In the present study, the equimolar reactions of dialkyltin (IV) dichlorides with Schiff base ligands have been processed, and from these reactions a series of new organotin complexes have formed with the general formula R2 SnL1–5 where R = Me, Bu and Ph and L = Schiff base ligands (obtained by the reaction of 5-phenylisoxazole-3-carboxylic acid hydrazide with salicylaldehyde and its derivatives) and the synthesized compounds were characterized by various spectroscopic techniques such as IR, 1H, 13C, 119Sn NMR, and mass spectrometry, and it was found that the Schiff bases bind to the metal center through ONO donor atoms in a tridentate manner. The synthesized ligands and the complexes were further tested for their antimicrobial properties against two Gram-positive bacteria (Bacillus subtilis (MTCC 441) and Staphylococcus aureus (MTCC 2901)), two Gram-negative bacteria (Escherichia coli (MTCC 732) and Pseudomonas aeruginosa (MTCC 424)), and two fungal strains (Candida albicans (MTCC 227) and Aspergillus niger (MTCC 9933)). The activity of the ligands becomes appreciably enhanced on binding with tin metal. Diorganotin complexes having diphenyl substrate showed better activity than the dibutyl and dimethyl substrates. Complex 17m, which has –NO2 and Ph2 Sn groups, found the most active one. The synthesized compounds were also tested for their antioxidant activity using DPPH free radical method and it was found that compound 20 showed the best result.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Antimicrobial diseases are seriously devastating to people’s health and the numbers of patients affected by antimicrobial diseases are increasing day by day. Curing bacterial or fungal diseases using antimicrobial drugs is getting bothersome and is becoming a global health concern because of the mounting resistance of the microbes toward the antimicrobial drugs [1,2,3]. The microbes are becoming more resistive to the developed drugs because of the mutations in their genetic material [4]. In order to overcome the unwholesomeness and the fatality caused by the pathogenic microbes there is a demand to explore more in this sector, and the synthesis of new antimicrobial medicines that have better action against pathogenic microbes is requisite; thus, divergent research has been going on to develop antimicrobial drugs with better action on a broad spectrum of pathogenic microbes.
Among all this, metal complexes are also increasing in recognition among mankind because of their appealing benefits in the field of pharmacology [5,6,7,8,9,10,11,12,13]. Metal-based drugs possess distinctive valuable properties and have been already used in the treatment of the several diseases; therefore, a large number of metal complexes have been synthesized and studied every year [14,15,16,17,18] and there is persistent research is going on worldwide to develop more metal-based complexes. Among them the Schiff base metal complexes maintain their significance and there is recommendable interest of researchers in the synthesis of Schiff base metal complexes because of their easily chelation [19, 20] and flexible structure. The Schiff bases easily bind to the metal center because their molecular structure consists of donor atoms such as nitrogen, oxygen, and sulfur. The organotin complexes of Schiff bases have their potential usefulness in the biological field [21,22,23,24,25,26,27,28] and they can be used as antimicrobial [29,30,31,32,33,34,35,36], antioxidant [37,38,39,40], and anticancer [41,42,43,44,45] agents. With the inspiration to do more in this area, a series of organotin complexes using Schiff bases as ligands have been synthesized and characterized using several spectroscopic techniques and studied for their biological application as antibacterial and antifungal agents.

Experimental
The used chemicals were purchased from Sigma Aldrich. All the chemicals were pure, checked by TLC, and hence they were used as such without purification. Solvents were dried before use and by a standard procedure. THF was used directly. Necessary precautionary measures have been taken to avoid the addition of impurities and moisture. All the apparatus and instruments used were clean and dry. The electrical melting point apparatus took melting points by the use of capillaries. The FTIR spectra were recorded with the help of pressed techniques, on that account a small amount of KBr and synthesized samples were uniformly powdered together to prepare pellets and spectra were recorded with the Shimadzu IR Affinity-I 8000 FT-IR spectrophotometer. The NMR spectra (1H, 13C, and 119Sn) of the ligands and their complexes were recorded on the multinuclear Bruker Avance II 400 MHz NMR spectrometer in the solvents DMSO-d6 and CDCl3 respectively. The references used were tetramethylsilane for 1H and 13C NMR and tetramethyltin for 119Sn NMR. The mass spectroscopy of ligands and their complexes was recorded on a SCIEX-QTOF mass spectrometer by using acetonitrile as a solvent.
Synthesis of Schiff base ligands (1–5)
The Schiff bases were synthesized by the reaction of the 5-phenylisoxazole-3-carboxylic acid hydrazide (3 mmol) dissolved in methanol with salicylaldehyde and its derivatives (3 mmol), which were also dissolved in methanol by refluxing or 1 h. and the status of the reactions was monitored from time to time by taking TLC. After completion of the reactions and by evaporating the solvents, yellow-colored solid products were obtained and the obtained crude solid products were purified by recrystallization techniques, which were used to prepare the complexes.
(N-(2-Hydroxybenzylidene)-5-phenylisoxazole-3-carbohydrazide (1): yield: 86%; light yellow solid; m.p. 218–222°C; IR (KBr pellets, cm-1): 3283 (vO–H), 3448 (vN–H), 1679 (vC=O), 1609 (vC=N). 1H NMR (400 MHz, DMSO-d6) δ: 12.56 (s, 1H, NH), 11.01 (s, 1H, OH), 8.75 (s, 1H, –N=CH), 8.00–7.97 (m, 2H, Ar-H), 7.60–7.56 (m, 4H, Ar-H), 7.52 (s, 1H, -OC=CH), 7.36–7.31 (m, 1H, Ar-H), 6.97–6.92 (m, 2H, Ar-H). 13C NMR (100 MHz, DMSO-d6) δ: 171.12 (CHONH), 159.10 (C=N–N), 157.96, 155.42, 150.22, 132.33, 131.48, 129.84, 129.75, 126.62, 126.34, 119.92, 119.13, 116.93 (aromatic carbons), 100.74 (–OC=C). MS: m/z (M+) for C17H13N3O3: 307.10; found: 308.12 (M+H) +.
N-(5-Chloro-2-hydroxybenzylidene)-5-phenylisoxazole-3-carbohydrazide (2): yield: 90%; light yellow solid; m.p. 225–228°C; IR (KBr pellets, cm-1): 3218 (vO–H), 3418 (vN–H), 1678 (vC=O), 1608 (vC=N). 1H NMR (400 MHz, DMSO-d6) δ: 12.60 (s, 1H, NH), 11.10 (s, 1H, OH), 8.73 (s, 1H,–N=CH), 7.98 (dd, J = 7.4, 1.9 Hz, 2H, Ar-H), 7.68 (d, J = 2.6 Hz, 1H, Ar-H), 7.60–7.55 (m, 3H, Ar-H), 7.53 (s, 1H,–OC=CH), 7.37–7.31 (m, 1H, Ar-H), 6.96 (t, J = 7.0 Hz, 1H, Ar-H). 13C NMR (100 MHz, DMSO-d6) δ: 171.08 (CHONH), 159.22 (C=N–N), 156.65, 155.74, 147.59, 131.65, 131.48, 129.86, 127.62, 126.64, 126.34, 123.49, 121.26, 118.77 (aromatic carbons), 100.79 (–OC=C) MS: m/z (M+) calc. for C17H12ClN3O3 : 341.06; found: 342.44 (M+H)+.
N-(5-Bromo-2-hydroxybenzylidene)-5-phenylisoxazole-3-carbohydrazide (3): yield: 91%; yellow solid; m.p. 248–251°C; IR (KBr pellets, cm-1): 3306 (vO–H), 3435 (vN–H), 1696 (_C=O), 1610 (vC=N). 1H NMR (400 MHz, DMSO-d6) δ: 12.65 (s, 1H, NH), 11.03 (s, 1H, OH), 8.73 (s, 1H,–N=CH), 8.01–7.97 (m, 2H, Ar-H), 7.82 (s, 1H, Ar-H), 7.61–7.56 (m, 3H, Ar-H), 7.55 (s, 1H, –OC=CH), 7.46 (dd, J = 8.7, 2.6 Hz, 1H, Ar-H), 6.92 (d, J = 8.8 Hz, 1H, Ar-H). 13C NMR (100 MHz, DMSO-d6) δ: 171.12 (CHONH), 159.09 (C=N-N), 156.92, 155.58, 147.39, 134.49, 131.50, 130.44, 129.86, 126.60, 126.34, 121.85, 119.19, 111.02 (aromatic carbons), 100.79 (–OC=C). MS: m/z (M+) calc. for C17H12BrN3O3 385.01; found: 386.12 (M+H)+.
N-(2-Hydroxy-5-nitrobenzylidene)-5-phenylisoxazole-3-carbohydrazide (4): yield: 92%; yellow solid; m.p. 254–258°C; IR (KBr pellets, cm-1): 3201 (vO–H), 3483 (vN–H), 1668 (vC=O), 1610 (vC=N). 1H NMR (400 MHz, DMSO-d6) δ: 12.73 (s, 1H, NH), 12.12 (s, 1H, –OH), 8.84 (s, 1H,–N=CH), 8.60 (d, J = 2.9 Hz, 1H, Ar-H), 8.20 (dd, J = 9.1, 2.9 Hz, 1H, Ar-H), 7.99 (dd, J = 7.4, 2.1 Hz, 2H, Ar-H), 7.60–7.55 (m, 3H, Ar-H), 7.56 (s, 1H, –OC=CH), 7.13 (d, J = 9.1 Hz, 1H, Ar-H). 13C NMR (100 MHz, DMSO-d6) δ: 171.17 (CHONH), 159.07 (C=N-N), 163.07, 155.72, 146.05, 140.42, 131.50, 129.85, 127.45, 126.59, 126.34, 123.75, 120.59, 117.99 (aromatic carbons), 100.82 (–OC=C). MS: m/z (M+) calc. for C17H12N4O5: 352.08; found: 353.26 (M+H)+.
N-(3-Ethoxy-2-hydroxybenzylidene)-5-phenylisoxazole-3-carbohydrazide (5): yield: 81%; light yellow solid; m.p. 150–155°C; IR (KBr pellets, cm-1): 3260 (vO–H), 3420 (vN–H), 1660 (vC=O), 1606 (vC=N). 1H NMR (400 MHz, DMSO-d6) δ: 12.59 (s, 1H, NH), 10.64 (s, 1H, OH), 8.76 (s, 1H,–N=CH), 8.01–7.97 (m, 2H, Ar-H), 7.60–7.56 (m, 3H, Ar-H), 7.55 (s, 1H, –OC=CH), 7.17 (dd, J = 7.9, 1.3 Hz, 1H, Ar-H), 7.05 (dd, J = 8.1, 1.2 Hz, 1H, Ar-H), 6.86 (m, 1H, Ar-H), 4.08 (q, J = 7.0 Hz, 2H, –OCH2CH3), 1.36 (t, J = 7.0 Hz, 3H,,–OCH2CH3). 13C NMR (100 MHz, DMSO-d6) δ: 171.11 (CHONH), 159.11 (C=N-N), 155.38, 150.19, 147.99, 147.56, 131.49, 129.86, 126.61, 126.33, 121.16, 119.64, 119.42, 115.90, (aromatic carbons), 100.79 (–OC=C), 64.62 (–OCH2CH3), 15.19 (–OCH2CH3). MS: m/z (M+) calc. for C19H17N3O4 : 351.12; found: 351.95 (M+H)+.
Synthesis of complexes (6–20)
The organotin(IV) complexes were synthesized by the reflux of ligands (0.5 mmol) dissolved in warm THF and dialkyltindichloride (0.5 mmol) dissolved in methanol with a few drops of triethylamine. The reactions were monitored by TLC and took 7–8 h for completion. The obtained products were cooled and solidified by evaporating the solvent under vacuum.
2,2-Dibutyl-4-(5-phenylisoxazol-3-yl)benzo[h][1,2,3, 5, 6]-dioxadiazastannonine (6): yield: 52%; yellow solid; m.p. 118–120°C; IR (KBr pellets, cm-1): 1610 (s, v-C=N), 684 (vSn-C), 569 (vSn-O), 429 (vSn-N), 1H NMR (400 MHz, CDCl3) δ: 8.79 (s, 1H,–N=CH), 7.86–7.77 (m, 4H, Ar-H), 7.56–7.41 (m, 5H, Ar-H), 6.95 (s, 1H, –OC=CH), 1.68–1.62 (m, 12H, Sn-Bu), 0.89 (t, J = 7.3 Hz, 6H, Sn-Bu). 13C NMR (100 MHz, CDCl3) δ: 170.66 (CHONH), 159.69 (C=N-N), 161.45, 161.02, 158.32, 139.09, 131.78, 130.15, 128.17, 126.12, 122.12, 121.45, 117.48, 115.52 (Ph-C), 98.95 (–OC=C), 26.78, 26.45, 23.01, 13.52 (Sn-Bu). 119Sn NMR (400 MHz, CDCl3) δ: -185.15. MS: m/z (M+) calc. for C25H29N3O3Sn: 539.12; found: 540.11 (M+H)+.
2,2-Dimethyl-4-(5-phenylisoxazol-3-yl)benzo[h][1,3,5,6,2]dioxadiazastannonine (7): yield: 65%; yellow solid; m.p. 125–127°C; IR (KBr pellets, cm-1): 1607 (v-C=N), 676 (vSn-C), 593 (vSn-O), 476 (vSn-N). 1H NMR (400 MHz, CDCl3) δ: 8.79 (s, 1H,–N=CH), 7.85 (dd, J=7.7, 1.6 Hz, 2H, Ar-H), 7.57–7.35 (m, 4H, Ar-H), 7.26–7.15 (m, 3H, Ar-H), 6.95 (s, 1H, –OC=CH), 0.98 (t, J = 7.3 Hz, 6H, Sn-Bu). 13C NMR (100 MHz, CDCl3) δ: 170.18 (CHONH), 159.45 (–C=N-N), 164.39, 161.98, 159.82, 153.87, 151.98, 148.55, 137.73, 136.60, 128.92, 126.43, 125.78, 118.57, 116.35 (aromatic carbon), 98.80 (–OC=C), 2.61 (Sn-Me). 119Sn NMR (400 MHz, CDCl3) δ: -146.68. MS: m/z (M+) calc. for C19H17N3O3Sn: 455.03; found: 456.14 (M+H)+.
2,2-Diphenyl-4-(5-phenylisoxazol-3-yl)benzo[h][1,3,5,6,2]dioxadiazastannonine (8): yield: 56%; yellow solid; m.p. 128–130°C; IR (KBr pellets, cm-1): 1612 (v-C=N), 690 (vSn-C), 576 (vSn-O), 417 (vSn-N). 1H NMR (400 MHz, CDCl3) δ: 9.05 (s, 1H,–N=CH), 8.59 (s, 1H, Ar-H), 7.94–7.64 (m, 4H, Ar-H), 7.60–7.31 (m, 10H, Ar-H), 7.12–7.00 (m, 2H, Ar-H), 6.99 (s, 1H,-OC=CH), 6.94 (dd, J = 17.9, 9.5 Hz, 1H, Ar-H), 6.79 (s, 1H, Ar-H). 13C NMR (100 MHz, CDCl3) δ: 171.02 (CHONH), 159.35 (C=N-N), 166.36, 161.13, 151.50, 146.14, 145.97, 143.00, 138.94, 136.70, 135.76, 134.41, 132.51, 130.53, 129.21, 129.10, 129.05, 128.33, 128.29, 128.24, 125.86, 125.51 (aromatic carbons), 99.39 (–OC=C), 119Sn NMR (400 MHz, CDCl3): δ –330.19. MS: m/z (M+) calc. for C29H21N3O3Sn: 579.06 found: 580.18 (M+H)+.
2,2-Dibutyl-9-chloro-4-(5-phenylisoxazol-3-yl)benzo-[h][1,3,5,6,2]dioxadiazastannonine (9): yield: 57%; yellow solid; m.p. 151–156°C; IR (KBr pellets, cm–1): 1609 (v-C=N), 668 (vSn-C), 553 (vSn-O), 440 (vSn-N). 1H NMR (400 MHz, CDCl3) δ: 8.71 (s, 1H, -N=CH), 7.88–7.84 (m, 2H, Ar-H), 7.54–7.46 (m, 3H, Ar-H), 7.32–7.28 (m, 1H, Ar-H), 7.16 (d, J = 2.7 Hz, 1H, Ar-H), 6.95 (s, 1H, –OC=CH), 6.76 (d, J = 9.0 Hz, 1H, Ar-H), 1.68–1.58 (m, 12H, Sn-Bu), 0.93 (s, 6H, Sn-Bu). 13C NMR (100 MHz, CDCl3) δ: 170.76 (CHONH), 159.67 (C=N-N), 162.48, 161.43, 160.34, 158.87, 137.87, 136.55, 131.52, 128.98, 127.87, 125.65, 122.84, 118.95, 116.72 (aromatic carbons), 98.96 (–OC=C), 26.71, 26.48, 23.06, 13.56 (Sn-Bu). 119Sn NMR (400 MHz, CDCl3): δ -188.17. MS: m/z (M+) calc. for C25H28ClN3O3Sn: 573.08; found: 574.25 (M+H)+.
9-Chloro-2,2-dimethyl-4-(5-phenylisoxazol-3yl)benzo-[h][1,3,5,6,2]dioxadiazastannonine (10): yield: 69%; yellow solid; m.p. 154–158°C; IR (KBr pellets, cm-1): 1609 (v-C=N), 669 (vSn-C), 557 (vSn-O), 464 (vSn-N). 1H NMR (400 MHz, CDCl3) δ: 8.71 (s, 1H, -N=CH), δ 7.88–7.82 (m, 2H, Ar-H), 7.55–7.46 (m, 3H, Ar-H), 7.34–7.27 (m, 1H, Ar-H), 7.17 (d, J = 2.7 Hz, 1H, Ar-H), 6.95 (s, 1H, –OC=CH), 6.75 (d, J = 9.0 Hz, 1H, Ar-H), 0.93 (s, 6H, Sn-Me). 13C NMR (100 MHz, CDCl3) δ: 170.86 (CHONH), 159.16 (C=N-N), 162.50, 135.89, 132.62, 130.50, 129.07, 127.07, 125.91, 123.55, 121.72, 116.91 (aromatic carbons), 98.82 (–OC=C), 2.11 (Sn-Me). 119Sn NMR (400 MHz, CDCl3): δ -148.09. MS: m/z (M+) calc. for C19H16ClN3O3Sn: 488.99; found: 489.18 (M+H)+.
9-Chloro-2,2-diphenyl-4-(5-phenylisoxazol-3-yl)benzo-[h][1,3,5,6,2]dioxadiazastannonine (11): yield: 61%; yellow solid; m.p. 158–160°C; IR (KBr pellets, cm-1): 1618 (v-C=N), 668 (vSn-C), 532 (vSn-O), 448 (vSn-N). 1H NMR (400 MHz, CDCl3) δ: 8.98 (s, 1H,–N=CH), 8.44 (d, J = 16.9 Hz, 2H, Ar-H), 7.92–7.75 (m, 3H, Ar-H), 7.70–7.36 (m, 8H, Ar-H), 7.20–7.12 (m, 4H, Ar-H), 7.00 (s, 1H,–OC=CH), 6.89 (m, 1H, Ar-H). 13C NMR (100 MHz, CDCl3) δ: 170.79 (CHONH), 159.24 (C=N-N), 163.56, 162.92, 157.71, 156.27, 151.51, 135.96, 135.76, 134.49, 132.63, 130.88, 130.62, 129.77, 129.34, 129.19, 129.08, 128.25, 125.99, 125.91, 125.52, 118.43 (aromatic carbons), 99.50 (–OC=C). 119Sn NMR (400 MHz, CDCl3) δ –332.14 MS: m/z (M+) calc. for C29H20ClN3O3Sn: 613.02; found: 614.26 (M+H)+.
9-Bromo-2,2-dibutyl-4-(5-phenylisoxazol-3-yl)benzo-[h][1,3,5,6,2]dioxadiazastannonine (12): yield: 54%; yellow solid; m.p. 150–152°C; IR (KBr pellets, cm-1): 1609 (v-C=N), 686 (vSn-C), 551 (vSn-O), 480 (vSn-N). 1H NMR (400 MHz, CDCl3) δ: 8.70 (s, 1H,–N=CH), 7.86 (dd, J = 7.8, 1.7 Hz, 2H, Ar-H), 7.55–7.39 (m, 4H, Ar-H), 7.31–7.27 (m, 1H, Ar-H), 6.95 (s, 1H, –OC=CH), 6.71 (d, J = 9.0 Hz, 1H, Ar-H) 1.78–1.47 (m, 12H, Sn-Bu), 0.89 (t, J = 7.3 Hz, 6H, Sn-Bu). 13C NMR (100 MHz, CDCl3) δ: 170.78 (CHONH), 159.25 (C=N-N), 166.48, 163.44, 162.14, 138.41, 135.72, 130.45, 129.06, 127.13, 125.91, 125.52, 123.87, 117.76, 108.00 (aromatic carbons), 98.94 (–OC=C), 26.72, 26.45, 22.92, 13.55 (Sn-Bu). 119Sn NMR (400 MHz, CDCl3): δ-189.27.MS: m/z (M+) calc. for C25H28BrN3O3Sn: 617.03; found: 617.25 (M+H)+.
9-Bromo-2,2-dimethyl-4-(5-phenylisoxazol-3-yl)benzo-[h][1,3,5,6,2]dioxadiazastannonine (13): yield: 64%; yellow solid; m.p. 248–250°C; IR (KBr pellets, cm-1): 1607 (v-C=N), 672 (vSn-C), 554 (vSn-O), 483 (vSn-N). 1H NMR (400 MHz, CDCl3) δ: 8.71 (s, 1H,–N=CH), 7.91–7.80 (m, 2H, Ar-H), 7.56–7.40 (m, 4H, Ar-H), 7.31 (d, J = 2.6 Hz, 1H, Ar-H), 6.95 (s, 1H, –OC=CH), 6.70 (d, J = 9.0 Hz, 1H, Ar-H), 0.93 (s, 6H, Sn-Me). 13C NMR (100 MHz, CDCl3) δ: 170.87 (CHONH), 159.15 (C=N-N), 162.43, 152.92, 151.05, 149.98, 138.55, 135.73, 130.50, 129.07, 127.06, 125.92, 125.52, 123.94, 117.71, 108.38 (aromatic carbons), 98.82 (–OC=C), 2.13 (Sn-Me).119Sn NMR (400 MHz, CDCl3) δ-145.16. MS: m/z (M+) calc. for C19H16BrN3O3Sn: 532.94; found: 533.08 (M+H)+.
9-Bromo-2,2-diphenyl-4-(5-phenylisoxazol-3-yl)benzo-[h][1,3,5,6,2]dioxadiazastannonine (14): yield: 58%; yellow solid; m.p. 250–252°C; IR (KBr pellets, cm-1): 1610 (v-C=N), 689 (vSn-C), 552 (vSn-O), 448 (vSn-N). 1H NMR (400 MHz, CDCl3) δ: 9.04 (s, 1H,–N=CH), 8.45–7.74 (s, 5H, Ar-H), 7.60–7.31 (m, 9H, Ar-H), 7.32–6.99 (m, 4H, Ar-H), 6.92 (s, 1H, –OC=CH). 13C NMR (100 MHz, CDCl3) δ: 171.04 (CHONH), 161.25 (C=N-N), 167.94, 166.45, 163.78, 161.67, 159.32, 156.89, 152.56, 135.32, 134.19, 131.12, 130.12, 129.56, 128.17, 127.83, 125.93, 123.84, 122.23, 121.58, 117.87, 115.78 (aromatic carbons), 99.37 (–OC=C), 119Sn NMR (400 MHz, CDCl3): δ–331.19. MS: m/z (M+) calc. for C29H20BrN3O3Sn: 656.97; found: 657.12 (M+H)+.
2,2-Dibutyl-9-nitro-4-(5-phenylisoxazol-3-yl)benzo-[h][1,3,5,6,2]dioxadiazastannonine (15): yield: 51%; yellow solid; m.p. 156–160°C; IR (KBr pellets, cm-1): 1610 (v-C=N), 671 (vSn-C), 561 (vSn-O), 452 (vSn-N). 1H NMR (400 MHz, CDCl3) δ: 8.85 (s, 1H,–N=CH), 8.28–8.20 (m, 2H, Ar-H), 7.87 (dd, J = 7.7, 1.8 Hz, 2H, Ar-H), 7.55–7.45 (m, 3H, Ar-H), 6.97 (s, 1H, –OC=CH), 6.86–6.79 (m, 1H, Ar-H), 1.82–1.57 (m, 12H, Sn-Bu), 0.90 (t, J =7.3Hz, 6H, Sn-Bu). 13C NMR (100 MHz, CDCl3) δ: 172.35 (CHONH), 158.96 (C=N-N), 171.00, 163.93, 162.12, 138.09, 131.43, 130.58, 130.32, 129.10, 127.01, 125.93, 122.61, 115.38 (aromatic carbons), 98.94 (-OC=C), 26.66, 26.43, 23.49, 13.52 (Sn-Bu). 119Sn NMR (400 MHz, CDCl3): δ-187.78. MS: m/z (M+) calc. for C25H28N4O5Sn: 584.11; found: 585.11 (M+H)+.
2,2-Dimethyl-9-nitro-4-(5-phenylisoxazol-3-yl)benzo-[h][1,3,5,6,2]dioxadiazastannonine (16): yield: 69%; yellow solid; m.p. 180–182°C; IR (KBr pellets, cm-1): 1610 (v-C=N), 668 (vSn-C), 567 (vSn-O), 428 (vSn-N). 1H NMR (400 MHz, CDCl3) δ: 8.86 (s, 1H,–N=CH), 8.24 (m, J = 2.6 Hz, 2H, Ar-H), 7.91–7.83 (m, 2H, Ar-H), 7.58–7.46 (m, 3H, Ar-H), 6.97 (s, 1H, –OC=CH), 6.82 (d, J = 8.8 Hz, 1H, Ar-H), 1.01 (s, 6H, Sn-Me). 13C NMR (100 MHz, CDCl3) δ: 171.70 (CHONH), 159.71 (C=N-N), 165.58, 162.43, 136.04, 136.00, 135.91, 135.67, 131.35, 130.61, 130.38, 129.14, 129.11, 126.96, 125.94 (aromatic carbons), 98.83 (–OC=C), 2.64 (Sn-Me). 119Sn NMR (400 MHz, CDCl3): δ-146.98. MS: m/z (M+) calc. for C19H16N4O5Sn: 500.01; found: 501.40 (M+H)+.
9-Nitro-2,2-diphenyl-4-(5-phenylisoxazol-3-yl)benzo-[h][1,3,5,6,2]dioxadiazastannonine (17): yield: 57%; yellow solid; m.p. 181–184°C; IR (KBr pellets, cm-1): 1609 (v-C=N), 676 (vSn-C), 572 (vSn-O), 429 (vSn-N). 1H NMR (400 MHz, CDCl3) δ: 9.01 (s, 1H,–N=CH), 8.41–7.83 (m, 7H, Ar-H), 7.81–7.34 (m, 11H, Ph), 6.98 (s, 1H, –OC=CH). 13C NMR (100 MHz, CDCl3) δ: 171.03 (CHONH), 160.17 (C=N-N), 163.64, 161.98, 157.28, 155.68, 150.67, 136.87, 135.56, 133.21, 131.69, 129.56, 128.23, 128.02, 127.49, 125.78, 121.89, 120.23, 119.57, 118.02, 116.44 (aromatic carbons), 99.48 (–OC=C). 119Sn NMR (400 MHz, CDCl3): δ-331.49. MS: m/z (M+) calc. for C29H20N4O5Sn: 624.05; found: 625.18 (M+H)+.
2,2-Dibutyl-11-ethoxy-4-(5-phenylisoxazol-3-yl)benzo-[h][1,3,5,6,2]dioxadiazastannonine (18): yield: 45%; yellow solid; m.p. 122–126°C; IR (KBr pellets, cm-1): 1605 (v-C=N), 686 (vSn-C), 570 (s, vSn-O), 481 (vSn-N). 1HNMR (400 MHz, CDCl3) δ: 8.79 (s, 1H,–N=CH), 7.89–7.82 (m, 2H, Ar-H), 7.54–7.47 (m, 3H, Ar-H), 7.02–6.97 (m, 1H, Ar-H), 6.95 (s, 1H, –OC=CH), 6.84 (dd, J = 8.0, 1.5 Hz, 1H, Ar-H), 6.68 (t, J = 7.8 Hz, 1H, Ar-H), 4.11 (q, J = 7.3 Hz, 2H, –OCH2CH3), 1.37 (t, J = 7.0 Hz, 3H, –OCH2CH3), 1.69–1.63 (m, 12H, Sn-Bu), 0.88 (t, J = 7.3 Hz, 6H, Sn-Bu). 13C NMR (100 MHz, CDCl3) δ: 170.64 (CHONH), 159.44 (HC=N), 163.64, 162.98, 158.93, 150.52, 130.38, 129.04, 127.22, 126.57, 125.90, 119.48, 116.58, 116.46 (aromatic carbons), 98.92 (–OC=CH), 65.07 (–OCH2CH3), 14.95 (–OCH2CH3), 26.74, 26.46, 23.05, 13.58 (Sn-Bu). 119Sn NMR (400 MHz, CDCl3): δ-186.65. MS: m/z (M+) calc. for C27H33N3O4Sn: 583.15; found: 583.91 (M+H)+.
11-Ethoxy-2,2-dimethyl-4-(5-phenylisoxazol-3-yl)benzo[h][1,3,5,6,2]dioxadiastannonine (19): yield: 58%; yellow solid; m.p. 180–182°C; IR (KBr pellets, cm-1): 1604 (v-C=N), 690 (vSn-C), 578 (vSn-O), 444 (vSn-N). 1H NMR (400 MHz, CDCl3) δ: 8.79 (s, 1H, -N=CH), 7.89–7.82 (m, 2H, Ar-H), 7.55–7.44 (m, 3H, Ar-H), 7.02–6.97 (m, 1H, Ar-H), 6.96 (m, 1H, –OC=CH), 6.85 (dd, J = 8.0, 1.5 Hz, 1H, Ar-H), 6.70 (dd, J = 10.1, 5.6 Hz, 1H, Ar-H), 4.13 (q, J = 7.0 Hz, 2H, –OCH2CH3), 1.42 (t, J = 7.0 Hz, 3H, –OCH2CH3), 0.97 (s, 6H, Sn-Me). 13C NMR (100 MHz, CDCl3) δ: 170.72 (CHONH), 159.36 (C=N-N), 164.48, 162.75, 150.46, 130.42, 129.05, 127.17, 126.44, 125.91, 125.52, 119.03, 116.79, 116.46 (aromatic carbons), 98.80 (–OC=C), 64.95 (–OCH2CH3), 14.85 (–OCH2CH3), 2.63 (Sn-Me). 119Sn NMR (400 MHz, CDCl3): δ -149.29. MS: m/z (M+) calc. for C21H21N3O4Sn: 499.06; found: 499.98 (M+H)+.
11-Ethoxy-2,2-diphenyl-4-(5-phenylisoxazol-3-yl)benzo[h][1,3,5,6,2]dioxadiaza stannonine (20): yield: 51%; yellow solid; m.p. 125–127°C; IR (KBr pellets, cm-1): 1606 (v-C=N), 688 (s, vSn-C), 581 (s, vSn-O), 434 (vSn-N). 1H NMR (400 MHz, CDCl3) δ: 9.05 (s, 1H,–N=CH), 8.47 (m, 2H, Ar-H), 7.95–7.77 (m, 3H, Ar-H), 7.59–7.34 (m, 7H, Ar-H), 7.16–7.01 (m, 3H, Ar-H), 7.00 (s, 1H, –OC=CH), 6.97 (d, J=9.4Hz, 1H, Ar-H), 6.87 (dd, J = 12.7, 4.9 Hz, 1H, Ar-H), 6.72 (d, J = 7.9 Hz, 1H, Ar-H), 4.14–4.06 (q, J = 7.0 Hz, 2H, –OCH2CH3), 1.39 (t, J = 7.0 Hz, 3H, –OCH2CH3). 13C NMR (100 MHz, CDCl3) δ: 171.01 (CHONH), 160.28 (HC=N), 164.21, 162.35, 161.95, 159.32, 157.01, 156.92, 145.59, 137.97, 136.21, 135.32, 129.23, 125.98, 124,56, 122.74, 121.28, 119.87, 118.65, 117.85 (aromatic carbons), 99.48 (–OC=CH), 65.01 (–OCH2CH3), 14.75 (–OCH2CH3). 119Sn NMR (400 MHz, CDCl3): δ -334.78. MS: m/z (M+) calc. for C31H25N3O4Sn: 623.09; found: 623.92 (M+H)+.
In vitro antimicrobial activity assay
The synthesized hydrazone ligands and their corresponding complexes were assessed for their antibacterial and antifungal activities against the Gram-positive bacteria, Bacillus subtilis and Staphylococcus aureus, two Gram-negative bacteria, Escherichia coli and Pseudomonas aeruginosa, and two fungal strains, Candida albicans and Aspergillus niger, by serial dilution method [46, 47] to obtain a minimum inhibitory concentration (MIC). For this, the nutrient broth and potato dextrose broth were freshly prepared, autoclaved to inoculate bacteria and fungi in the respective media, and incubated at 37 ± 1°C for 24 h (all bacteria), 25 ± 1°C for 7 days (A. niger) and 37 ± 1°C for 48 h (C. albicans). The stock solution of 100 μg/mL of the ligands and complexes were prepared by dissolving 1 mg of the synthesized compounds in 10 mL of DMSO and further diluted to get the concentrations of 50, 25, 12.5, 6.25, 3.12, and 1.56 μg/mL. The bacteria and fungi were transferred into each set of test tubes and incubated at a favorable temperature. The standard drugs used for the comparison of the results were ciprofloxacin (for antibacterial study) and fluconazole (for antifungal study). The microbial growths were observed visually and MIC values were calculated. The experiment was repeated three times with the same procedure and each time the growth of the microbes were observed visually in same serially diluted test tube; thus, only the MIC values have been reported with no standard deviation. The data for the microbial study are listed in Table 1.
Antioxidant activity
The synthesized ligands and their corresponding complexes were screened for their antioxidant strength using the conventional DPPH free radical method. This method is primarily based on the percentage free radical scavenging activity of the synthesized compound by DPPH. For this, initially, a solution of 12.5, 25, 50, 100, or 200 μg/mL was prepared in DMSO and then 1 mL of each solution was transferred into a different test tube, with 1 mL of the DPPH, and kept in the dark for 30 min. After that, the absorbance was recorded as 517 nm. Ascorbic acid was used as the reference compound whereas the DPPH in DMSO was used as the blank. The percentage scavenging activity for each compound was calculated using the formula
where, A0 is the absorbance of the control whereas A is the absorbance of the sample at 517 nm.
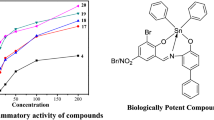
The experiment was repeated three times with the same procedure and IC50 was reported as the average of three replicates. The IC50 values were calculated by plotting the graph of percentage scavenging activity against the concentration of the compounds, as depicted in Fig. 1. The IC50 values of the ligands and their corresponding complexes are listed in Table 2.
Results and Discussion
The synthetic route started with the preparation of Schiff bases, which were synthesized by the reactions of 5-phenylisoxazole-3-carboxylic acid hydrazide with salicylaldehyde and its derivatives. All reactions gave a good yield and these Schiff bases were further reacted with the dialkyltin dichlorides to prepare the organotin complexes (Scheme 1).

Scheme 1
IR spectra
A comparative analysis of the IR spectra of the ligands and their complexes was carried out. The peaks appeared around 3306–3201 cm-1 owing to –OH of the salicylaldehydic part of the ligands being entirely dissipated in the complexes, which indicates that the proton of –OH had been removed completely and oxygen had bonded to the tin atom. The –C=O peaks, which appeared between 1690 and 1660 cm-1 and the –NH peaks, which were between 3483 and 3418 cm-1, that were present in the ligands, were also absent in the complexes indicate that the ligands bonded to the tin metal through its enolic form. The formation of the complexes were also established by the appearance of the new peaks, which were observed at 690–668 cm-1, 593–532 cm-1, and 481–417 cm-1 owing to (vSn-C), (vSn-O), and (vSn-N) [48, 49].
NMR spectral Analysis
1H NMR Spectra
Structures of Schiff base ligands and their respective tin complexes were further confirmed by analyzing and comparing their proton NMR spectra. In the proton NMR, the peak appeared between δ 12.73 and 12.56 ppm owing to the –NH group in the Schiff base ligands, which were absent in the complexes. The –OH peak of the salicylaldehydic part of the ligands assigned at δ 12.12–10.64 ppm had also completely disappeared in the complexes, which also strongly supports the formation of the complexes. The peak of the azomethine hydrogen (CH=N) assigned at δ 8.84–8.73 ppm were shifted downfield, indicating its coordination to the tin atom because of the electron density shifted toward the tin atom. Additional new peaks also appeared in the complexes between δ 1.79 and 0.88 ppm because of the dibutyl, between δ 1.01 and 0.93 ppm because of the dimethyl part, and δ 8.47 and 6.72 ppm because of the phenyl part of the complexes, which were not present in the ligands [50,51,52].
13C NMR Spectra
The structure formations were investigated by the study of the 13C NMR spectra. In the 13C NMR, the signal appeared in the region δ 171.17–170.18 ppm owing to the carbon atoms of the CHONH and δ 161.25–159.07 ppm owing to C=N-N carbon atoms. New peaks observed in the aliphatic region of the complexes between δ 26.78 and 26.66, 26.48 and 26.43, 23.49 and 22.92, and δ 13.58 and 13.52 ppm because of the butyl group, and between δ 2.64 and 2.11 ppm because of the methyl group and 171 and 108 ppm because of the phenyl group present in the complexes, which were not there in the ligands [53].
119Sn NMR Spectra
The 119Sn NMR spectra exhibit the coordination number and geometry of the tin atom in complexes. The tin NMR showed the intense singlet peak at δ -149.29 to -145.16 ppm for methyl, δ -189.27 to -185.15 ppm for butyl and δ -334.78 to -330.19 ppm for phenyl complexes. All these peaks indicate the formation of the pentacoordinated geometry around the tin atom [27, 54].
Mass Spectra
The mass spectra further verified the synthesis of the ligands and their corresponding complexes. In the ligands the (M+H)+ peak appeared at the calculated values and in the complexes also molecular ion peaks (M+H)+ appeared at their calculated value but additionally one more peak appeared at the gap of 2 m/z units; this is due to the presence of the different isotopic abundance of the tin atom, one is due to 119Sn and another due to 117Sn. The mass spectroscopy indicates the mononuclear nature of the ligands and they bind to the metal in a 1:1 ratio.
Antimicrobial Studies
The synthesized Schiff base ligands and their corresponding diorganotin complexes have been studied for their antibacterial and antifungal potency. They have been tested against Gram-positive bacterial strains Bacillus subtilis and Staphylococcus aureus, two Gram-negative bacterial strains Escherichia coli and Pseudomonas aeruginosa for their antibacterial activity by using ciprofloxacin as a standard drug, whereas Candida albicans and Aspergillus niger were used for the antifungal study of synthesized compounds 1–20 by selecting fluconazole as the standard drug. The MIC values of all the tested compounds are listed in Table 1.
-
1.
The general trend followed by the Schiff base ligands as 4 > 3 > 5 > 2 > 1 against B. subtilis, E. coli, and A. niger whereas the trend followed by S. aureus, P. aeruginosa, and C. albicans was 3 > 4 > 5 > 2 > 1. Thus, the Schiff base ligands, having groups such as –NO2 and –Br showed better activity than other Schiff base ligands, having groups such as –Cl, –OC2H5 , and –H. Therefore, when the target molecule consists of groups such as –NO2 and –Br, then it was more selective against the chosen microbes.
-
2.
The presence of the R’’2Sn group with the target molecule helps to enhance its antimicrobial activity. As depicted from the MIC value, the activity of the Schiff base ligands enhanced as they coordinated with R’’2Sn groups, or it can also be stated that the diorganotin complexes showed better activities than their corresponding Schiff base ligands. This can be explained by the chelation theory because as the ligand chelates to the metal center there has been sharing of the positive charge of the metal with the ligands and hence, the lipophilicity of the metal complexes increases and they can easily penetrate through the cell membrane of the microbes, bind to the DNA of the microorganism, and work more efficiently [27, 55].
-
3.
The results of antimicrobial activity signified that the diphenyl substituted complexes showed better activity than dimethyl and dibutyl substituted complexes against the chosen microbes. Thus, the substitution of the Ph2Sn group with the target molecule helps to increase the antimicrobial activity of the molecule.
-
4.
The complexes 14, 17, and 20 showed promising antibacterial activity but none of the compounds displayed better antibacterial activity than the standard drug ciprofloxacin (MIC value = 0.0047 μg/mL).
-
5.
Among all the synthesized compounds, compound 17, having both –NO2 and diphenyltin groups, showed the best antibacterial activity, with an MIC value of 0.0100 μg/mL against all the chosen bacterial strains. Therefore, the presence of the diphenyltin and –NO2 groups with the target molecule helps to strengthen the antibacterial activity of the target molecule. Complex 17 was found to be most selective against all the chosen bacterial strains.
-
6.
Compounds 8 and 11 (MIC values = 0.108 and 0.102 μg/mL) specifically showed very good activity against S. aureus, and their MIC values were around 2-fold higher than the standard drug ciprofloxacin (MIC value = 0.0047 μg/mL).
-
7.
The antifungal activity results demonstrated that Compound 17 was the most potent antifungal compound and it showed better activity than the standard drug fluconazole, with an MIC value of 0.0100 μg/mL against both fungal strains.
-
8.
Compound 14 also showed some good antifungal activity results and MIC values were around two-fold higher than the standard drug fluconazole.

From the structure–activity relationship (SAR), as discussed above, it can be concluded that the antimicrobial activity of the target molecule has been greatly influenced by different group substitutions. The presence of groups such as –NO2, –Br, and Ph2Sn with the target molecule helped to enhance its antimicrobial activity. Therefore, when the target molecule had the –NO2 and Ph2Sn groups (in Compound 17), then the maximum selectivity was observed against the chosen bacterial and fungal strains. Of the bacterial and fungal strains, the compounds showed more selectivity toward fungal strains.
Antioxidant Activity
The synthesized compounds 1–20 were tested against DPPH free radical scavenging activity using ascorbic acid as the standard compound. From the IC50 value as depicted in Table 2, it can be concluded that:
-
1.
The antioxidant activity of the Schiff base ligands enhances as the electron-donating group is attached to the ligands [56] and the antioxidant activity strength trend is followed by the ligands as such 5 1 > 2 > 3 > 4. This can be explained as such after the donation of the hydrogen radical to the DPPH, the Schiff base ligands, having electron-donating groups, become more stabilized than the Schiff base ligands, which have electron-withdrawing groups.[PF1] So when the –OC2H5 group is attached to the target molecule, it becomes more selective against DPPH free radical scavenging activity.
-
2.
Attachment of the R’’2Sn group to the target molecule further helps to enhance the antioxidant activity of the target molecule. The result of the DPPH free radical scavenging activity signified that the complexes showed better activity than their corresponding parent Schiff base ligands. The better results of the complexes can be explained based on the proton-donating ability of the complexes being better than their corresponding Schiff base ligands [57].
-
3.
Among the various R’’2Sn group substitutions with the target molecule, when the R’’ was the phenyl group it showed better activity than when R’’ was butyl and methyl. Therefore, the attachment of the diphenyltin group with the target molecule made it more selective against the DPPH free radical scavenging activity.
-
4.
Compounds 6, 8, and 18 showed good activity and their IC50 value was around 1.5-fold more than the standard compound ascorbic acid.
-
5.
Compound 20 (IC50 = 53.2 μg/mL), which has electron-donating group (–OC2H5) and has two phenyl groups attached to tin, showed better activity than the reference compound ascorbic acid (IC50 = 59.19 μg/mL).

From the SAR of the DPPH free radical scavenging activity, it can be deduced that when groups such as –OC2H5 and Ph2Sn are attached to the target molecule, they help to boost the free radical scavenging activity of the target molecule, whereas the attachment of groups such as –NO2 and Me2Sn to the target molecule diminishes its free radical scavenging activity. Therefore, Compound 20 showed the most selectivity against the DPPH free radical scavenging activity
Conclusion
A series of new diorganotin (IV) complexes had synthesized successfully from the Schiff base ligands, which were formed by the reaction of 5-phenylisoxazole-3-carboxylic acid hydrazide with the salicylaldehyde and its derivatives. The synthesized compounds were characterized by the various spectroscopic techniques such as IR, 1H NMR, 13C NMR, 119Sn NMR, and mass spectroscopy. From the spectroscopy analysis it was concluded that the ligands had bonded to the tin metal in a tridentate manner. The Schiff base ligands bonded with the oxygen atom of the hydroxyl group of the salicylaldehyde part, the nitrogen atom of the azomethine group, and the oxygen atom of the –OH of the hydrazide group that was involved in the enolization. The tin atom exhibited the pentacoordinated geometry. All the synthesized ligands and their corresponding complexes were tested for their biological efficiency against microbes (bacteria and fungi). The result obtained indicated that the activity of the Schiff bases has increased magnificently on binding with the tin metal. Some of the complexes showed promising activity and it was found that the different group substitutions greatly affected the antimicrobial activity. The phenyl-substituted complexes showed better activity followed by the butyl- and then methyl-substituted complexes. Complex 17 showed the best activity. The antioxidant results explained that the ligands that have an electron-donating group acted better as antioxidant agents and the antioxidant activity of the complexes were better than that of the ligands.
References
C. A. Michael, D. Dominey-Howes and M. Labbate, Front. Public Health, 2, 145 (2014).
P. Dadgostar, Infect. Drug Resist., 12, 3903 – 3910 (2019).
D. Jasovský, J. Littmann, A. Zorzet, and O. Cars, Ups. J. Med. Sc., 121, 159 – 164 (2016).
F. C. Tenover, Am. J. Med., 119, S3-S10 (2006).
G. Stochel, A. Wanat, E. Kulioe, and Z. Stasicka, Coord. Chem. Rev., 171, 203 – 220 (1998).
C.-M. Che and F.-M. Siu, Curr. Opin. Chem. Biol., 14, 255 – 261 (2010).
R. Bakhtiar and E.-I. Ochiai, Gen. Pharmacol., 32, 525 – 540 (1999).
J. Karges, R. W. Stokes and S. M. Cohen, Trends Chem., 3, 523 – 534 (2021).
P. C. A Bruijnincx and P. J. Sadler, Curr. Opin. Chem. Biol., 12, 197 – 206 (2008).
D. M. Yufanyi, H. S. Abbo, S. J. J. Titinchi, and T. Neville, Coord. Chem. Rev., 414, 213285 (2020).
Y. Deswal, S. Asija, D. Kumar, et al., Res. Chem. Intermed., 48(2), 703 – 729 (2022).
Y. Deswal, S. Asija, A. Dubey, et al., J. Mol. Struct., 1253, 132266 (2022).
S. Saroya, S. Asija, N. Kumar, and Y. Deswal, J. Indian Chem. Soc., 99, 100379 (2022).
D. J. Hayne, S. Lim and P. S. Donnelly, Chem. Soc. Rev., 43, 6701 – 6715 (2014).
C. X. Zhang and S. J. Lippard, Curr. Opin. Chem. Biol., 7, 481 – 489 (2003).
D.-L. Ma, C. Wu, L. Guodong, et al., J. Mater. Chem. B, 8, 4715 – 4725 (2020).
S. P. Fricker, R. M. Mosi, B. R. Cameron, et al., J. Inorg. Biochem., 102, 1839 – 1845 (2008).
H. Sakurai, Y. Yoshikawa and H. Yasuib, Chem. Soc. Rev., 37, 2383 – 2392 (2008).
M. S. Refat, S. A. El-Korashy, D. N. Kumar, and A. S. Ahmed, Spectrochim. Acta. A. Mol. Biomol. Spectrosc., 70, 898 – 906 (2008).
S. Thamizharasi and V. R. Reddy, Eur. Polym. J., 28, 119 – 123 (1992).
E. N. Md. Yusof, M. A. M. Latif, M. I. M. Tahir, et al., J. Mol. Struct., 1205, 127635 (2020).
J. M. Galván-Hidalgo, D. M. Roldán-Marchán, A. González-Hernández, et al., Med. Chem. Res., 29, 2146 – 2156 (2020).
K. Singh, Dharampal and S. S. Dhiman, J. Iran. Chem. Soc., 7, 243–250 (2010).
H. L. Singh, Phosphorus Sulfur Silicon Relat. Elem., 184, 1768 – 1778 (2008).
H. L. Singh, J. B. Singh and K. P. Sharma, Res. Chem. Intermed., 38, 53 – 65 (2012).
A. Ahlawat, V. Singh and S. Asija, Chem. Pap., 71, 2195 – 2208 (2017).
P. Khatkar, A. Ahlawat, S. Asija, and V. Singh, Phosphorus Sulfur Silicon Relat. Elem., 196, 133 – 145 (2021).
K. S. Prasad, L. S. Kumar, S. Chandan, et al., Spectrochim. Acta. A. Mol. Biomol. Spectrosc., 81, 276 – 282 (2011).
T. S B. Baul, Appl. Organomet. Chem., 22, 195 – 204 (2008).
A. K Mishra, N. Manav and N. K Kaushik, Spectrochim. Acta. A. Mol. Biomol. Spectrosc., 61, 3097 – 3101 (2005).
A. Ahlawat, P. Khatkar, V. Singh, and S. Asija, Res. Chem. Intermed., 44, 4415 – 4435 (2018).
W. Rehman, M. K. Baloch and A. Badshah, Eur. J. Med. Chem., 43, 2380 – 2385 (2008).
Z. Messasma, D. Aggoun, S. Houchi, et al., J. Mol. Struct., 1228, 129463 (2021).
P. Khatkar, S. Asija, A. Ahlawat and V. Singh, Monatshefte für Chemie-Chemical, 150, 207 – 218 (2019).
S. Asijaa, N. Malhotra and R. Malhotra, Phosphorus Sulfur Silicon Relat. Elem., 187, 1510 – 1520 (2012).
P. Khatkar and S. Asija, Phosphorus Sulfur Silicon Relat. Elem., 192, 446 – 453 (2017).
A. Ramírez-Jiménez, R. Luna-García, A. Cortés-Lozada, et al., J. Organomet. Chem., 738, 10 – 19 (2013).
Z. Messasma, D. Aggoun and S. Houchi, J. Mol. Struct., 1228, 129463 (2021). The same as [33]!!!!!
T. A. Antonenko, D. B. Shpakovsky and M. A. Vorobyov, Appl. Organomet. Chem., 32, e4381 (2018).
M. Sirajuddin, S. Ali, F. A. Shah, et al., J. Iran. Chem. Soc., 11, 297 – 313 (2014).
Y. X. Tan, Z. -J. Zhang, Y. Liu, et al., J. Mol. Struct., 1149, 874 – 881 (2017).
A. Saxena and J. P. Tandon, Cancer Lett., 19, 73 – 76 (1983).
H. Ullah, V. Previtali, H. B. Mihigo, et al., Eur. J. Med. Chem., 181, 111544 (2019).
Q. Liu, B. Xie, S. Lin, et al., J. Chem. Sci., 131, 73 (2019).
M. Hong, H. Yin, X. Zhang, et al., J. Organomet. Chem., 724, 23 – 31 (2013).
L. Deswal, V. Verma, D. Kumar, et al., Future Med. Chem., 13(11), 975 – 991 (2021).
L. Deswal, V. Verma, D. Kumar, et al., Arch. Pharm., 353(9), 2000090 (2020).
Y. Yang, M. Hong, L. Xu, et al., J. Organomet. Chem., 804, 48 – 58 (2016).
S. Yadav, I. Yousuf, M. Usman, et al., RSC Adv., 5, 50673 – 50690 (2015).
L. S. Mun, M. A. Hapipah, S. K. Shin, et al., Appl. Organomet. Chem., 26, 310 – 319 (2012).
S. M. Lee, H. M. Ali, K. S. Sim, et al., Inorganica Chim. Acta, 406, 272 – 278 (2013).
L. Xu, M. Hong, Y. Yang, et al., J. Coord. Chem., 69, 2598 – 2609 (2016).
S. Shujha, A. Shah, N. Muhammad, et al., Eur. J. Med. Chem., 45, 2902 – 2911 (2010).
P. Khatkar, S. Asija and N. Singh, J. Serb. Chem. Soc., 82, 13 – 23 (2017).
T. Sedaghat, L. Tahmasbi, H. Motamedi, et al., J. Coord Chem., 66, 712 – 724 (2013).
A. G. Al-Sehemi and A. Irfan, Arab. J. Chem., 10, S1703-S1710 (2017).
A. Corona-Bustamante, J. M. Viveros-Paredes, A. Flores-Parra, et al., Molecules, 15, 5445 – 5459 (2010).
Acknowledgements
The authors are very thankful to the Dr. APJ Abdul Kalam Central Instrumentation Laboratory and Department of Chemistry, Guru Jambheshwar University of Science and Technology, Hisar for providing instrumentation facilities.
Conflicts of Interest: The authors declare that they have no conflicts of interest.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Saroya, S., Asija, S., Deswal, Y. et al. Synthesis, Characterization, and In Vitro Antimicrobial and Antioxidant Study of the Pentacoordinated Organotin(IV) Complexes Based on Acid Hydrazide Schiff Base Ligands. Pharm Chem J 57, 1372–1383 (2023). https://doi.org/10.1007/s11094-023-03000-1
Received:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11094-023-03000-1