig. 1). Phylogenetic analysis revealed that OsSIPP2C1 was grouped into group A PP2C proteins [3], and close to ZmPP2C1 and SbPP2C1 but far from another rice PP2C protein OsBIPP2C1 [22]. As OsSIPP2C1 is more close to AtPP2CA [26, 27], HAI1 [29], HAI2/AtAIP1 [18], and HAI3 than ABI1 and ABI2 (Fig. 2), OsSIPP2C1 may be functionally related to AtPP2CA and HAIs.
Phylogenetic analysis of plant PP2C proteins. The tree was constructed with MEGA program with amino acid sequences of plant PP2C proteins. Branch numbers represent a percentage of the bootstrap values in 1,000 sampling replicates, and the scale bar indicates the branch length. The accession numbers for the sequences are as follows: Oryza sativa OsSIPP2C1(AK063334) and OsBIPP2C1(AY603974); Arabidopsis thaliana ABI1(At4g26080), ABI2(At5g57050), AtPP2CA(At3g11410), HAI1(At5g59220), HAI2/AtAIP1(At1g07430), HAI3(At2g29380), HAB1(At1g72770), HAB2(At1g17550), AHG1(At5g51760) and AtPPH1(At4g27800); Zea mays ZmPP2C(AAT40439), ZmPP2C1(NP_001146047), and ZmPP2C2(ABA41456); Sorghum bicolor SbPP2C1(XP_002460055); Fagus sylvatica FsPP2C1(CAB90633); and Vitis vinifera VvPP2C1(CBI16058)
Cis-acting Element Analysis in OsSIPP2C1 Promoter
The 1,200-bp promoter sequence of OsSIPP2C1 was analyzed for cis-acting elements. The promoter sequence contains some putative stress-related cis-acting elements, such as dehydration responsive element (DRE), MYC recognition sites (MYCS), multiple ABA responsive element (ABREs), and MYB recognition sites (MYBSs) (Table 1). These cis-acting elements may be responsive for stress-regulated expression of OsSIPP2C1. In addition, a MADS-box protein binding site (ttaaaCCTAcaatagaaagtt, −889 to −869) also exists in the OsSIPP2C1 promoter region.
Expression Analysis of OsSIPP2C1 Under Stress Conditions
Based on the microarray data, it was found that OsSIPP2C1 was induced by various abiotic stresses, such as high salt, PEG6000, drought and biotic stresses including rice blast pathogen Magnaporthe grisa and bacterial blight pathogen Xanthomonas oryzae pv. oryzae (Xoo) (Fig. 3). These results suggest that OsSIPP2C1 is involved in both abiotic and biotic stress responses. Furthermore, OsSIPP2C1 is induced by exogenous ABA treatment but not markedly by cold or H2O2. To confirm the expression of OsSIPP2C1 in rice under abiotic stress, semi-quantitative RT-PCR was employed to analyze OsSIPP2C1 expression in rice seedlings under salt, PEG6000, cold, and ABA treatments. As shown in Fig. 4, the transcripts of OsSIPP2C1 accumulated at 1 h after salt treatment and reached the maximum at 24 h. For PEG treatment, the OsSIPP2C1 mRNA showed an increase at 1 h of treatment and decreased to the baseline after 24 h. For cold, the expression of OsSIPP2C1 was not markedly altered upon cold from 1 to 24 h. The presence of 0.1 mM exogenous ABA as a signal molecule induced OsSIPP2C1 expression to peak at 12 h. In addition, our data showed that the expression of OsSIPP2C1 was not significantly changed under normal growth condition (Fig. 4). Together, OsSIPP2C1 is induced by salt and osmotic stresses as well as exogenous ABA, but not markedly by cold.
In silico expression analysis of OsSIPP2C1, the Log2 expression data were derived from Genevestigator platform (https://www.genevestigator.com/) except expression values of OsSIPP2C1 in rice seedlings under drought (20 % PEG6000), cold (4 °C), salt (150 mM NaCl), and ABA (0.1 mM) treatments that are from our unpublished microarray data
Expression analysis of OsSIPP2C1 in rice under abiotic stress. The rice seedlings grown in Yoshida’s culture solution were treated with H2O, 150 mM NaCl, 20 % PEG6000, 4 °C, or 0.1 mM ABA. The rice shoots were collected for gene expression analysis after the treatments of different time intervals indicated in the figure
Expression of OsSIPP2C1 in Rice abl1 Mutant Under Abiotic Stress
The rice ABI5-Like1 (ABL1) deficiency mutant, abl1, shows suppressed ABA responses [23]. To analyze whether OsSIPP2C1 is involved in ABL1 signaling, we analyzed OsSIPP2C1 gene expression in abl1 and wild-type rice under salt and drought stresses. Interestingly, OsSIPP2C1 expression was enhanced in abl1 mutant under both normal and stress conditions (Fig. 5), suggesting that ABL1 probably negatively regulates OsSIPP2C1 gene expression.
Tissue-specific Expression Analysis of OsSIPP2C1
Tissue-specific expression of OsSIPP2C1 was analyzed by semi-quantitative RT-PCR. Under normal growth conditions, OsSIPP2C1 was detected only in the developing panicles (Fig. 6a). It was found that OsSIPP2C1 was weakly detected in the panicles at the length of 2 mm and gradually accumulated with the elongation of panicles. The highest expression level of OsSIPP2C1 was observed in the 5–10 cm panicles. Although OsSIPP2C1 expression was not detected in the leaves, roots, and culms under normal condition (Fig. 6a), we found that application of ABA could significantly induce expression of OsSIPP2C1 in those tissues (Fig. 6b).
Tissue-specific expression of OsSIPP2C1. a Expression of OsSIPP2C1 in rice tissues under normal growth condition. The roots and young culms from two-week-old seedlings, the panicles at different lengths, the leaves, culms, nodes, and anthers from adult plants were used for tissue-specific expression of OsSIPP2C1. b Expression of OsSIPP2C1 in rice tissues under 0.1 mM ABA treatment. The semi-quantitative RT-PCR was employed to analyze OsSIPP2C1 expression in different rice tissues as indicated in the figure
OsSIPP2C1 Predominately Locates in the Nucleus
To further explore OsSIPP2C1 function, we analyzed its subcellular localization using rice protoplast cells. As shown in Fig. 7, GFP-OsSIPP2C1 fusion protein was localized predominately in the nucleus, and this was also indicated by DAPI staining. In contrast, the control GFP protein was detected throughout the cell (Fig. 7).
Discussion
When suffering stresses, such as high salt, drought and cold, plants activate adaptive responses including changes in physiology, metabolism, and gene expression. The stress-derived signal transduction through protein phosphorylation and dephosphorylation plays an essential role in these processes [3]. The type 2C protein phosphatase has an important role in the signal transduction of stress response, especially in the ABA-dependent signaling pathway in Arabidopsis. Besides in Arabidopsis, PP2C genes have been less reported in other plant species. In the present study, a new PP2C gene OsSIPP2C1 was cloned and characterized in rice.
The gene expression analysis indicated OsSIPP2C1 was significantly induced upon salt and PEG stresses but not markedly by cold, implying that OsSIPP2C1 is mainly involved in signal transduction induced by salt or osmotic stress. Considering that the ABA-dependent signaling is mostly associated with salt and drought stress [1, 2], it is deduced that OsSIPP2C1 plays an important role in signal transduction of salt and drought responses through an ABA-dependent pathway. Phylogenetic analysis indicated that OsSIPP2C1 was similar to Arabidopsis PP2C protein AtPP2CA and HAIs (Fig. 2). AtPP2CA negatively regulates ABA signal transduction in Arabidopsis as its disruption mutants displayed strong ABA hypersensitivity [25]. Based on the fact that OsSIPP2C1 was induced by salt, PEG6000 and ABA treatments, it might play a similar role as AtPP2CA does in ABA signaling. To further study the position of OsSIPP2C1 in ABA-dependent pathway, we studied its expression in the rice ABL1 deficiency mutant, abl1, under salt and PEG conditions. ABL1 is a basic region/leucine zipper motif transcription factor positively regulating ABRE-containing gene expression [23]. The abl1 mutant showed suppressed ABA responses and the decreased expression of ABRE-containing genes. The expression of OsSIPP2C1 was enhanced in abl1 mutant under normal, salt, and PEG conditions, suggesting that ABL1 is a negative regulator of OsSIPP2C1. The studies for PP2C members such as AtPP2CA and FsPP2C1, the Arabidopsis and Fagus sylvatica orthologues of OsSIPP2C1, have suggested that PP2Cs are negative regulators of ABA signaling [26, 27, 30, 31]. We hypothesize here under stress conditions, the plant initiates an ABA-dependent pathway. This in turn activates expression of ABA responsive genes. Expression of ABL1 is then up-regulated, and subsequently ABL1 positively regulates expression of ABRE-containing genes. As OsSIPP2C1 probably functions in the repression of ABA responses, ABL1 may negatively control the abundance of OsSIPP2C1 to avoid OsSIPP2C1-dependent repression of ABA signaling. Therefore, OsSIPP2C1 may function in modulating ABA responses in plants.
It is interesting that OsSIPP2C1 is also responsive to biotic stresses including rice blast pathogen Magnaporthe grisa and bacterial blight pathogen Xoo. Two rice PP2C genes OsBIPP2C1 and OsBIPP2C2a were induced by disease resistance inducers and pathogen infections to confer disease resistance [20, 21]. Another rice PP2C gene Xb15 was shown to negatively regulate XA21-mediated innate immune response [22], suggesting that similar machinery involving PP2Cs may exist in both abiotic and biotic stress responses. Mutation of an Arabidopsis PP2C gene AP2C1 produced more jasmonate upon wounding and was more resistant to phytophagous mites while increase of AP2C1 levels reduced ethylene production and compromised innate immunity against Botrytis cinerea [32]. Further studies with OsSIPP2C1 transgenic plants are required to confirm the positive or negative role of OsSIPP2C1 in biotic stress response.
The expression of OsSIPP2C1 was not detected in most of tissues but regulated by panicle development in rice under normal growth condition. There are few reports connecting the role of PP2Cs with plant development [33]. With development of rice panicles, OsSIPP2C1 expression gradually increased, suggesting that OsSIPP2C1 might play an essential role in the early panicle development. During the early stage of panicle development, OsSIPP2C1 accumulates and may negatively regulate ABA signaling. Thereafter, expression of OsSIPP2C1 decreases at the late stage of panicle development to insure ABA functioning in the promotion of panicle maturing. However, this hypothesis needs further functional validation using OsSIPP2C1 knock-out mutant. The MADS-box transcription factors are very important in reproductive development in plants. For example, PANICLE PHYTOMER2, encoding a SEPALLATA subfamily MADS-box protein, positively determines the spikelet meristem identity in rice [34]. A MADS-box protein binding site in the promoter region of OsSIPP2C1 suggests that OsSIPP2C1 may participate in the early panicle development under the regulation of MADS-box transcription factors. Although the role of OsSIPP2C1 in panicle development is not understood yet, it provides an interesting clue for further study of the functions of PP2Cs in reproductive development in plants.
In summary, OsSIPP2C1 is a nuclear PP2C protein involved in abiotic stress and early panicle development. Moreover, OsSIPP2C1 was negatively regulated by ABL1. Our data provide the new clues in the understanding of the functions of PP2Cs in stress responses and reproductive development in plants.
References
Chinnusamy, V., Schumaker, K., & Zhu, J. K. (2004). Molecular genetic perspectives on cross-talk and specificity in abiotic stress signalling in plants. Journal of Experimental Botany, 55, 225–236.
Fujita, Y., Fujita, M., Shinozaki, K., & Yamaguchi-Shinozaki, K. (2011). ABA-mediated transcriptional regulation in response to osmotic stress in plants. Journal of Plant Research, 124, 509–525.
Schweighofer, A., Hirt, H., & Meskiene, I. (2004). Plant PP2C phosphatases: Emerging functions in stress signaling. Trends in Plant Science, 9, 236–243.
Cohen, P. T. (1997). Novel protein serine/threonine phosphatases: Variety is the spice of life. Trends in Biochemistry Science, 22, 245–251.
Cohen, P. T. (1989). The structure and regulation of protein phosphatases. Annual Review of Biochemistry, 58, 453–508.
Kerk, D., Bulgrien, J., Smith, D. W., Barsam, B., Veretnik, S., & Gribskov, M. (2002). The complement of protein phosphatase catalytic subunits encoded in the genome of Arabidopsis. Plant Physiology, 129, 908–925.
Leung, J., Merlot, S., & Giraudat, J. (1997). The Arabidopsis ABSCISIC ACID-INSENSITIVE2 (ABI2) and ABI1 genes encode homologous protein phosphatases 2C involved in abscisic acid signal transduction. Plant Cell, 9, 759–771.
Merlot, S., Gosti, F., Guerrier, D., Vavasseur, A., & Giraudat, J. (2001). The ABI1 and ABI2 protein phosphatases 2C act in a negative feedback regulatory loop of the abscisic acid signaling pathway. Plant Journal, 25, 295–303.
Sun, H. L., Wang, X. J., Ding, W. H., Zhu, S. Y., Zhao, R., Zhang, Y. X., et al. (2011). Identification of an important site for function of the type 2C protein phosphatase ABI2 in abscisic acid signaling in Arabidopsis. Journal of Experimental Botany, 62, 5713–5725.
Ludwików, A., Kierzek, D., Gallois, P., Zeef, L., & Sadowski, J. (2009). Gene expression profiling of ozone-treated Arabidopsis abi1td insertional mutant: Protein phosphatase 2C ABI1 modulates biosynthesis ratio of ABA and ethylene. Planta, 230, 1003–1017.
Ma, Y., Szostkiewicz, I., Korte, A., Moes, D., Yang, Y., Christmann, A., et al. (2009). Regulators of PP2C phosphatase activity function as abscisic acid sensors. Science, 324, 1064–1068.
Park, S. Y., Fung, P., Nishimura, N., Jensen, D. R., Fujii, H., Zhao, Y., et al. (2009). Abscisic acid inhibits type 2C protein phosphatases via the PYR/PYL family of START proteins. Science, 324, 1068–1071.
Santiago, J., Rodrigues, A., Saez, A., Rubio, S., Antoni, R., Dupeux, F., et al. (2009). Modulation of drought resistance by the abscisic acid receptor PYL5 through inhibition of clade A PP2Cs. Plant Journal, 60, 575–588.
Santiago, J., Dupeux, F., Betz, K., Antoni, R., Gonzalez-Guzman, M., Rodriguez, L., et al. (2012). Structural insights into PYR/PYL/RCAR ABA receptors and PP2Cs. Plant Science, 182, 3–11.
Umezawa, T., Sugiyama, N., Mizoguchi, M., Hayashi, S., Myouga, F., Yamaguchi-Shinozaki, K., et al. (2009). Type 2C protein phosphatases directly regulate abscisic acid-activated protein kinases in Arabidopsis. Proceedings of the National Academy of Sciences USA, 106, 17588–17593.
Fujita, Y., Nakashima, K., Yoshida, T., Katagiri, T., Kidokoro, S., Kanamori, N., et al. (2009). Three SnRK2 protein kinases are the main positive regulators of abscisic acid signaling in response to water stress in Arabidopsis. Plant Cell Physiology, 50, 2123–2132.
Soon, F. F., Ng, L. M., Zhou, X. E., West, G. M., Kovach, A., Tan, M. H., et al. (2012). Molecular mimicry regulates ABA signaling by SnRK2 kinases and PP2C phosphatases. Science, 335, 85–88.
Lim, C. W., Kim, J. H., Baek, W., Kim, B. S., & Lee, S. C. (2012). Functional roles of the protein phosphatase 2C, AtAIP1, in abscisic acid signaling and sugar tolerance in Arabidopsis. Plant Science, 187, 83–88.
Xue, T., Wang, D., Zhang, S., Ehlting, J., Ni, F., Jakab, S., et al. (2008). Genome-wide and expression analysis of protein phosphatase 2C in rice and Arabidopsis. BMC Genomics, 9, 550.
Hu, X., Zhang, H., Li, G., Yang, Y., Zheng, Z., & Song, F. (2009). Ectopic expression of a rice protein phosphatase 2C gene OsBIPP2C2 in tobacco improves disease resistance. Plant Cell Reports, 28, 985–995.
Hu, X., Song, F., & Zheng, Z. (2006). Molecular characterization and expression analysis of a rice protein phosphatase 2C gene, OsBIPP2C1, and overexpression in transgenic tobacco conferred enhanced disease resistance and abiotic tolerance. Physiologia Plantarum, 127, 225–236.
Park, C. J., Peng, Y., Chen, X., Dardick, C., Ruan, D., Bart, R., et al. (2008). Rice XB15, a protein phosphatase 2C, negatively regulates cell death and XA21-mediated innate immunity. PLoS Biology, 6, e231.
Yang, X., Yang, Y. N., Xue, L. J., Zou, M. J., Liu, J. Y., Chen, F., et al. (2011). Rice ABI5-Like1 regulates abscisic acid and auxin responses by affecting the expression of ABRE-containing genes. Plant Physiology, 156, 1397–1409.
Sun, S. J., Guo, S. Q., Yang, X., Bao, Y. M., Tang, H. J., Sun, H., et al. (2010). Functional analysis of a novel Cys2/His2-type zinc finger protein involved in salt tolerance in rice. Journal of Experimental Botany, 61, 2807–2818.
Sun, H., Huang, X., Xu, X., Lan, H., Huang, J., Zhang, H.S. (2012). ENAC1, a NAC transcription factor, is an early and transient response regulator induced by abiotic stress in Rice (Oryza sativa L.). Molecular Biotechnology. doi:10.1007/s12033-011-9477-4.
Yoshida, T., Nishimura, N., Kitahata, N., Kuromori, T., Ito, T., Asami, T., et al. (2006). ABA-hypersensitive germination3 encodes a protein phosphatase 2C (AtPP2CA) that strongly regulates abscisic acid signaling during germination among Arabidopsis protein phosphatase 2Cs. Plant Physiology, 140, 115–126.
Kuhn, J. M., Boisson-Dernier, A., Dizon, M. B., Maktabi, M. H., & Schroeder, J. I. (2006). The protein phosphatase AtPP2CA negatively regulates abscisic acid signal transduction in Arabidopsis, and effects of abh1 on AtPP2CA mRNA. Plant Physiology, 140, 127–139.
Bork, P., Brown, N. P., Hegyi, H., & Schultz, J. (1996). The protein phosphatase 2C (PP2C) superfamily: Detection of bacterial homologues. Protein Science, 5, 1421–1425.
Antoni, R., Gonzalez-Guzman, M., Rodriguez, L., Rodrigues, A., Pizzio, G. A., & Rodriguez, P. L. (2012). Selective inhibition of clade A phosphatases type 2C by PYR/PYL/RCAR abscisic acid receptors. Plant Physiology, 158, 970–980.
Lorenzo, O., Rodríguez, D., Nicolás, G., Rodríguez, P. L., & Nicolás, C. (2001). A new protein phosphatase 2C (FsPP2C1) induced by abscisic acid is specifically expressed in dormant beechnut seeds. Plant Physiology, 125, 1949–1956.
González-García, M. P., Rodríguez, D., Nicolás, C., Rodríguez, P. L., Nicolás, G., & Lorenzo, O. (2003). Negative regulation of abscisic acid signaling by the Fagus sylvatica FsPP2C1 plays a role in seed dormancy regulation and promotion of seed germination. Plant Physiology, 133, 135–144.
Schweighofer, A., Kazanaviciute, V., Scheikl, E., Teige, M., Doczi, R., Hirt, H., et al. (1997). The PP2C-type phosphatase AP2C1, which negatively regulates MPK4 and MPK6, modulates innate immunity, jasmonic acid, and ethylene levels in Arabidopsis. Plant Cell, 19, 2213–2224.
Singh, A., Giri, J., Kapoor, S., Tyagi, A. K., & Pandey, G. K. (2010). Protein phosphatase complement in rice: genome-wide identification and transcriptional analysis under abiotic stress conditions and reproductive development. BMC Genomics, 11, 435.
Kobayashi, K., Maekawa, M., Miyao, A., Hirochika, H., & Kyozuka, J. (2010). PANICLE PHYTOMER2 (PAP2), encoding a SEPALLATA subfamily MADS-box protein, positively controls spikelet meristem identity in rice. Plant Cell Physiology, 51, 47–57.
Acknowledgments
We thank Prof. Hongwei Xue from institute of plant physiology and ecology, SIBS, for kindly providing rice abl1 seeds. This work was supported by National Natural Science Foundation of China (nos. 30971556, 31071397), Jiangsu Agriculture Science and Technology Innovation Fund [no. CX(12)1003], State Key Laboratory of Rice Biology (no. 110101), and Fundamental Research Funds for the Central Universities (no. KYZ201137).
Author information
Authors and Affiliations
Corresponding author
Additional information
Yu-Sheng Li and Hui Sun contributed equally to this work.
Rights and permissions
About this article
Cite this article
Li, YS., Sun, H., Wang, ZF. et al. A Novel Nuclear Protein Phosphatase 2C Negatively Regulated by ABL1 is Involved in Abiotic Stress and Panicle Development in Rice. Mol Biotechnol 54, 703–710 (2013). https://doi.org/10.1007/s12033-012-9614-8
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12033-012-9614-8