Abstract
Developing brain cells express many proteins but little is known of how their protein composition responds to chronic exposure to alcohol and/or how such changes might relate to alcohol toxicity. We used cultures derived from embryonic rat brain (previously shown to contain mostly neural stem cells; rat NSC, rNSC), exposed them to ethanol (25–100 mM) for up to 96 h and studied how they reacted. Ethanol (50 and 100 mM) reduced cell numbers indicating either compromised cell proliferation, cytotoxicity or both. Increased lipid peroxidation was consistent with the presence of oxidative stress accompanying alcohol-induced cytotoxicity. Proteomics revealed 28 proteins as altered by ethanol (50 mM for 96 h). Some were constituents of cytoskeleton, others were involved in transcription/translation, signal transduction and oxidative stress. Nucleophosmin (NPM1) and dead-end protein homolog 1 (DND1) were further studied by immunological techniques in cultured neurons and astrocytes (derived from brain tissue at embryonic ages E15 and E20, respectively). In the case of DND1 (but not NPM1) ethanol induced similar pattern of changes in both types of cells. Given the critical role of the protein NPM1 in cell proliferation and differentiation, its reduced expression in the ethanol-exposed rNSC could, in part, explain the lower cells numbers. We conclude that chronic ethanol profoundly alters protein composition of rNSC to the extent that their functioning—including proliferation and survival—would be seriously compromised. Translated to humans, such changes could point the way towards mechanisms underlying the fetal alcohol spectrum disorder and/or alcoholism later in life.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Development and maturation of brain tissue includes both pre- and postnatal neurogenesis [1,2,3]. The rate of neurogenesis varies as a function of brain maturity [4] but may also be influenced by anxiety, depression as well as by drug and alcohol intake [5].
Neural stem cells (NSC) can generate neurons, astrocytes and oligodendrocytes in the central nervous system (CNS; [6]). NSC first migrate to form specific brain regions while, later in the life, during the process of adult neurogenesis, may respond to functional demands or serve as a replacement for damaged cells. The normal cell generation may become compromised if the damage to the tissue is too great or the initial insult continues such as in an ongoing stress. The latter would include chronic drug use and alcoholism and, indeed, animal models have demonstrated that alcohol is toxic to the neuron-generating regions in brain [7]. Even the blood ethanol levels not exceeding 330 mg/dl (app. 72 mM) have been shown to decrease neurogenesis in adolescent rats [8, 9]. In this context, it is of interest to note that blood alcohol level in Indian-American (indigenous) mothers who gave birth to offspring with fetal alcohol syndrome (fetal alcohol spectrum disorder; FASD) was reported at 80 mM [10]; it is known that 100 mM alcohol can significantly influence human neurogenesis [11]. In another study, moderate doses of ethanol were shown to reduce the number of new neurons by two-thirds and significantly increased the rate of cell death in the dentate gyrus [9]. Administration of ethanol to 35–40 days old rats indicated that the neurogenesis in the adolescent brain is particularly sensitive to ethanol [12]. Moreover, prenatal exposure to ethanol may further compromise postnatal neurogenesis and this may have an additional impact on mental health later in life [13, 14].
Production of neurons proceeds in four stages; proliferation of NSC, differentiation, migration and selective neuronal death/survival. Certain proportion of cells will succumb to programmed cell death (apoptosis); this is an integral part of the differentiation process and may accompany even the adult neurogenesis. Ethanol interferes mainly at the stages one and four [15, 16]; it inhibits the NSC proliferation and increases the cell death. Longer exposures (a 4-day binge model) result in reduced cell proliferation and impaired survival [8, 17]. Moreover, ethanol at 50 mM concentration can affect both the proliferation of NSC and gliogenesis (glial phenotype); ethanol can alter intrinsic cellular mechanisms of NSC, eventually impacting on the structural and functional characteristics of both neurons and glial cells [18]. Such changes occur in prefrontal cortex, nucleus accumbens, and striatum [19, 20] thus potentially contributing to alcohol-induced functional deficits in those regions [21,22,23].
Protein composition is of key importance to the understanding of normal cellular functions and their changes in disease. The technique of proteomics can thus serve as a convenient indicator of the state of health of a biological system. Specifically, proteomic analyses of human brain tissue have identified changes in 238 proteins as associated with alcoholic disorders [24,25,26,27,28,29,30,31].
Proteomic analysis of human NSC detected thousands of proteins at the differentiation stage (review: [32]) but little is known of how this proteome is influenced by ethanol. The aim of the present study is to investigate in vitro changes in the proteome of rat embryonic neural stem cells (rNSC) following a chronic exposure to the concentrations of alcohol which could be encountered in vivo during heavy (or very heavy) drinking. Firstly, we looked at whether ethanol applied for 96 h at concentrations 25, 50 and 100 mM can influence the numbers of cultured rNSC and whether such changes are accompanied by an increased lipid peroxidation indicating the presence of oxidative stress. Secondly, we used a proteomic approach to study the protein composition of rNSC exposed to 50 mM ethanol for 96 h. Four of the proteins found to be significantly changed (two increased and two decreased) by the exposure to ethanol were estimated by Western blotting to verify the validity of the proteomic analysis. Additionally, two of the proteins strongly affected by the ethanol exposure (nucleophosmin; NPM1 and dead end homolog1; DND1; both classified as “nuclear proteins” and therefore deemed as potentially involved in the regulation of protein synthesis as well as cell proliferation and cell survival) were selected for further studies by both Western blotting and immunocytochemistry, using primary cultures of neurons and astrocytes derived from rat brains.
The data we present may contribute to a better understanding of how heavy alcohol drinking (particularly during pregnancy) alters the brain structure and function.
Materials and Methods
Neural Stem Cell Culture
Neural stem cells (rNSC) were prepared from 14-day-old (E14) rat embryos as described earlier [15, 33]. Animal experimental procedures were approved by the The University of Sydney Animal Ethics Committee (AEC Protocol Number: 2013/5742). The telencephalon was separated and blood vessels and meningae were removed while the tissue was submerged in ice-cold Hank’s balanced salt solution (HBSS; Invitrogen, CA USA). After sectioning, the tissue was incubated with 0.05% trypsin solution for 30 min, triturated with a glass pipette and filtered through 70 µm nylon cell strainer (Falcon, USA). The resulting cell suspension was centrifuged (300 g for 5 min at 4 °C). The viability of the cells in the pellet was assessed by trypan blue (Invitrogen). The cells were seeded in complete neurobasal medium [(NBM, Invitrogen) + 2% B27 supplement (Invitrogen), + 0.5 mM L-glutamine + 20 ng/mL recombinant human fibroblast growth factor 2 (FGF-2) + 20 ng/mL recombinant human epidermal growth factor (EGF, Life Tech)] at a density of 1 × 106 cells per mL in culture dishes coated with poly-L-ornithine (Sigma). The dishes were maintained for a week at 37 °C in a 5% CO2 atmosphere. For differentiation, the cells were washed and cultured in differentiating media (all components of complete neurobasal media except FGF-2 and EGF).
Neurons and Astrocytes
The brain tissue was obtained at the embryonic age (E) of either E15 (neurons) or E20 (astrocytes) and treated as above. Following recommendations by available culture protocols (e.g. https://www.thermofisher.com/au/en/home/references/protocols/cell-culture) the cells were cultured either in neurobasal medium (NBM, Invitrogen) + 2% B27 supplement (Invitrogen), + 0.5 mM L-glutamine with antibiotics (neurons) or in serum/antibiotics containing DMEM/F-12 (astrocytes) for 2 weeks at 37 °C in humidified 5% CO2/95% air conditions. While most of the protocols usually recommend brain tissue harvested at embryonic ages E17 or E18 for the neuronal cultures and brains collected at later embryonic stages or from neonatal rats for the preparation of astrocytic cultures [34] we used E15 for the neuronal cultures (cf. also [35]) and E20 for the astrocytic culture with satisfactory results, as indicated both by the appearance of the cells and expression of the characteristic markers (Figs. 3, 4).
Ethanol Treatment and Sample Collection
The cultured cells were exposed to ethanol (0, 25, 50 and 100 mM) added directly to the differentiation neurobasal media. In order to reduce the loss of ethanol by evaporation [36], we used a closed system (closed 25 mL vessel) and changed the media every 24 h [36, 37]. The ethanol exposure lasted for up to 96 h. For proteomics, the cells were washed twice with ice-cold phosphate-buffered saline (PBS) and collected in 200 µL solubilising buffer (7 M urea, 2 M thiourea, 1% C7bZO and 40 mM Tris–HCl, pH 10.4; Sigma). After sonication (3 times for 5 s each) the samples were stored at -80 °C [38, 39].
Estimation of Cell Numbers and Immunofluorescence
Cells on coverslips were fixed with 4% paraformaldehyde for 15 min, followed by washing with PBS and exposure to 0.3% Triton-X100 for 5 min. After washing with PBS, the cells were incubated (blocked) with 5% normal horse serum for 1 h, followed by anti-microtubule-associated-protein 2 antibody (MAP2, mouse, 1:1000; Sigma) for 2 h. The cells were further incubated in mouse IgG FITC labelling reagent (1:1000, Invitrogen) for 45 min and subsequently washed with PBS. Following the application of DAPI (for 10 min) cells were observed under Leica SPE2 confocal microscope (Leica Microsystem). The images were captured and the cells with DAPI-labelled nuclei were counted (about 150 per image) using Image J software (manual option). Whole coverslip contained, typically, 200 to 300 cells or fewer in ethanol treated cultures. (cf. legend of Fig. 1 for further details).
For immuno-histochemistry of neurons and astrocytes, the cells were fixed, treated with Triton X-100 and blocked with horse serum as described above (immunofluorescence) and incubated with a mixture of neuron- or astrocyte-specific marker antibodies. For neurons, anti-microtubule-associated-protein 2 (MAP2, mouse and rabbit, 1:1000; Sigma) was used as a marker while anti-glial fibrillary acidic protein (GFAP, mouse and rabbit, 1:1000; Sigma) was applied to label astrocytes. The cells were further incubated with anti-rabbit nucleophosmin (NPM1, 1:500; Santa Cruz Biotech, Australia) or anti-mouse dead end protein homolog 1 (DND1, 1:500; Santa Cruz Biotech, Australia) for 2 h. Anti- rabbit and/or anti-mouse secondary antibodies (always diluted at 1:1000) conjugated with CF-568 and/or CF488 were applied for 45 min and then washed with PBS. The cells were observed under confocal microscope (Leica SPE2) and the intensity of the fluorescence was quantified by image J software [40, 41].
Lipid Peroxidation Assay
Cells were homogenised in 0.5 mL of 1% KCl solution and supernatant was collected after centrifugation at 16,000 g x 10 min at 4 °C. Aliquots of 0.25 mL of the homogenates were mixed with double volume (0.50 mL) of 15% acetic acid containing thiobarbituric acid (0.5% [42]). The mixture was heated at 95 °C for 15 min and, after cooling, centrifuged at 5000 g x 5 min to remove any precipitated matter. The absorbance of the supernatant was measured at 532 nm. The malondialdehyde concentration was calculated from the following formula: absorbance/1.56 × 105; (where 1.56 × 105 M− 1cm− 1 is the malondialdehyde extinction coefficient).
Statistical Analyses and Presentation of Data
Statistical evaluation of differences between alcohol-free (controls) and alcohol-treated cells was done by one way ANOVA followed by post-hoc Tukey multiple comparisons where a significant effect was found. Both the statistics and the graphical representation of data (Figs. 1, 3, 4) was done using GrapPad Prism software version 7.02 except for Fig. 2 where BioRad software was used. For details see the figure legends.
Proteomics
The cells (exposed to ethanol for 96 h; cf. the section on “Ethanol treatment and sample collection” above) were trypsinized, extracted with urea buffer (7 M urea, 2 M thiourea and 1% C7bZO and 40 mM tris-HCl) and the proteomics performed as detailed earlier [30, 33, 38, 39, 43, 44]. The extracted samples were pelleted at 16,000 g for 20 min at 15 °C; the supernatant was reduced and alkylated with 5 mM tributyl phosphine and 10 mM acrylamide monomer at room temperature for 2 h. The reaction was quenched by 10 mM dithiothreitol (DTT). The citric acid was used to acidify the samples to approximately pH 5. The acetone-precipitated pellets were air dried and resuspended in 0.3 mL of buffer consisting of 7 M urea, 2 M thiourea and 1% C7bZO. The final extracts were stored at − 80 °C before isoelectric focusing.
The protein was quantified [45] using BSA (Sigma–Aldrich, Castle Hill, NSW, Australia) as a standard. Details of isoelectric focusing, electrophoresis and staining have been published [30, 33, 38, 39, 43, 44]. A total of 12 gels (duplicates for each sample, n = 3) were scanned using a flatbed scanner (UMAX). The images were analysed by Phoretix 2D Expression software (Nonlinear). Averaged gels were created for each alcohol treated/control group and averaging parameters were set at 70%. Protein spots were evaluated as volumes (spot area × optical density) with the image analysis software and compared between groups with statistical analysis (ANOVA, P < 0.05). The false discovery rate (FDR) was calculated as described by Storey [46]. The protein spots of interest were cut from the gel, de-stained using 25 mM NH4HCO3/50% (v/v) acetonitrile (ACN) for 3 min × 15 min at 37 °C and digested with 12.5 ng/mL trypsin (Roche, sequencing grade), The peptides were purified using C-18 purification tips (Eppendorf). Matrix (5 mg/mL solution of α-cyano-4-hydroxycinnamic acid in 70% (v/v) ACN/0.1% (v/v) TFA) mixed samples were analysed by Qstar XL Excll Hybride MS system (AB Applied Biosystems) in positive reflector mode, with delayed extraction.
The spectra from MALDI-TOF were searched against the Swissprot protein databases using the MASCOT search engine (http://www.matrixscience.com/). Positive protein identification was performed based on a MOWSE score (> 54, rattus database) with matched isoelectric pH (pI) and molecular weight (MW) values (estimated from 2D gels) and sequence coverage.
Western Blot Analysis
Western blot analysis was performed as described previously [29,30,31]. Proteins separated by SDS–PAGE were transferred to PVDF (polyvinylidene difluoride) membranes [30] blocked with 5% skim milk and incubated with primary antibodies [NPM, DND1, heat shock protein, heterogeneous nuclear ribonucleoproteins-C (hnRNP-C), enolase and synaptosome associated protein-29 (SNAP), all diluted at 1:500] solution. Secondary antibodies (anti-mouse and anti-rabbit IgG, Sigma, diluted at 1:1000) were added and the spots were visualized using an ECL (enhanced chemiluminescence) full spell system (GE Healthcare, Australia). The intensity of the spots was quantified by Biorad software (Bio-Rad, Sydney, Australia).
Results
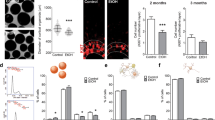
Exposure (96 h) to the lowest ethanol concentration (25 mM) had no significant (P > 0.05) impact on either cell numbers or morphology but the higher concentrations (50 or 100 mM) reduced cell growth (Fig. 1a). MAP-2 positive cells appeared to be reduced in numbers at 50 and 100 mM ethanol relative to control (Fig. 1a). The reduction in cell numbers was quantified by counting the cell nuclei stained by DAPI. DAPI-positive cells were reduced in numbers by 38% and 52% (P < 0.05) at 50 and 100 mM ethanol relative to control (Fig. 1b).
Immunofluorogenic visualization of rat neural stem cells treated with various concentration of alcohol at 96 h of incubation (a). a displays MAP-2 positive cells. b shows the numbers of DAPI stained nuclei and the extent of lipid peroxidation in the cells after 96 h of ethanol exposure. The scales are in arbitrary units and the columns are means ± SD (n = 3). The data were analysed by one way ANOVA followed by post-hoc Tukey multiple comparisons and found significantly different from control (0 mM ethanol) at *P < 0.05, **P < 0.01 and ***P < 0.001
Lipid peroxidation analysis (Fig. 1b) revealed that the exposure of the cells to ethanol induced oxidative stress compared to controls (at 50 mM about 1.6-fold and at 100 mM about 2.1-fold).
In the proteomics, typical 2-DE gels displayed 550 spots (six gels from controls and six gels from ethanol-treated samples) and more than 92% of all spots were matched across the two groups. Analysis revealed that 40 protein spots were differentially expressed in ethanol-exposed cells relative to the ethanol-free controls (P < 0.05; ANOVA); 17 protein spots (~ 43%) showing an increased expression while the remainder were decreased. From the differentially expressed protein spots, we have positively identified 28 proteins using MALDI-TOF (Table 1) while the remaining ones were not identifiable because of low expression of proteins. Functionally, the identified proteins could be classified as structural (two proteins, ~ 7%), intracellular metabolism (six proteins, ~ 21%), nuclear (seven proteins, ~ 25%), related to oxidative stress (six proteins, ~ 21%), signal transduction (four proteins, ~ 14%) and no specific class (three proteins, ~ 11%). Among the identified proteins, nine were increased (including two out of six metabolic, three out of seven nuclear and three out of six oxidative stress-related proteins) while the remaining ones were decreased (including both structural, four out of six metabolic, four out of seven nuclear, three out of six oxidative stress-related and all four signalling proteins).
Heat shock protein-70 (HSP-70), synaptosomal associated protein-29 (SNAP-29), enolase (α) and heterogeneous nuclear ribonucleoprotein C (hnRNP C) which were altered in ethanol-exposed rNSC (Table 1) were assessed by Western blotting (Fig. 2; all primary antibodies were purchased from Santa Cruz Biotech, Australia). These analyses did not contradict the outcomes of the estimations obtained by the proteomic studies (Table 1) and were not pursued further. Nucleolar phosphoprotein (nucleophosmin, NPM1) and dead end homolog1 (DND1) which were selected for immunocytochemical analyses in cultured neurons and astrocytes were also estimated by Western blotting in those preparations (Figs. 3, 4).
Total protein was extracted from the neural stem cells from both control and 50 mM alcohol-exposed cells and western blotting was performed on selected proteins identified as altered by alcohol-exposure by proteomics. MAP-2 was used as an internal control. Bands were digitized and evaluated by Bio-Rad software. b represents average values of two bands
Immunofluorescence indicated somewhat higher expressions of NPM1 at 25 mM ethanol, especially in neurons (cf. also Western blotting in Fig. 3d, f), but a lower expression in astrocytes exposed to 100 mM ethanol (Fig. 3e). The immunofluorescence was detected particularly over the nuclear area though we also noticed the NPM1 staining in cytosolic area at the lowest concentration (25 mM) of ethanol (Fig. 3c).
Immunocytochemical visualization of the expression of NPM1 in neurons (a) and astrocytes (c) exposed to various concentrations of alcohol for 96 h. The green colour of the Fig. 3a is MAP-2 and red colour is NPM protein; in astrocytes (c), green is NPM and red is GFAP. Intensity of fluorescence was estimated for 9 or 10 cells by Image J software (e). The scales (e) are in arbitrary units and the columns are means ± SD (n = 9–10). The data were analysed by one way ANOVA followed by post-hoc Tukey multiple comparisons; those significantly different from control (0 mM ethanol) are marked by asterisks as **P < 0.01 and ***P < 0.001. Total protein was extracted from cells exposed to 50 mM alcohol for 96 h and subjected to western blotting using an antibody against NPM1 protein (neurons: b, astrocytes: d). Bands were analysed by Bio-rad software; f shows average of the two bands shown in (b) and (d), normalized so as the control (0 mM ethanol) equals 100
In the case of DND1, immunocytochemical analysis revealed that the expression significantly increased both in neurons and astrocytes at 25, 50 and 100 mM ethanol (Fig. 4). Western blotting was consistent with the immunocytochemistry, indicating that DND1 protein expression increased in both neurons and astrocytes at the higher (50 and 100 mM) concentration of ethanol (Fig. 4).
Immunocytochemical visualization of the expression of DND1 in neurons (a) and astrocytes (c) exposed to various concentrations of alcohol for 96 h. The green colour of the Fig. 4a is MAP2 and red colour is DND1 protein; in astrocytes, green is DND1 and red is GFAP. Intensity of fluorescence was estimated for 10 or 11 cells by Image J software (e). The scales (e) are in arbitrary units and the columns are means ± SD (n = 10–11). The data were analysed by one way ANOVA followed by post-hoc Tukey multiple comparisons; those significantly different from control (0 mM ethanol) are marked by asterisks as *P < 0.05, **P < 0.01 and ***P < 0.001. Total protein was extracted from cells exposed to 50 mM alcohol for 96 h and subjected to western blotting using an antibody against DND1 protein (neurons: b, astrocytes: d). Bands were analysed by Bio-rad software; f shows average of the two bands shown in b and d, normalized so as the control (0 mM ethanol) equals 100
Discussion
Inhibition of glutamate NMDA receptors [47] and/or activation of a subtype of GABA receptors [48,49,50] parallel immediate effects of alcohol-drinking and probably account for much of the short-term response and consequences (acute alcohol intoxication followed by “morning after”) [51, 52]. In contrast to the receptor-mediated acute effects, longer exposures to ethanol may trigger additional mechanisms leading to profound (and probably irreversible) changes in the living cells and tissues. Apart from resulting in cytotoxicity, chronic alcohol can specifically influence protein synthesis and cell proliferation, particularly in the developing brain [53, 54]. The main finding of the present study is the major changes in the protein composition of cells cultured from developing rat brain following a four-day exposure to high concentrations of ethanol. In addition, we noted reduced cell numbers and found evidence for neurotoxic events in the alcohol-exposed cultures.
As we did not specifically investigate neural stem cell functions and properties; the “rNSC” culture represents, strictly speaking, merely a primary culture of fetal brain tissue harvested at certain stage of development. However, the methodology that we used is known to produce > 95% NSC [33] and the results in Fig. 1a are, therefore, consistent with significant cytotoxic effects of alcohol on the fetal rat brain-derived stem cells. This is further underscored by the increased lipid peroxidation in the presence of alcohol (Fig. 1b). Lipid peroxidation has been used as a marker of oxidative stress associated with cytotoxicity in a range of brain cells and tissues [55,56,57]. Therefore, the current data suggest that oxidative damage is a potentially significant component of the ethanol-related cytotoxicity leading to the loss of cells cultured from embryonic (E15) rat brain tissue.
The remainder of this section will try to evaluate possible consequences of the detected changes in protein composition following the alcohol exposure; given that ethanol interferes with the NSC proliferation and increases the cell death [15, 16] the focus will be on proteins potentially involved in the cell proliferation/differentiation, oxidative stress and nucleus-associated mechanisms.
Cell Proliferation and Differentiation
Hepatoma-derived growth factor (HDGF) was significantly decreased in ethanol exposed cells (Table 1). HDGF has been linked to hepatocellular carcinoma (HCC) as a multi-functional protein involved in cell proliferation, angiogenesis and anti-apoptosis mechanisms [58]. HDGF overexpression promotes the proliferation of HCC [59] and/or gliomas [60] while the reduction in HDGF expression inhibits the proliferation of HCC cells. Knocking down of nuclear HDGF expression in the cells of human glioblastoma multiformis induces apoptosis [61]. By analogy, the decreased expression of HDGF in the ethanol-exposed rNSC could explain lower proliferation rates of brain cells early in the development.
Two isoforms of dihydropyrimidinase-related protein-2 and 3 (DRP-2 and DRP-3) were reported to be lower in human alcoholic brain tissue [62]. The DRP family of proteins, also called the collapsin response mediator proteins (CRMP), has been implicated in the development of the CNS particularly in axon guidance and cell migration. Lower DRP-2 could, therefore, disturb neural development and plasticity [63] and this is supported by reports of DRP-2 being decreased in other conditions involving abnormal development such as schizophrenia and Down’s syndrome [64]. Alternatively, the decreased DRP-2 levels observed in the present study could be a result of proteolysis activated by cytosolic Ca2+ increased by the ethanol-exposure ([65]; cf. also the effect of ethanol on Ca2+-permeable NMDA receptors [51]).
ADP-ribosylarginine hydrolase (ADPRH) removes mono-ADP-ribose moieties from arginines in cellular proteins. Mutation in the ADPRH gene alters G1 stage of the cell-cycle and the reduced expression ADPRH (Table 1) could lead to altered G1-S phase thus decreasing the cell proliferation [66] which could contribute to the lower cell numbers.
NPM1, a nucleolar and nucleoplasma-localized protein [67,68,69], may bind to unduplicated centrosomes thus altering Cyclin E/CDK2 activity which, in turn, regulates G1/S transition [70]. NPM1 has been shown to foster survival of proliferating cells [71, 72], therefore the lower cell numbers in the present study may have been, in part, caused by the lower expression of NPM1 induced by ethanol and/or its metabolites (see also below in the section on “Nuclear Activities and Protein Synthesis”).
Changes in Oxidative Stress Defence Pathways
Ethanol may generate free radicals (review [65]) in vivo either through its metabolism (e.g. via the oxidation to aldehyde) or by interfering with radical scavenging systems. Indeed, long-term chronic ethanol induced lipid peroxidation and depleted GSH levels in human striatum [31]. Reduced expression of glutathione transferase (GST) in the present study is also reminiscent of results obtained in human post-mortem brain tissue of heavy drinkers [24, 27, 29,30,31]. The ethanol-produced reactive oxygen species are eliminated by GST activity [73] and low expression of GST reduces the protection of neurons against oxidative insults.
The glycolytic enzyme α-enolase also acts as a neurotrophic factor [74], a heat-shock protein (HSP48) and a hypoxic stress protein (review: [75]). Glycolytic enzymes including α-enolase are enriched in apoptotic cells and α-enolase deficit (Table 1) could compromise the hypoxia tolerance (which may include nonglycolytic mechanisms [75, 76]).
Changes in SNARE complex have been associated with mental disease [77] and the major increase in the level SNAP-29 (also a part of the SNARE system) may indicate activation of various intracellular mechanisms such as transport of proteins [78], which might have been activated as an adaptation to the effects of ethanol [79].
Heat shock protein 70 (hsp70) expression is regulated by environmental stresses including ethanol exposure [80, 81]. Upregulations of hsp70 mRNA, hsp70 protein and heat shock factor-1 (HSF1) have all been reported in the liver of mice exposed to acute ethanol [79, 82]. Acute and chronic ethanol induces HSP genes such as hsp70, hsp90, and glucose-regulated protein 78 (GRP-78), via HSF-1 activation in neuronal cells [83]. HSP contributes to cell survival via binding to other proteins thus preventing the protein misfolding [84, 85]. The observed increases in the HSP’s expression (Table 1) could reflect activation of such defence mechanisms.
Nuclear Activities and Protein Synthesis
The heterogeneous nuclear protein (hnRNP) corresponds to a family of multifunctional RNA-binding nuclear proteins (review: [86]). Two of these proteins, hnRNP H and hnRNP C were significantly reduced in the ethanol-exposed rNSC (Table 1). The protein hnRNP H may be linked to human cancers while protein C of the hnRNP family is best known for its role in pre-mRNA alternative splicing [86]. Alternative splicing is of interest in alcoholism since the glutamate transporter EAAT1 (GLAST), reported as severely affected by chronic drinking (review: [87] but see [88, 89]), exists in many alternate splicing forms [90]. EAAT1 (GLAST) is a key protein in glutamatergic neurotransmission which is known to be perturbed by ethanol [48]; the conflicting data ([88, 89] v. [91, 92]) could be explained by a changed pattern of its alternate splicing in ethanol-affected tissue. The protein hnRNP C has been linked to breast cancer [93] but, perhaps more importantly in the context of FASD, its abnormal expression has been associated with birth defects in mammals including humans [94, 95].
“Helicase” is a group of enzymes which can unwind double helices of nucleic acids [96]. Spliceosome RNA helicase Ddx39b is involved in the nuclear export of mRNA and its reduced expression in the ethanol-exposed rNSC (Table 1) could have a severe impact on the RNA translation [97].
The DND1 (increased by ethanol in both neurons and astrocytes, Fig. 4) can block microRNA-mediated gene suppression. DND1 plays a key role in the primordial germ cell (PGC) survival and migration [98] and its defects also cause sperm sterility, inducing testicular germ cell tumours in mice [99, 100]. DND1 inactivation in mice leads to sterility at birth stage [101]. DND1 interacts with several mRNAs including mRNAs of both anti- and pro-apoptotic factors, BCLX and BAX, respectively. Transcripts of pluripotency factors, cell cycle regulators and apoptotic factors associated with DND1 have been previously identified using DND1 recombinant study in human embryonic stem cells [100].
Exposure to high concentrations of ethanol decreased NPM1 in rNSC and perturbed its expression in both neurons and astrocytes (Table 1; Fig. 3). This might in part explain the lower cell numbers of the ethanol-exposed cells (Fig. 1) since NPM1 is involved in DNA replication, recombination, transcription and repair [67, 102] as well as elsewhere [103, 104]. This interpretation is also in agreement with reports that the knockout of NPM1 in mice leads to unrestricted centrosome duplication, genomic instability and impaired ribosome biogenesis [67, 105]. In addition, NPM-induced p21 expression activates gene transcription by de-repressing p300–CREBBP (CREB-binding protein; [106, 107] and can regulate p53-mediated apoptosis under the conditions of cellular stress [108]. Thus, the normal level of NPM1 expression could very well be a critical factor in determining the healthy proliferation and differentiation (including the regulation of the normal rate of cell death) of the rNSC (cf. also above under “Cell Proliferation and Differentiation”).
Summary and Concluding Statements
The main finding of the study is the large extent of changes in the expression of proteins in cultured embryonic cells (including neural stem cells) exposed to alcohol. Altered expression of so many proteins that are crucial for the normal cell proliferation and survival, should it occur following a similar exposure to alcohol in vivo, would not be without profound consequences for brain development.
More specifically, the present study resulted in the first observation of changes in DND1 and NPM1 expressions linked to ethanol exposure. Given the roles of NPM1 in cell proliferation, the lower expression of NPM1 could, in part, account for the reduction in neural stem cell numbers following the exposure to ethanol; the changes in the NPM1 expression may have a lesser impact on the cultured neurons and astrocytes, though, where NPM1 was actually increased at 25 mM ethanol and only moderately decreased at the highest (100 mM) ethanol concentration.
The present observations may help to identify novel mechanisms by which chronic alcohol exposures in utero exert their cytotoxic effects. It should be obvious that the existence of such mechanisms does not rule out the role of GABA(A) and/or NMDA receptors in the actions of alcohol including the neurotoxicity [48,49,50,51,52]. Ethanol would interact with the neurotransmitter receptors regardless of the length of the exposure and, as recent studies with the anaesthetics propofol, sevoflurane and ketamine suggest, such interactions could be cytotoxic for brain stem cells [109,110,111]. Our data merely suggest that additional mechanisms involving many more protein species (particularly those 28 protein molecules identified by the present study) could contribute to the process.
The findings of the study are of particular significance for the understanding of mechanisms underlying alcoholism-related conditions such as the foetal alcohol spectrum disorder, especially in relation to alcohol interference with the earliest stages of brain development. Given that neurogenesis may occur in adult brains, too, the present results could also be relevant for the understanding of changes in adult alcoholic brains.
References
Giedd JN, Blumenthal J, Jeffries NO, Castellanos FX, Liu H, Zijdenbos A, Paus T, Evans AC, Rapoport JL (1999) Brain development during childhood and adolescence: a longitudinal MRI study. Nat Neurosci 2(10):861–863
Andersen SL, Thompson AT, Rutstein M, Hostetter JC, Teicher MH (2000) Dopamine receptor pruning in prefrontal cortex during the periadolescent period in rats. Synapse 37(2):167–169
Andersen SL, Teicher MH (2004) Delayed effects of early stress on hippocampal development. Neuropsychopharmacology 29(11):1988–1993
Dennis CV, Suh LS, Rodriguez ML, Krill JJ, Sutherland GT (2016) Human adult neurogenesis across the ages: an immunohistochemical study. Neuropathology Appl Neurobiol 42(7):621–638. https://doi.org/10.1111/nan.12337
Cowen DS, Takase LF, Fornal CA, Jacobs BL (2008) Age-dependent decline in hippocampal neurogenesis is not altered by chronic treatment with fluoxetine. Brain Res 1228(4):14–19 doi:. https://doi.org/10.1016/j.brainres.2008.06.059
McKay R (1997) Stem cells in the central nervous system. Science 276(5309):66–67
Geil CR, Hayes DM, McClain JA, Liput DJ, Marshall SA, Chen KY, Nixon K (2014) Alcohol and adult hippocampal neurogenesis: Promiscuous drug, wanton effects. Prog Neuro Psychopharmacol Biol Psychiatry 54:103–113. https://doi.org/10.1016/j.pnpbp.2014.05.003
Nixon K, Crews FT (2002) Binge ethanol exposure decreases neurogenesis in adult rat hippocampus. J Neurochem 83(5):1087–1093
Herrera DG, Yague AG, Johnsen-Soriano S, Bosch-Morell F, Collado-Morente L, Muriach M, Romero FJ, Garcia-Verdugo JM (2003) Selective impairment of hippocampal neurogenesis by chronic alcoholism: protective effects of an antioxidant. Proc Nat Acad Sci USA 100(13):7919–7924
Kvigne VL, Randall B, Simanton EG, Brenneman G, Welty TK (2012) Blood alcohol levels for American Indian mothers and newborns. Pediatrics 130(4):e1015–e1018. https://doi.org/10.1542/peds.2011-1400
Louis LK, Gopurappilly R, Surendran H, Dutta S, Pal R (2017) Transcriptional profiling of human neural precursors post alcohol exposure reveals impair neurogenesis via dysregulation of ERK signaling and miR-145. J Neurochem. https://doi.org/10.1111/jnc.14155
Crews FT, Mdzinarishvili A, Kim D, He J, Nixon K (2006) Neurogenesis in adolescent brain is potently inhibited by ethanol. Neuroscience 137(2):437–445 https://doi.org/10.1016/j.neuroscience2005.08.090
Redila VA, Olson AK, Swann SE, Mohades G, Webber AJ, Weinberg J, Christie BR (2006) Hippocampal cell proliferation is reduced following prenatal ethanol exposure but can be rescued with voluntary exercise. Hippocampus 16(3):305–311. https://doi.org/10.1002/hipo.20164
Klintsova AY, Helfer JL, Calizo LH, Dong WK, Goodlett CR, Greenough WT (2007) Persistent impairment of hippocampal neurogenesis in young adult rats following early postnatal alcohol exposure. Alcoholism Clin Exp Res 31(12):2073–2082
Tateno M, Ukai W, Hashimoto E, Ikeda H, Saito T (2006) Implication of increased NRSF/REST binding activity in the mechanisms of ethanol inhibition of neuronal differentiation. J Neural Transm 113:283–293
Campbell JC, Stipcevic T, Flores RE, Perry C, Kippin TE (2014) Alcohol exposure inhibits adult neural stem cell proliferation. Exp Brain Res 232(9):2775–2784
Morris SA, Eaves DW, Smith AR, Nixon K (2010) Alcohol inhibition of neurogenesis: a mechanism of hippocampal neurodegeneration in an adolescent alcohol abuse model. Hippocampus 20(5):596–607
Vemuri MC, Chetty CS (2005) Alcohol impairs astrogliogenesis by stem cells in rodent neurospheres. Neurochem Internat 47(1–2):129–135
MacDonald JL, Roskams AJ (2009) Epigenetic regulation of nervous system development by DNA methylation and histone deacetylation. Prog Neurobiol 88(3):170–183
Witt ED (2010) Research on alcohol and adolescent brain development: opportunities and future directions. Alcohol 44(1):119–124
Liyanage VR, Jarmasz JS, Murugeshan N, Del Bigio MR, Rastegar M, Davie JR (2014) DNA modifications: function and applications in normal and disease states. Biology 3(4):670–723
Stragier E, Martin V, Davenas E, Poilbout C, Mongeau R, Corradetti R, Lanfumey L (2015) Brain plasticity and cognitive functions after ethanol consumption in C57BL/6J mice. Translat Psychiatry 5::e696. https://doi.org/10.1038/tp.2015.183
Bonnaud EM, Suberbielle E, Malnou CE (2016) Histone acetylation in neuronal (dys)function. Biomol Concepts 7(2):103–116. https://doi.org/10.1515/bmc-2016-0002
Kashem MA, James G, Harper C, Wilce P, Matsumoto I (2007) Differential protein expression in the corpus callosum (splenium) of human alcoholics: a proteomics study. Neurochem Internat 50(2):450–459
Matsuda-Matsumoto H, Iwazaki T, Kashem MA, Harper C, Matsumoto I (2007) Differential protein expression profiles in the hippocampus of human alcoholics. Neurochem Int 51(6–7):370–376
Matsumoto I, Alexander-Kaufman K, Iwazaki T, Kashem MA, Matsuda-Matsumoto H (2007) CNS proteomes in alcohol and drug abuse and dependence. Expert Rev Proteomics 4(4):539–552
Kashem MA, Harper C, Matsumoto I (2008) Differential protein expression in the corpus callosum (genu) of human alcoholics. Neurochem Internat 53(1–2):1–11. https://doi.org/10.1016/j.neuint.2008.04.003
Hargreaves GA, Quinn H, Kashem MA, Matsumoto I, McGregor IS (2009) Proteomic analysis demonstrates adolescent vulnerability to lasting hippocampal changes following chronic alcohol consumption. Alcoholism Clin Exp Res 33(1):86–94. https://doi.org/10.1111/j.1530-0277.2008.00814.x
Kashem MA, Etages HD, Kopitar-Jerala N, McGregor IS, Matsumoto I (2009) Differential protein expression in the corpus callosum (body) of human alcoholic brain. J Neurochem 110(2):486–495. https://doi.org/10.1111/j.1471-4159.2009. 06141.x
Kashem MA, Ahmed S, Sarker R, Ahmed EU, Hargreaves GA, McGregor IS (2012) Long-term daily access to alcohol alters dopamine-related synthesis and signaling proteins in the rat striatum. Neurochem Internat 61(8):1280–1288. https://doi.org/10.3389/fphar.2013.00086
Kashem MA, Ahmed S, Sultana N, Ahmed EU, Pickford R, Rae C, Sery O, McGregor IS, Balcar VJ (2016) Metabolomics of neurotransmitters and related metabolites in post-mortem tissue from the dorsal and ventral striatum of alcoholic human hrain. Neurochem Res 41(1–2):385–397. https://doi.org/10.1007/s11064-016-1830-3
van Hoof D, Krijgsveld J, Mummery C (2012) Proteomic analysis of stem cell differentiation and early development. Cold Spring Harbor Persp Biol 4:a008177. https://doi.org/10.1101/cshperspect.a008177
Kashem MA, Ummehany R, Ukai W, Hashimoto E, Saito T, McGregor IS, Matsumoto I (2009) Effects of typical (haloperidol) and atypical (risperidone) antipsychotic agents on protein expression in rat neural stem cells. Neurochem Internat 55(7):558–565. https://doi.org/10.1016/j.neuint.2009.05.007
Hansson E (1984) Cellular composition of a cerebral hemisphere primary culture. Neurochem Res 9(2):153–171
Dichter MA (1978) Rat cortical neurons in cell culture: culture methods, cell morphology, electrophysiology and synapse formation. Brain Res 149(2):279–293
Rodriguez FD, Simonsson P, Alling C (1992) A method for maintaining constant ethanol concentrations in cell media. Alcohol Alcoholism 27(3):309–313
Eysseric H, Gonthier B, Soubeyran A, Bessard G, Saxod R, Barret L (1997) There is no simple method to maintain a constant ethanol concentration in long-term cell culture, keys to a solution applied to the survey of astrocytic ethanol absorption. Alcohol 14(2):111–115
Kashem MA, Sarker R, Des Etages H, Machaalani R, King N, McGregor IS, Matsumoto I (2009) Comparative proteomics in the corpus callosal sub-regions of postmortem human brain. Neurochem Internat 55(7):483–490. https://doi.org/10.1016/j.neuint.2009.04.017
Ahmed EU, Ahmed S, Ukai W, Matsumoto I, Kemp A, McGregor IS, Kashem MA (2012) Antipsychotic induced alteration of growth and proteome of rat neural stem cells. Neurochem Res 37(8):1649–1659. https://doi.org/10.1007/s11064-012-0768-3
Shin JW, Nguyen KTD, Pow DV, Knight T, Buljan V, Bennett MR, Balcar VJ (2009) Distribution of glutamate transporter GLAST in membranes of clutired astrocytes in the presence of glutamate transport substrates and ATP. Neurochem Res 34(10):1758–1766. https://doi.org/10.1007/s11064-009-9996-6
Nguyen KTD, Buljan V, Else PL, Pow DV, Balcar VJ (2010) Cardiac glycosides and digoxine interfere with the regulation of glutamate trabsporter GLAST in astrocytes cultured from neonatal rat brain. Neurochem Res 35(12):2062–2069. https://doi.org/10.1007/s11064-010-0274-4
Garcia YJ, Rodríguez-Malaver AJ, Peñaloza N (2005) Lipid preoxidation measurement by thiobarbituric acid assay in rat cerebellar slices. J Neurosci Methods 144(1):127–135
Tang S, Machaalani R, Kashem MA, Matsumoto I, Waters KA (2009) Intermittent hypercapnic hypoxia induced protein changes in the piglet hippocampus identified by MALDI-TOF-MS. Neurochem Res 34(12):2215–2225. https://doi.org/10.1016/j.resp.2011.10.004
van Nieuwenhuijzen PS, Kashem MA, Matsumoto I, Hunt GE, McGregor IS (2010) A long hangover from party drugs: residual proteomic changes in the hippocampus of rats 8 weeks after gamma-hydroxybutyrate (GHB), 3,4-methylenedioxymethamphetamine (MDMA) or their combination. Neurochem Intern 56(8):871–877. https://doi.org/10.3389/fphar.2013.00086
Bradford MM (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Analyt Biochem 72:248–254
Storey JD, Tibshirani R (2003) Statistical significance for genome wide studies. Proc Nat Acad Sci USA 100(16):9440–9445
Lovinger DM, White G, Weight FF (1990) Ethanol inhibition of neuronal glutamate inhibition of neuronal of glutamate receptor function. Ann Med 22(4):247–252
Spanagel R (2009) Alcoholism: a system approach from molecular physiology to addicitive behavior. Physiol Rev 89(2):649–705
Rae CD, Davidson JE, Maher AD, Rowlands BD, Kashem MA, Nasrallah FA, Rallapalli SK, Cook JM, Balcar VJ (2014) Ethanol, not detectably metanolized in brain, significantly reduces brain metabolism, probably via action at specific GABA(A) receptors and has measurable metabolic effects at very low concentrations. J Neurochem 129(2):304–314. https://doi.org/10.1111/jnc.12634
Rae CD, Balcar VJ (2014) A metabolomic multivariate statistical approach for obtaining data-driven information in neuropharmcological research. Recept Clin Investig 1(3):153–156. https://doi.org/10.14800/rci.143
Davidson M, Shanley B, Wilce P (1995) Increased NMDA-induced excitability during ethanol withdrawal: a behavioural study. Brain Res 674(1):91–96
Hoffman PL (1995) Glutamate receptors in alcohol withdrawal-induced neurotoxicity. Metab Brain Dis 10:73–79
Berg DA, Belnoue L, Song H, Simon S (2013) Neurotransmitter-mediated control of neurogenesis in the adult vertebrate brain. Development 140(1):2548–2561
Ming GL, Song H (2011) Adult neurogenesis in the mammalian brain: significant answers and significant questions. Neuron 70(4):687–702. https://doi.org/10.1016/j.neuron
Šťastný F, Lisý V, Mareš V, Lisá V, Balcar VJ, Santamaria A (2004) Quinolinic acid induces NMDA receptor-mediated lipid peroxidation in rat brain microvessels. Redox Rep 9(4):229–233
Garcia E, Limon D, Perez-De La Cruz V, Giordano M, Diaz-Munñz M, Maldonado PD, Herrera-Mundo MN, Pedraza-Chaverri J, Santamaría A (2008) Free Radical Res 42(10):892–902. https://doi.org/10.1080/10715760802506356
Rangel-López E, Colín-Gonzales AL, Paz-Loyola AL, Pinzón E, Torres I, Serratos IN, Castellanos P, Wajner M, Souza DO, Santamaría A (2015) Cannabinoid receptor agonists reduce the short-term mitochondrial dysfunction and oxidative stress linked to excitotoxicity in the rat brain. Neuroscience 285:97–106
Enomoto H, Nakamura H, Liu W, Nishiguchi S (2015) Hepatoma-derived gowth factor: Its possible involvement in the progression of hepatocellular carcinoma. Int J Mol Sci 16(6):14086–14097. https://doi.org/10.3390/ijms160614086
Enomoto H, Nakamura H, Liu W, Iwata Y, Nishikawa H, Takata R, Yoh K, Hasegawa K, Ishii A, Takashima T, Sakai Y, Aizawa N, Ikeda N, Iijima H, Nishiguchi S (2015) Down-regulation of HDGF Inhibits the Growth of Hepatocellular Carcinoma Cells In Vitro and In Vivo. Anticancer Res 35(12):6475–6479
Zhang A, Long W, Guo Z, Cao BB (2012) Downregulation of hepatoma-derived growth factor suppresses the malignant phenotype of U87 human glioma cells. Onc Reports 28(1):62–68
Hsu SS, Chen CH, Liu GS, Tai MH, Wang JS, Wu JC, Kung ML, Chan EC, Liu LF (2012) Tumorigenesis and prognostic role of hepatoma-derived growth factor in human gliomas. J Neuro Oncol 107(1):101–109. https://doi.org/10.1007/s11060-011-0733-z
Alexander-Kaufman K, James G, Sheedy D, Harper C, Matsumoto I (2006) Differential protein expression in the prefrontal white matter of human alcoholics: a proteomics study. Mol Psychiatry 11(1):56–65. https://doi.org/10.1038/sj.mp.4001741
Nakata K, Ujike H, Sakai A, Takaki M, Imamura T, Tanaka Y, Kuroda S (2003) The human dihydropyrimidinase-related protein 2 gene on chromosome 8p21 is associated with paranoid-type schizophrenia. Biol Psychiatry 53(7):571–576
Johnston-Wilson NL, Sims CD, Hofmann JP, Anderson L, Shore AD, Torrey EF, Yolken RH (2000) Disease-specific alterations in frontal cortex brain proteins in schizophrenia, bipolar disorder, and major depressive disorder. Stanley Neuropathol Consort Mol Psychiatry 5(2):142–149
Luo J (2014) Autophagy and ethanol neurotoxicity. Autophagy 10(12):2099–2108. https://doi.org/10.4161/15548627.2014.981916
Kato J, Zhu J, Liu C, Stylianou M, Hoffmann V, Lizak MJ, Glasgow CG, Moss J (2011) ADP-ribosylarginine hydrolase regulates cell proliferation and tumorigenesis. Cancer Res 71(15):5327–5335. https://doi.org/10.1158/0008-5472.CAN-10-0733
Grisendi S, Pandolfi PP (2005) NPM mutations in acute myelogenous leukemia. New Engl J Med 352(3):291–292
Qing Y, Yingmao G, Lujun B, Shaoling L (2008) Role of Npm1 in proliferation, apoptosis and differentiation of neural stem cells. J Neurol Sci 266(1–2):131–137
Pfister JA, D’Mello SR (2015) Insights into the regulation of neuronal viability by nucleophosmin/B23. Exp Biol Med (Maywood) 240(6):774–786. https://doi.org/10.1177/1535370215579168
Okuda M, Horn HF, Tarapore P, Tokuyama Y, Smulian AG, Chan PK, Knudsen ES, Hofmann IA, Snyder JD, Bove KE, Fukasawa K (2000) Nucleophosmin/B23 is a target of CDK2/cyclin E in centrosome duplication. Cell 103(1):127–140
Lindstrom MS (2011) NPM1/B23: a multifunctional chaperone in ribosome biogenesis and chromatin remodeling. Biochem Res Int 2011:195209. https://doi.org/10.1155/2011/195209
Box JK, Paquet N, Adams MN, Boucher D, Bolderson E, O’Byrne KJ, Richard DJ (2016) Nucleophosmin: from structure and function to disease development. BMC Mol Biol 17(1):19. https://doi.org/10.1186/s12867-016-0073-9
Peter N, Chiramel KJ, A RS (2013) Effect of alcohol withdrawl on glutathione S-transferase, total antioxidant capacity and amylase in blood and saliva of alcohol-dependent males. J Clin Diag Res 7(5):797–800. https://doi.org/10.7860/JCDR/2013/4658.2942
Takei N, Kondo J, Nagaike K, Ohsawa K, Kato K, Kohsaka S (1991) Neuronal survival factor from bovine brain is identical to neuron-specific enolase. J Neurochem 57(4):1178–1184
Diaz-Ramos A, Roig-Borrellas A, Garcia-Melero A, Lopez-Alemany R (2012) Alpha-Enolase, a multifunctional protein: its role in pathophysiological situations. J Biomed Biotech 2012:156795. https://doi.org/10.1155/2012/156795
Subramanian A, Miller DM (2000) Structural analysis of alpha-enolase. Mapping the functional domains involved in down-regulation of the c-myc protooncogene. J Biol Chem 275(8):5958–5965
Fatemi SH, Earle JA, Stary JM, Lee S, Sedgewick J (2001) Altered levels of the synaptosomal associated protein SNAP-25 in hippocampus of subjects with mood disorders and schizophrenia. Neuroreport 12(15):3257–3262
Hohenstein AC, Roche PA (2001) SNAP-29 is a promiscuous syntaxin-binding SNARE. Biochem Biophys Res Comm 285(2):167–171
Varodayan FP, Pignataro L, Harrison NL (2011) Alcohol induces synaptotagmin 1 expression in neurons via activation of heat shock factor 1. Neuroscience 193:63–71. doi:_10.1016/j.neuroscience.2011.07.035
Mandrekar P, Catalano D, Jeliazkova V, Kodys K (2008) Alcohol exposure regulates heat shock transcription factor binding and heat shock proteins 70 and 90 in monocytes and macrophages: implication for TNF-alpha regulation. J Leukocyte Biol 84(5):1335–1345. doi:_10.1189/jlb.0407256
Gorini G, Roberts AJ, Mayfield RD (2013) Neurobiological signatures of alcohol dependence revealed by protein profiling. PLoS ONE 8(12):e82656. https://doi.org/10.1371/journal.pone.0082656
Nanji AA, Griniuviene B, Yacoub LK, Sadrzadeh SM, Levitsky S, McCully JD (1995) Heat-shock gene expression in alcoholic liver disease in the rat is related to the severity of liver injury and lipid peroxidation. Proc Soc Exp Biol Med 210(1):12–19
Tunici P, Schiaffonati L, Rabellotti E, Tiberio L, Perin A, Sessa A (1999) In vivo modulation of 73 kDa heat shock cognate and 78 kDa glucose-regulating protein gene expression in rat liver and brain by ethanol. Alcoholism Clin Exp Res 23(12):1861–1867
Borges JC, Ramos CH (2005) Protein folding assisted by chaperones. Prot Pept Lett 12(3):257–261
Verghese L, Abrams J, Wang Y, Morano KA (2012) Biology of the heat shock response and protein chaperones: budding yeast (Saccharomyces cerevisiae) as a model system. Microbiol Mol Biol Rev 76(2):115–158. https://doi.org/10.1128/MMBR.05018-11
Geuens T, Bouhy D, Timmermann V (2016) The hnRNP family: insights into their role in health and disease. Hum Genet 135(8):851–867. https://doi.org/10.1007/s00439-016-1683-5
Šerý O, Sultana N, Kashem MA, Pow DV, Balcar VJ (2015) GLAST but not least–distribution, function, genetics and epigenetics of L-glutamate transport inbrain–focus on GLAST/EAAT1. Neurochem Res 40(12):2461–2472. https://doi.org/10.1007/s11064-015-1605-2
Alshehri FS, Althobaiti YS, Sari Y (2017) Effects of administered athanol and amphetamine on glial glutamate transporters in rat striatum and Hippocampus. J Mol Neurosci 61(3):343–350 doi:_10.1007/s12031-016-0859-8
Kryger R, Wilce PA (2010) The effects of alcoholism on the human basolateral amygdala. Neuroscience 167(2):361–371. https://doi.org/10.1016/j.neuroscience.2010.01.061
Lee A, Pow DV (2010) Astrocytes: Glutamate transport and alternate splicing of transporters. Int J Biochem Cell Biol 42(12):1901–1906
Rimondini R, Arlinde C, Sommer W, Heilig M (2002) Long-lasting increase in voluntary ethanol consumption and transcriptional regulaton in the rat brain after intermittent exposure to alcohol. FASEB J 16(1):27–35
Flatscher-Bader T, Wilce PA (2005) Impact of alcohol abuse on protein expression of midkine and excitatory amino acid transporter 1 in the human prefrontal cortex. Alcoholism Clin Exp Res 32(10):1849–1858
Anantha RW, Alcivar AL, Ma J, Cai H, Simhadri S, Ule J, Konig J, Xia B (2013) Requirement of heterogeneous nuclear ribonucleoprotein C for BRCA gene expression and homologous recombination. PloS ONE 8(4)::e61368. https://doi.org/10.1371/journal.pone.0061368
Zhang Y, Zhang YL, Feng C, Wu YT, Liu AX, Sheng JZ, Cai J, Huang HF (2008) Comparative proteomic analysis of human placenta derived from assisted reproductive technology. Proteomics 8(20):4344–4356. https://doi.org/10.1002/pmic.200800294
Sari Y, Zhang M, Mechref Y (2010) Differential expression of proteins in fetal brains of alcohol-treated prenatally C57BL/6 mice: a proteomic investigation. Electrophoresis 31(3):483–496. https://doi.org/10.1002/elps.200900385
Umate P, Tuteja N, Tuteja R (2011) Genome-wide comprehensive analysis of human helicases. Comm Integr Biol 4(1):118–137. https://doi.org/10.4161/cib.4.1.13844
Schwer B, Meszaros T (2000) RNA helicase dynamics in pre-mRNA splicing. Eur Mol Biol J 19(23):6582–6591
Kedde M, Agami R (2008) Interplay between microRNAs and RNA-binding proteins determines developmental processes. Cell Cycle 7(7):899–903
Youngren KK, Coveney D, Peng X, Bhattacharya C, Schmidt LS, Nickerson ML, Lamb BT, Deng JM, Behringer RR, Capel B, Rubin EM, Nadeau JH, Matin A (2005) The Ter mutation in the dead end gene causes germ cell loss and testicular germ cell tumours. Nature 435(7040):360–364
Zhu R, Iacovino M, Mahen E, Kyba M, Matin A (2011) Transcripts that associate with the RNA binding protein, DEAD-END (DND1), in embryonic stem (ES) cells. BMC Mol Biol 12::37
Bhattacharya C, Aggarwal S, Zhu R, Kumar M, Zhao M, Meistrich ML, Matin A (2007) The mouse dead-end gene isoform alpha is necessary for germ cell and embryonic viability. Biochem Biophys Res Comm 355(1):194–199
Colombo E, Bonetti P, Lazzerini Denchi E, Martinelli P, Zamponi R, Marine JC, Helin K, Falini B, Pelicci PG (2005) Nucleophosmin is required for DNA integrity and p19Arf protein stability. Mol Cell Biol 25(20):8874–8886
Okuwaki M, Matsumoto K, Tsujimoto M, Nagata K (2001) Function of nucleophosmin/B23, a nuceleolar acidic protein, as a histone chaperone. FEBS Lett 506(3):272–276
Abe M, Lin J, Nagata K, Okuwaki M (2018) Selective regulation of type II interferon-inducible genes by NPM1/nucleophosmin. FEBS Lett 595(2):244–255. https://doi.org/10.1002/1873-3468.12952
Chiarella S, De Cola A, Scaglione GL, Carletti E, Graziano V, Barcaroli D, Lo Sterzo C, Di Matteo A, Di Ilio C, Falini B, Arcovito A, De Laurenzi V, Federici L (2013) Nucleophosmin mutations alter its nucleolar localization by impairing G-quadruplex binding at ribosomal DNA. Nucl Acids Res 41(5):3228–3239. https://doi.org/10.1093/nar/gkt001
Lill NL, Grossman SR, Ginsberg D, DiCaprio J, Livingston DM (1997) Binding and modulation of p53 by p300/CBP coactivators. Nature 387:823–827
Kasper LH, Thomas MC, Zambetti GP, Brindle PK (2011) Double null cells reveal that CBP and p300 are dispensable for p53 targets p21 and Mdm2 but variably required for target genes of other signaling pathways. Cell Cycle 10(2):212–221
Abbas T, Dutta A (2009) P21 in cancer: intricate networks and multiple activities. Nat Rev Cancer 9(6):400–414. https://doi.org/10.1038/nrc2657
Sikker W Jr, Liu F, Rainosk SW, Patterson TA, Sadovova N, Hanig JP, Paule MG, Wang C (2015) Ketamine-induced toxicity in neurons differntiated from neural stem cells. Mol Neurobiol 52:959–969 https://doi.org/10.1007/s12035-015-9248-5
Wang JW, Cheng WW, Xu T, Yang ZY (2015) Propofol nduces apoptosis and inhibits the proliferation of rat embryonic stem cells via gamma-aminobutyric acid type A receptor. Genet Mol Res 14(4):14920–14928 doi. https://doi.org/10.4238/2015.November.18.57 org/
Qiu J, Shi P, Mao W, Zhao Y, Liu W, Wang Y (2015) Effect of apoptosis in neural stem cells treated with sevoflurane. BMC Anesthesiology 15:25. https://doi.org/10.1186/s12871-015-0018-8
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Kashem, M.A., Sultana, N. & Balcar, V.J. Exposure of Rat Neural Stem Cells to Ethanol Affects Cell Numbers and Alters Expression of 28 Proteins. Neurochem Res 43, 1841–1854 (2018). https://doi.org/10.1007/s11064-018-2600-1
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11064-018-2600-1