Abstract
This paper presents an automatic diagnosis system for the tumor grade classification through magnetic resonance imaging (MRI). The diagnosis system involves a region of interest (ROI) delineation using intensity and edge magnitude based multilevel thresholding algorithm. Then the intensity and the texture attributes are extracted from the segregated ROI. Subsequently, a combined approach known as Fisher+ Parameter-Free BAT (PFreeBAT) optimization is employed to derive the optimal feature subset. Finally, a novel learning approach dubbed as PFree BAT enhanced fuzzy K-nearest neighbor (FKNN) is proposed by combining FKNN with PFree BAT for the classification of MR images into two categories: High and Low-Grade. In PFree BAT enhanced FKNN, the model parameters, i.e., neighborhood size k and the fuzzy strength parameter m are adaptively specified by the PFree BAT optimization approach. Integrating PFree BAT with FKNN enhances the classification capability of the FKNN. The diagnostic system is rigorously evaluated on four MR images datasets including images from BRATS 2012 database and the Harvard repository using classification performance metrics. The empirical results illustrate that the diagnostic system reached to ceiling level of accuracy on the test MR image dataset via 5-fold cross-validation mechanism. Additionally, the proposed PFree BAT enhanced FKNN is evaluated on the Parkinson dataset (PD) from the UCI repository having the pre-extracted feature space. The proposed PFree BAT enhanced FKNN reached to an average accuracy of 98% and 97.45%. with and without feature selection on PD dataset. Moreover, solely to contrast, the performance of the proposed PFree BAT enhanced FKNN with the existing FKNN variants the experimentations were also done on six other standard datasets from KEEL repository. The results indicate that the proposed learning strategy achieves the best value of accuracy in contrast to the existing FKNN variants.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
1 Introduction
Brain image classification has been an active area of research for the past few years. For categorization of the abnormal brain images into low or high-grade (LG/HG), malignant/benign, etc., many feature extraction approaches have been devised in the last few years [56], and among them, the second order statistics like co-occurrence matrices and fractal attributes are most prevalent [19, 20, 28, 32, 55, 80, 83].
Zhang et al. [91] designed a mechanism for classification of the magnetic resonance (MR) brain images into normal and abnormal categories. The mechanism comprised of the stages of the feature extraction using 2D- discrete wavelet transform (DWT), followed by dimensionality reduction using principal component analysis (PCA), and classification using particle swarm optimized (PSO) Kernel support vector machine (KSVM). The parameters C and σ of the KSVM classifier were fined tuned using the PSO algorithm. Experimenting on 90 MR images from the Harvard Repository, the obtained sensitivity (Se), Specificity (Sp), and Accuracy (Acc) were 98.12%, 92%, and 97.78% using 1024 DWT coefficients and following a 5-fold cross-validation (CV) partitioning scheme. The attained measures were better than those obtained using Back Propagation-Neural Network (BP-NN) and Radial Basis Function (RBF)-Neural Network (RBF-NN) for which the Acc values were 86.22% and 91.33%. The limitation of the work was that it required a huge feature space.
Sachdeva et al. [68] developed a computer-aided diagnosis (CAD) system for the automatic segmentation and the classification of the brain tumors. The system was tested on two datasets. First comprised of 428 post-contrast T1-weighted images collected from 55 patients. The second consist of 260 MR images taken from publically available Surgical Planning Laboratory (SPL) database. The tumor regions were marked using content-based active contour (CBAC) model. From the segmented regions of interest (SROIs) intensity and texture feature set were extracted. Genetic Algorithm (GA) was used to select the optimal features. For classification, two different machine learning algorithms were used namely SVM and artificial neural network (ANN). The test results on both datasets indicated that the use of GA for the feature selection has significantly increased the classification accuracy. It has surged from 79.3% to 91.7% using SVM and using ANN it has increased to 94.9% from 74.6% for the first dataset. Similar results are reported for the second dataset where an increase from 80.8% to 89% was noticed for SVM and for the ANN the increase was from 77.5% to 94.1%. These measures were attained using a feature count of 71 and following a leave one out method (LOOM) for partitioning.
Yang et al. [88] developed a novel wavelet-energy(WE) based approach for automated classification of MR brain images as normal or abnormal using the KSVM classifier. Biogeography-based optimization (BBO) was used to optimize the weights of the SVM, i.e., regularization parameter C, scaling factor σ. The validation was done on 90 Images (5 Healthy (H), 85 Abnormal (Ab)) from the Harvard repository(http://www.med.harvard.edu/aanlib/). The attained Se, Sp, and Acc were 98.12%, 92.00%, and 97.78% using a total of 10 features (wavelet energies from the 10 sub-bands decomposed using Haar wavelet) and following 5 × 5 fold CV partitioning scheme. The attained metrics were better than that obtained using WE and KSVM without optimization for which the Se, Sp, and Acc were 93.18%, 68.00%, and 91.78%. The performance was sensitive to the choice of the mother wavelet and the number of decomposition levels. Better value of the accuracy measure can be expected by employing efficient feature descriptors, such as scale-invariant features and using some advanced pattern recognition techniques.
Skogen et al. [72] designed a mechanism for distinguishing HG from LG glioma based on the exploration of MR image texture information. The mechanism encompasses demarcation of the tumorous region manually tailed by the utilization of the filtration histogram method. This method subjects the segmented region to a filtering process by applying a Laplacian of Gaussian band-pass filter that is then employed to excerpt the attributes at the different size of the spatial scale filter. Evaluation on 67 HG and 27 LG glioma subjects indicated that standard deviation at the fine texture scale was the best distinguishing feature resulting in a Se of 81% and a Sp of 93%.
Lu et al. [52] proposed a novel approach for MR image classification that employed 2D-DWT and calculated the entropies of the sub-bands that served as features. Thereafter, a BAT algorithm optimized ELM (BA-ELM) was trained to classify pathological brains from healthy controls. BAT algorithm was employed to fine-tune the weights/biases and the parameters of the hidden layer. The experimentation was done on 132 MR brain images Harvard Medical School Repository that comprised of 18 H and 114 Ab MR Images. Using a total of 7 features and following a 10 × 10 fold CV, the obtained Se, Sp, and Acc were 99.04%, 93.89%, and 98.33%. The results were better than that obtained by other classifiers like SVM, Naive Bayes, and RBFNN for which the Acc values were 95%, 92.60%, and 91.33%. The drawback of the approach was that it required empirical tuning of the number of the hidden neurons and the type of wavelet used for the decomposition.
Lahmiri [38] compared three automatic diagnosis schemes for the categorization of the normal subjects from glioma patients. The MR images used for categorization were obtained from the Harvard Medical School Repository. The images comprised of 50 T2-w MR scans in axial orientation out of which 20 were abnormal brains affected with glioma and 30 were normal brains. The diagnosis scheme involved four stages: Firstly, the conventional PSO, Darwinian PSO (DPSO), or fractional order DPSO (FODPSO) were used for the segmentation of the MR image. Secondly, for the segmented regions directional spectral distribution (DSD) was calculated. Thirdly, multifractals of the DSD (also called as multi-scale analysis (MSA)) were computed that constituted the feature vector. Lastly, the categorization was done using SVM with LOOM partitioning scheme. The FODPSO+DSD + MSA + SVM scheme provided the finest results using 6 features. This scheme resulted in an Acc value of 99.18% with Se and Sp of 100% and 97.95% respectively.
Bahadure et al. [4] devised CAD technology for the automatic segmentation and classification of tumorous MR images. The CAD consists of stages of pre-processing that involved contrast enhancement and skull stripping, segmentation, feature extraction, feature selection, and classification. For segmentation, different techniques were adopted that involved watershed segmentation, Fuzzy C Means, Discrete Cosine Transformation, and Berkley Wavelet Transform. Feature extraction involved extracting first order statistics, features from gray level co-occurrence matrix(GLCM), and the tumor area. The features were reduced using the GA algorithm, and the tumor classification was done using adaptive neuro-fuzzy inference system, K-Nearest Neighbors (K-NN), and GA based approach. The experimentation involved 135 tumorous and 67 normal images. A comparative analysis was also done using the aforesaid segmentation and classification approaches. Best results were obtained by employing a Berkley Wavelet Transform for segmentation and adopting a GA based classification scheme. The experimental results achieved Se of 92.36%, Sp of 91.42%, and Acc of 92.03% for the task of normal versus tumor classification.
Hemanth et al. [26] devised modified GA for the classification of the abnormal MR brain tumor images. The reproduction operators of GA algorithm were modified to generate new offspring. Thereafter the modified versions were used for feature selection. The validation was done 450 T2-w images collected from the private scan center comprising of the categories of meningioma, metastase, glioma, and astrocytoma. Gray level Difference matrix was used to extract features from these images. A total of 14 features were extracted which were further reduced to 9 using GA1. These reduced features were fed to the Back Propagation neural network for classification. The obtained Se, Sp, and Acc values were 96%, 98%, and 98.1% using a fixed partition scheme (120: Training, 330: Testing).
Gupta et al. [22] developed a glioma detection system based on the texture and the morphological features coupled with ensemble learning. The system worked at three different levels. At the first level, the tumor was segmented using adaptive thresholding or canny edge detection technique followed by morphological operation. At the second level, its location was analyzed and it was classified as super-tentorial or Infra-tentorial. At the third level of classification, the tumor type was identified based on the morphological and other inherent characteristics like area, perimeter, solidity, and the orientation as features through an ensemble of SVM, KNN, and Naïve Bayes (NB) classifiers. Experimentation was done on BRATS 2012 dataset, taking 120 HG, 80 LG images in all the four sequences. The obtained Acc using a 10-fold CV was 94.40/94% for LG/HG using T1 and T1-CE Images. Using FLAIR and T2-w images, following a 10-fold CV, the attained Acc was 96.5/97%. The limitation of the developed system was that it needs preprocessing stages of cropping and enhancement.
Optimization techniques have always played a vital role in the design of the CAD system be it be feature selection or for tuning the model parameters of the classifier [4, 26, 52, 68, 72, 88, 91]. Empirically tuning the model parameters of the classifier is a computationally complex task. To ease this complexity and to improvise the accuracy, optimization techniques have been often used to fine-tune the model parameters of the machine learning algorithms especially for medical diagnosis [50, 52, 71, 73, 88, 91]. Improper model or kernel parameters lead to poor generalization performance of the algorithms.
Prominently, the algorithms like PSO, BBO, and BA have been often employed to optimize the model parameters of the KSVM, i.e., C and σ or weights/biases of the hidden layer of the ELM algorithm and thereafter use it for MR image classification [52, 88, 91]. But none of the algorithms have used PFree Bat Algorithm for optimizing the neighborhood size k and the fuzzy strength parameter m of the Fuzzy K-nearest neighbor (FKNN) classifier.
Moreover, most of the CAD system dealing with tumor classification (either normal versus abnormal or high grade/low grade or multiclass classification) discussed above have validated their system on either the benchmark dataset from the Harvard repository or on their own self-collected private datasets [4, 26, 38, 52, 68, 72, 88, 91]. Very few works like that in [22] have addressed the tumor grade classification using the BRATS 2012 dataset.
The work is motivated by the excellent performance achieved by the FKNN classifier on disease diagnosis like thyroid [50] and the Parkinson [8, 73]. In this paper, an attempt has been made to investigate the efficacy of PFree BAT enhanced FKNN classifier in constructing an automatic system for diagnosis of the abnormality like tumors in MR images.
In this present work, we have designed PFree enhanced FKNN classifier by adaptively tuning the neighborhood size k and the fuzzy strength parameter m of the FKNN classifier using the PFree BAT optimization algorithm. PFree BAT algorithm is an improvised version of the BAT algorithm that combines the major strengths of the BAT and the parameter Free PSO algorithm. Thereafter, the PFree enhanced FKNN classifier is integrated with the initial stages of segmentation, feature extraction, and the feature selection to form a CAD system. The CAD system consists of four major modules.
In the first module segregation of the complete tumor is done by the intensity and the edge magnitude based multilevel thresholding approach [34]. The delineated tumor regions are saved as segmented regions of interest (SROIs). In the second module, intensity and texture features are extracted from the SROIs [33, 35]. In the third module, feature selection is done using Fisher and the PFree BAT optimization algorithm [35]. The selected features are then used as inputs to the PFree BAT enhanced FKNN classifier whose model parameters are fine-tuned using the PFree BAT optimization algorithm. Finally, the classification module consists of classifying brain tumor class as HG or LG by using the enhanced FKNN model.
The designed CAD has been extensively validated on tumor images taken from the BRATS 2012 dataset and on three versions of datasets taken from the Harvard Repository.
As the work primarily focuses on the optimizing the neighborhood size k and the fuzzy strength parameter m of the FKNN classifier through PFree BAT optimization algorithm, so the performance is validated on the benchmark Parkinson dataset from the UCI machine learning repository and six other standard datasets from KEEL repository having pre-extracted feature space. Such experimentation has been explicitly undertaken to highlight the strength of the PFree enhanced FKNN classifier in the absence of the previous stages like segmentation, feature extraction, and selection. The proposed scheme is compared with standard KNN and different optimized version of the FKNN. The comparison is statistically validated using the paired t-test and the Friedman statistical significance test. Furthermore, the comparison with the existing state of the art has also been done.
To summarize the key contributions of the paper are as follows:
-
1.
Firstly, the main contribution of the paper is proposing a new FKNN parameter tuning scheme that uses PFree BAT algorithm. The full potential of the FKNN classifier is explored by the use of the PFree BAT algorithm that determines the optimal value of parameters k, and m in FKNN. Integrating PFree BAT with FKNN enhances the classification capability of FKNN.
-
2.
Secondly, a simple and effective solution is proposed for grading of glioma into low and the high-grade categories by integrating an automatic segmentation method [34], feature extraction approach [33, 35], feature selection algorithm [35], and the proposed PFree enhanced FKNN model.
-
3.
Extensive experimentations have been conducted on the different datasets like those from BRATS 2012, Harvard Repository (3 Versions of the datasets), Parkinson data set from UCI machine learning repository, and six benchmark datasets from KEEL repository. Experimentation on the datasets from the UCI and KEEL repository have been explicitly undertaken to validate the performance of PFree BAT Enhanced FKNN on the pre-extracted feature space. The proposed scheme is compared with standard KNN and different optimized version of the FKNN. The comparison is statistically validated using the paired t-test and the Friedman statistical significance test. Moreover, for every dataset comparison with the recent existing state of art methods is also done.
Paper organization: The paper is structured as follows; Section 2 provides the mathematical background of the related methodologies, Section 3 describes the proposed methodology, Section 4 details the datasets and the evaluation criteria, Section 5 gives the experimental results, Section 6 provides the discussion, and finally, the conclusion is given in the last section.
2 Mathematical backgrounds of the related methodologies
The work presented in this paper is primarily focused on the optimizing the neighborhood size k and the fuzzy strength parameter m of the FKNN classifier through PFree BAT optimization algorithm. This section is devoted to the theoretical background of FKNN and the PFree BAT optimization algorithm.
2.1 Fuzzy k-nearest neighbor method (FKNN)
Fuzzy K-NN adds the concept of the fuzzy logic into the conventional K-NN classifier. Rather than assigning a crisp value of 0 or 1 to a sample vector, it assigns membership to the sample vector. The basis of the FKNN algorithm is to assign the membership as a function of the sample vector’s distance from its K-nearest neighbors and those neighbors’ class memberships in the possible classes.
The fuzzy memberships for the sample vectors are assigned to different classes as per the following formula [36]:
Where j = 1,2,……,k, and i = 1,2……..C, with k as number of nearest neighbors and C as the number of classes, m denotes the fuzzy strength parameter that decides how heavily the distance is weighted when computing the contribution of each neighbor to the membership value. The ‖x − xj‖is the Euclidean distance between x and its jth nearest neighborxj. The uij denotes the membership degree of the pattern xjof the training set to the class i. There are different ways in which uijcan be defined. One works by assigning crisp membership to each training pattern, i.e., the training patterns possess whole membership in their particular class and non-membership in all other classes. The other way works by assigning constrained fuzzy membership, i.e., the k nearest neighbors of each training sample vector are found, and the membership of xk in each class is assigned as [36]:
In eq. (2) nj specify the number of the neighbors belonging to the jth class. The membership computed via eq. (2) must satisfy the equations as follows [36].
Where
In our preliminary experimentation, the first mechanism, i.e., the fuzzy classifier with crisp initialization had resulted in improved classification accuracy than with the constrained initialization.
For any query sample, the membership value ui is computed for all the classes under consideration, and after that, it is allocated to the class to which it possesses the highest membership, i.e.,
2.2 Parameter free BAT (PFree BAT) optimization algorithm
Bat Algorithm (BAT) introduced by Yang [84] is the latest meta-heuristic optimization algorithm inspired by the echolocation skill of the microbats which directs them on their foraging behavior. The BAT is an amalgamation of the major strengths of the PSO and Harmony Search algorithm.
Every bat follows certain procedures for the position (xi) and velocity (vi) updating at each time step, t, in accordance with the pulse frequencyQi. The details regarding the conventional BAT algorithm can be found in the works by [84]. The newer solutions are generated by following a set of equations as given below [84]:
The exploitation and exploration capabilities of the conventional BAT algorithm are indigent [89]. To counteract with this limitation, the modification structure was proposed, inspired by the study in [5, 64]. The bat locations are directly updated through a new position update equation as illustrated below [35]:
The improved version of position update equation guides the bat under the influence of the x∗and the previous best solution (pbesti). As the modified Bat algorithm eliminates the velocity update equation, so it is termed as PFree BAT Algorithm. Better performance of the proposed PFree BAT optimization algorithm has been ascertained by experimenting on standard benchmark functions as reported in our previous works [35].
3 Proposed methodology
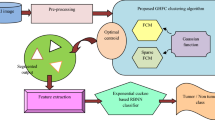
The flowchart of the CAD system incorporating the proposed PFree BAT enhanced FKNN classifier for tumor grade classification (HG/LG) is shown in Fig. 1. The step by step procedure of how the CAD operates is briefed as Algorithm 1.

The CAD system as shown in Fig. 1 primarily comprises of Tumor segmentation, Slice selection, Feature extraction, Feature Selection, and the Classification using the proposed PFree BAT enhanced FKNN classifier.
3.1 Tumor segmentation
The complete tumor region is segmented from the Fluid-attenuated inversion recovery (FLAIR) images using the Intensity and edge magnitude-based multilevel thresholding [34] technique. This technique exploits both intensity and edge magnitude information present in image histogram and GLCM to compute the multiple thresholds through an optimization procedure which are then used for segmentation.
3.2 Slice selection
Following, the process of tumor segmentation, the segregated tumor region was mapped onto the T1-Contrast and the T1-w images. Thereafter, the difference image was generated. Those slices were selected for further processing that exhibited maximum contrast difference [33].
3.3 Feature extraction
A total of 52 features were extracted from the difference image by applying the standard texture models. These include the First-Order Statistics, Gray Level Co-occurrence Matrix (GLCM) [24, 79], Gray Level Run Length Matrix (GLRM) [76], Gray Tone Difference Matrix (GTDM) [1], Law’s Texture Features (LTF) [77], Fractal [12], Gabor Filters [29], Gabor Wavelet [31], and the Empirical mode decomposition (EMD) [11] based features. After extracting the feature vector, normalization was done to standardize all the features to the same level, i.e., in the range between [0 1].
3.4 Feature selection
After extraction of these 52 attributes, feature selection method formulated by fusion of Fisher and the PFree BAT optimization algorithm was applied resulting in the selection of the most informative attributes [35]. The parameters for the PFree BAT algorithm have been chosen after experimenting on the training data that results in maximum classification accuracy in the minimum amount of time with a minimal number of features.
3.5 Classification
The selected features were then fed to the classifier for the final label prediction. The two parameters, i.e., k, m of the FKNN classifier greatly affect the classification accuracy. However, the selection of the appropriate values of these parameters is a tedious task. Therefore, PFree BAT optimization algorithm is used to search for the optimal combinations of the parameters of FKNN. So a PFree BAT enhanced FKNN classifier is designed in the present work in which k, m of the FKNN classifier are fine-tuned by PFree BAT algorithm.
The individual bat positions in PFree BAT enhanced FKNN classifier contains a value of k, and m and they are initialized in the range between 1 to 10. The average classification accuracy measure is taken into consideration while constructing the fitness function and is mathematically given as
where mean(Accuracy) in the above expression for g indicates the average testing accuracy attained by the FKNN classifier via k-fold CV, where k = 3. A 3-fold CV has been employed for the model parameter selection that is different from the outer loop of 5-fold CV, that is utilized for performance computation. Such an explicit combination has been taken by empirically computing the value of the performance metrics for the different combination of folds in the outer and the inner loop. The pseudocode for the inner parameter optimization is given as Algorithm 2 that comprehensively elaborates how the proposed PFree enhanced FKNN classifier computes the optimal values for the k, and m.
The PFree BAT obtains the optimal solution after a series of iterative computation. The detailed iterative procedure is given as a flowchart in Fig. 2. In the PFree BAT algorithm, the following values are initialized as population size (N) = 10, no. of generation of bats = 10, loudness(Ai) = 0.2, emitting sound pulse rate (ri) =0.4, minimum frequencyQmin=0 and maximum frequencyQmax=2. To obtain the best classification results for characterization of the tumors or abnormal MR Images, the overall accuracy has to be maximized.

4 Datasets and the evaluation criteria
4.1 Datasets
The proposed CAD system employing the PFree BAT enhanced FKNN classifier has been validated on tumor images procured from BRATS 2012 database [58]. The dataset encompassed of the real low and high-grade glioma volumes. From the training volumes, after the segmentation process, a slice selection procedure was applied. In the slice selection procedure, the complete abnormal region that has been delineated from the FLAIR set of images [34] was then mapped onto the T1 and T1- CE images. From the mapped out area, an image was created by differencing T1 and T1-CE intensity levels [33]. Only those segmented slices were taken for the further processing that exhibited a significant contrast difference. In the present work a total of 120 such difference images were taken (60 HG and 60 LG).
The latest datasets, i.e., BRATS 2013/2015 also contain the cases continued from BRATS 2012 with slight variations in the labelling of the ground truth (more labels are included in the ground truth of BRATS 2013 dataset) and reduction in the number of image slices (For the same case the slices are reduced in BRATS 2015 dataset).
To the best of the author’s knowledge, only limited works have addressed tumor grade classification problem using the BRATS 2012 dataset [22]. In order to facilitate fair comparison with other recent works on brain image classification, experiments were also conducted on images obtained from the Harvard medical school repository apart from solely employing the BRATS 2012 Dataset. Three versions of the dataset from the Harvard repository have been used. The Version 1 [38] comprises of 50 T2-weighted MR brain images including 30 images of normal (healthy) brains and 20 of abnormal (unhealthy) brains affected with glioma tumor. Version 2 [59] consist of 66 T2-weighted MR brain images out of which 18 were normal, and 48 were abnormal containing cases of Glioma, Alzheimer, Metastatic bronchogenic carcinoma, Herpes encephalitis, and Multiple sclerosis. Version 3 [59] encompasses 160 T2-weighted MR brain images out of which 20 were normal and 140 were abnormal. The abnormal class has images of glioma, meningioma, Alzheimer’s disease, Alzheimer’s disease plus visual agnosia, Pick’s disease, sarcoma, and Huntington’s disease.
Additionally, the versatility of the proposed PFree BAT enhanced FKNN model has been validated on benchmark Parkinson dataset (PD) from the UCI repository (http://archive.ics.uci.edu/ml/machine-learning-databases/parkinsons/, last accessed: September 2016) having fixed set extracted feature space. This dataset consists of biomedical voice measurements from 31 people out of which 23 had PD. Each column entry in the data file denotes a distinct voice measure (comprising of a total of 22 features), and each row corresponds to one of 195 voice recording. The main objective of the data is to distinguish healthy subjects from those having PD, according to the “Status” column that is set to 0 for healthy and 1 for PD.
Moreover, the performance of the proposed PFree BAT enhanced FKNN model has been tested on six benchmark datasets taken from KEEL repository (http://www.keel.es/datasets.php) solely to contrast the performance ability of the proposed PFree BAT enhanced FKNN model with other existing FKNN variants as reported in the works by Derrac et al. [16]. The table below details the datasets employed in the present work. (Table 1).
4.2 Evaluation criteria
The well-known classification metrics of Sensitivity(Se), Specificity(Sp), and Accuracy(Acc) have been chosen to measure the proficiency of the proposed diagnosis system. These measures are widely utilized for the application of the disease diagnosis. These measures comprise of True Negative (TN), False positive (FP), True Positive (TP), and False Negative (FN). TN means the cases of the low-grade glioma that are correctly categorized as low-grade, FP is the number of the low-grade cases categorized as high-grade. TP is the number of the high-grade tumors correctly categorized as high-grade, and FN is the number of high-grade tumors incorrectly categorized as low-grade. Numerically, Se and Sp are defined as [40].
Thereafter, the Acc which reflects the complete detection rate is computed as.
In addition to the Se, Sp, and Acc, the area under the receiver operating characteristic (ROC) curve, i.e., Auc and the Mathew correlation coefficient (Coeff) are also computed. For computing Auc, the method proposed in [18] was used. The Coeff was directly calculated by the formula given below
5 Experimental results
The result section has been primarily focused on the analysis of the brain tumor images from BRATS 2012 dataset using the quantitative performance measures. It is supplemented by comparison with conventional KNN and several optimized FKNN models. The quantitative comparison is tailed by statistical verification by the paired t-test. The section also highlights the classification performance of the proposed PFree BAT enhanced FKNN with and without the feature selection mechanism.
5.1 Comparison of the proposed CAD with different optimized FKNN models
Table 2 reports the results using the various performance metrics, i.e., Se, Sp, Acc, Auc, and Coeff on BRATS 2012 brain tumor dataset [58] using different optimization algorithms like Real-Coded Genetic Algorithm (RGA), PSO, biogeography-based optimization (BBO), Cuckoo Search [86, 87], Firefly Algorithm, Improved Harmony Search [54], BAT Algorithm [84], Novel BAT with Habitat Selection [57], Differential Evolution (DE) [10, 78], Time-variant PSO [7], and the proposed PFree Bat algorithm. The standard deviation values accompany these metrics in the brackets.
These metrics were computed after feature selection and the parameter optimization stage. For the feature selection, the optimization algorithm was used in conjunction with Fisher criteria to choose the most discriminatory subset of attributes. The reduced feature set was then given as training input to the FKNN classifier whose parameters were optimized using the various meta-heuristic algorithms as detailed in Table 2. The MATLAB source codes for the optimization algorithms, i.e., Real-Coded GA, PSO, BBO, DE, and Firefly Algorithm, were taken from the yarpiz.com (http://yarpiz.com/ last accessed: August 2016). The parameters of the optimization algorithms used for the comparison purpose were taken in accordance with their original papers.
As seen from the entries the Se, Sp, Acc, Auc, and Coeff using the adaptive FKNN are larger than the traditional KNN when employing any of the algorithms at the parameter optimization stage. Moreover, the best results were attained using the PFree BAT algorithm at the attribute selection and the parameter optimization stage. The obtained average values for Sp, Acc, Auc, and Coeff were 100% each for the proposed approach that is higher than that equal to 98%, 98.95%, 99% and 98% obtained using the traditional KNN. Better results are accredited to the usage of the PFree Bat algorithm at both stages. The proposed variant of the BAT algorithm has increased the diversity of the search space via which optimal features were selected at the first stage and the best values of ‘k’ and ‘m’ i.e. (k_opt and m_opt) at the later stage.
Moreover, the results attained using the proposed approach are statistically better than the competing algorithms. For statistical verification paired t-test is used that compares the results produced by the proposed algorithm with other competing algorithms. The test assumes the null hypothesis that the accuracy of the two algorithms is the same, against the alternative that the accuracy of the proposed algorithm is higher than the comparison method. If the result of the t-test is 1, then it specifies the rejection of the null hypothesis at a 5% significance level and the acceptance of the alternate hypothesis. If the result is 0, then the null hypothesis cannot be rejected at 5% significance level. The last column of Table 2 reports the statistical significance level of the difference of accuracy value for the proposed algorithm w.r.t. to other meta-heuristic algorithms. For every pairwise comparison, a p value less than 0.05 was obtained. Typically, in contrast to the time-variant PSO, the p value was equal to 0.0035 signifying that accuracy for the proposed algorithm is significantly better than the competing optimized FKNN variant for every fold.
Table 3 summarizes the attained values of ‘k_opt’ and ‘m_opt’ for each value of the outer fold. The attained values for ‘k_opt’ and ‘m_opt’ at each fold were equal to 1, 2, 2, 2, 1 (values were rounded as they signify the nearest neighbors) and 0.4132, 1.7826, 0.9320, 1.9565 and 0.2134 using the proposed PFree Bat algorithm. At each fold of the data, the PFree Bat algorithm adaptively specifies these values. With this optimal combination of model parameters, i.e., ‘k_opt’ and ‘m_opt’; FKNN obtained the best classification accuracy in each fold of data.
5.2 Performance evaluation of the proposed CAD with original extracted feature space (without feature selection)
The classification capability of PFree BAT Enhanced FKNN without feature selection is also investigated, and the findings are summarized in Table 4. The PFree Enhanced FKNN model operating on the complete extracted feature space reaches to an average Se, Sp, Acc, Auc, and Coeff values of 91.11%, 92%, 91.58%, 92.67%, and 85.74%. The obtained value of k_opt, and m_opt are 1,3,1,3,1 and 0.6481, 2.2786, 0.6497, 2, and 0.3820 respectively.
From the tabular findings it is concluded that with feature selection, the PFree BAT enhanced FKNN shows an improvement of 9.76%, 8.7%, 9.19%, 7.91% and 16.63% in Se, Sp, Acc, Auc, and Coeff in contrast to PFree BAT Enhanced FKNN working on the complete feature set. Moreover, this improvement is statistically significant (p value = 0.0017).
6 Discussions
This section is dedicated to the comparison of the proposed CAD with the existing state of the artworks. Firstly, the comparison is done with the works reported on brain tumor categorization. Secondly, the performance is contrasted with existing brain image classification schemes for Version 2 and 3 of the dataset from Harvard Medical Repository. Thirdly, performance comparison is done with existing classification schemes for PD dataset taken from the UCI Machine learning repository having a fixed set of the extracted feature space. Lastly, the validation is done on six benchmark datasets from KEEL repository having a fixed set of the extracted feature space solely to contrast the performance with existing FKNN variants.
6.1 Comparison with existing state-of-art works reported in the domain of brain tumor categorization
Table 5 compares the performance of the proposed CAD with the recent state of artworks dealing with the classification of the glioma images [4, 26, 27, 40, 67, 68, 72, 75, 81, 90, 93]. The proposed CAD outperformed the competing algorithms by achieving 100% Se, Sp, and Acc. Therefore, the proposed method is fully capable of detecting the glioma grade i.e. low or high than the other presented approaches which is highly desirable in clinical applications like therapy planning. This is entirely attributed to the designed CAD system incorporating the proposed PFree BAT enhanced FKNN classifier that has proven to be highly decisive for classification. As the works reported in [4, 26, 27, 40, 67, 68, 72, 75, 81, 90, 93], also deals with the glioma classification, the objective comparison is difficult to draw since the results have been obtained on their collected private datasets. However, for the method in [38] as the database was available from the Harvard Medical school, so the proposed approach has been duly validated on this dataset also. On this dataset also, the proposed approach yielded 100% Sp and Acc which is better than the average value equal to 99.18% and 97.95% reported in [38].
Just for indication the works by [4, 26, 27, 68, 75, 90] dealing with the collected private tumor dataset, have reached to an Acc values equal to 88%, 93.60%, 94.9%, 91.67%, 92.03%, and 98.1%.
Although, not directly comparable the results for the CAD system employing the proposed PFree BAT enhanced FKNN model are better than the reported works in terms of the Se, Sp, and Acc that were 100% for the BRATS 2012 dataset. Recently, the work by [22] has addressed the tumor grade classification problem using BRATS 2012 dataset via the design of a CAD system consisting of phases of preprocessing, segmentation, feature extraction, and classification. The authors attained 94% and 97% accuracy (T1 and the FLAIR images) on a dataset comprising of 120 HG and 80 LG images. Our proposed work is comparable to [22] with the advantage that there was no need for any preprocessing stage of cropping and the image enhancement. The proposed CAD provides automatic classification without using any pre-processing on images.
Furthermore, the proposed CAD using PFree BAT enhanced FKNN model offers multiple benefits in contrast to similar works reported in this field as indicative from Table 5. First, it provides high accuracy not only for BRATS 2012 dataset but also for the dataset reported in the works by [38]. Second, the proposed PFree BAT enhanced FKNN classifier gives this performance using least number of features, i.e., just a single feature in contrast to existing works like [4, 26, 38, 40, 68, 90] that have utilized a feature space of 7, 20, 71, 6, 11, and 9 respectively for classifier training. Lastly, the proposed CAD does not involve any pre-processing stages of cropping and enhancement as reported in the existing work by [22].
6.2 Performance comparison with existing brain image classification schemes for version 2 and 3 of the dataset from Harvard medical repository [25]
Table 6 reports the performance comparison of the proposed CAD with the existing works on brain image classification using Version 2 and version 3 of the dataset from the Harvard Repository. From the Table 6, it is seen that most the works have employed the optimization techniques like PSO [91], ABC-PSO [82], BBO [88], and BAT [52] etc. for the tuning of the model parameters of the classifier.
From Table 6, it is inferred that the proposed CAD that employs the PFree BAT algorithm for FKNN parameter tuning have attained 100% accuracy for Version 2 and 3 of the dataset. It is superior to the value equal to 97.78%, 99.75%, 99.30%, 97.78%, 98.33%, and 99.75% obtained by the techniques devised in works by [52, 59, 82, 88, 91, 92]. Most of the approaches have attained 100% accuracy on the Version 2; their performance declines when moving over to bigger datasets [52, 59, 82, 88, 91, 92]. While few of the approaches given in Table 6 have reached to 100% accuracy both for Version 2 and 3 of the dataset like those presented in [60] but they use a feature count of 3. The prime advantage of the proposed CAD in contrast to [60] is that the proposed PFree enhanced FKNN classifier gives a ceiling value of the accuracy using just a single feature rather than employing a larger number of feature count.
6.3 Performance comparison with existing classification schemes for PD dataset taken from the UCI machine learning repository having a fixed set of the extracted feature space
To validate the versatility of the proposed PFree BAT enhanced FKNN algorithm on the fixed set extracted feature space, this section reports the results on the PD dataset from the UCI repository. For the fair comparison with existence works on the PD dataset, the results are computed without and with feature selection mechanism.
6.3.1 Classification using the proposed PFree BAT enhanced FKNN model with whole original feature space
In this experiment, we evaluated the effectiveness of PFree BAT enhanced FKNN model on the original feature space. The detailed results attained by the PFree BAT enhanced FKNN classifier are shown in Table 7. From, the Table 7, it is seen that PFree BAT enhanced FKNN classifier had reached to average Se, Sp, Acc, Auc, and Coeff values equal to 97.24%, 98%, 97.45%, 97.63%, and 93.58%. The attained values are better than the works reported in [9] in which they obtained the values equal to 96.93%, 88.78%, 94.63%, and 92.85% for Se, Sp, Acc, and Auc.
In addition, it is seen from the Table 7 that the value of the k_opt and m_opt are adaptively specified by PFree BAT optimization algorithm for each run which is the probable reason that the proposed adaptive FKNN classifier performed better than [9].
6.3.2 Classification using the proposed PFree BAT enhanced FKNN model with feature selection
In order to investigate whether feature selection can further improve the detection performance for PD, the PFree BAT enhanced FKNN model was applied to the reduced feature space. Fisher criterion was used in conjunction to rank the features [35]. The top 1, 5, 10, 15, and 20 features are given in Table 8. Table 9 lists the detailed results of PFree BAT enhanced FKNN in terms of Se, Sp, Acc, Auc, and Coeff. From Table 9, it is seen that the performance of the PFree BAT enhanced FKNN model built with feature subset size of 15 is better than one built with all the features. Typically for a feature count equal to 15 the achieved average Se, Sp, Acc, Auc, and Coeff were equal to 97.33%, 100%, 98%, 98.60%, and 97.30% which is higher than the comparison algorithms presented in [9] that reported a mean value of 97.27%, 91.11%, 95.49%, and 94.19% for Se, Sp, Acc, Auc, and Coeff. At each feature count (1, 5, 10, 15, 20, 22) the obtained Se, Sp, Acc and Auc attained via the adaptive FKNN is much higher than the comparison works reported in the literature [9]. Moreover, from the tabular results it is seen that with the utility of the feature selection, the PFree enhanced FKNN has improved performance. The Se, Sp, Acc, Auc, and Coeff measures improvise by 0.09%, 2.04%, 0.56%, 1.74% and 2.1% in contrast to without feature selection. Moreover, this improvement is statistically significant (p value = 0.0429).
The optimal value of ‘k’ and ‘m’ obtained via the PFree enhanced FKNN classifier working on the reduced feature space, i.e., for a typical feature count of 15, at each subsequent fold are given in Table 10. The increase in the value of the performance metrics is attributed to the generation of ‘k_opt’, and ‘m_opt’ by the application of the PFree BAT enhanced FKNN classifier on the reduced dataset.
For the sake of comparison, the classification accuracies attained by the existing approaches on the benchmark PD dataset are given in Table 11. As indicated in the table, accuracy rates ranging between 74% and 96% have been achieved by the other existing works. The proposed approach surpasses the competing algorithms by achieving the mean value of accuracy as 98% and the best value as 100% at a feature count of 15. The increase has been equal to 2% in the average accuracy and 4% in the best accuracy value with feature selection in contrast to the existing work in [9]. Moreover, in contrast to [6], the increase has been 0.8% via the proposed PFree BAT enhanced FKNN classifier using all the 22 features.
6.4 Performance comparison with existing FKNN variants for the six benchmark datasets from KEEL repository having a fixed set of the extracted feature space
This section compares the proficiency of the proposed PFree BAT enhanced FKNN system over six benchmark datasets from KEEL repository. This dataset has been specially chosen to compare the performance of the proposed technique with other FKNN variants as reported in the works by Derrac et al. [16]. Just for the comparative purpose, the results have been reported under the same test conditions as given in [16]. Three sets of test conditions were taken as given below, and the Acc was computed as given in Table 12
-
1.
Fixed k and optimized m
-
2.
Fixed k, optimized m, and Feature Selection
-
3.
Optimized k, optimized m, and Feature Selection
The results under the three test conditions show how the uncertainty in the selection of the k value is reduced by the optimization algorithm and the feature selection process through which an increase in the average accuracy was achieved.
The results are supplemented by the nonparametric statistical tests, i.e., Friedman Shaffer with the post-hoc procedure. The best results for Acc were obtained for the third test condition followed by second and first.
The results presented in this section indicate that the proposed method is scalable to classification problems having a larger sample size and varying dimensionality. The proposed method is validated on eleven benchmark datasets with the sample size varying from 66 to 569, and feature space varying from 6 to 52. The attained accuracy values for most of these datasets were in the range from 88 to 100%. Moreover, the average run time for the datasets is in seconds. Just for indication, for the Parkinson dataset, using all the features the time taken by the PFree BAT enhanced FKNN classifier is 2 s and for tumor dataset, it is 3.54 s. Accuracy and the runtime value on different datasets with varying sizes and dimensionality are indicative that the PFree enhanced FKNN model is scalable in handling an increasing amount of data with larger feature space with minimum computation cost.
Future works will be focused on applying the proposed CAD for the task of the face and palmprint recognition [41,42,43,44,45,46]. Moreover, some advance feature extraction, feature selection, and learning methods like those presented in [47, 51] will be explored.
7 Conclusion
In this work, an automatic brain tumor diagnosis system has been developed. The core component of the system is the adaptive FKNN classifier whose maximum potential is explored by the PFree-BAT optimization algorithm resulting in the generation of the optimal value of neighborhood size and fuzzy strength parameter. Better classification accuracy obtained on the brain tumor dataset has proven that the proposed system can distinguish well enough between the patients with the different grades of glioma, i.e., Low-grade and High-grade and also healthy groups from the glioma patients. It was observed that PFree-Bat enhanced FKNN classifier while working on the reduced feature set achieved 100% recognition rate via fivefold cross-validation. Additionally, the versatility of the method was also proven by experimenting on the standard MR brain image dataset, Parkinson data set and the KEEL data sets. The promising results of the proposed method have shown that our designed system is also capable of distinguishing abnormal subjects from healthy groups. Extensive experimentation on benchmark data sets from KEEL repository also validates that the proposed mechanism is superior to the existing FKNN variants existing in the literature.
References
Amadasun M, King R (1989) Texural features corresponding to textural properties. IEEE Trans Syst Man Cybern 19:1264–1274
Arif M, Akram MU, others (2010) Pruned fuzzy K-nearest neighbor classifier for beat classification. J Biomed Sci Eng 3:380.
Astrom F, Koker R (2011) A parallel neural network approach to prediction of Parkinson’s disease. Expert Syst Appl 38:12470–12474
Bahadure NB, Ray AK, Thethi HP (2018) Comparative approach of MRI-based brain tumor segmentation and classification using genetic algorithm. J Digit Imaging 31:477–489
Bakwad KM, Pattnaik SSSS, Sohi BS, et al (2009) Hybrid bacterial foraging with parameter free PSO. In: Nat. Biol. Inspired Comput. 2009. NaBIC 2009. World Congr. Ieee, pp 1077–1081
Cai Z, Gu J, Wen C et al (2018) An intelligent Parkinson’s disease diagnostic system based on a chaotic bacterial foraging optimization enhanced fuzzy KNN approach. Comput Math Methods Med 2018:1–24
Chen H-L, Yang B, Wang G et al (2011) A novel bankruptcy prediction model based on an adaptive fuzzy k-nearest neighbor method. Knowledge-Based Syst 24:1348–1359
Chen H-L, Huang C-C, Yu X-G et al (2013) An efficient diagnosis system for detection of Parkinson’s disease using fuzzy k-nearest neighbor approach. Expert Syst Appl 40:263–271
Chen H-L, Wang G, Ma C et al (2016) An efficient hybrid kernel extreme learning machine approach for early diagnosis of Parkinson′s disease. Neurocomputing 184:131–144
Cheng M, Hoang N (2014) Groutability estimation of grouting processes with microfine cements using an evolutionary instance-based learning approach. J Comput Civ Eng 28:04014014
Colominas MA, Schlotthauer G, Torres ME (2014) Improved complete ensemble EMD: a suitable tool for biomedical signal processing. Biomed Sign Proc Control 14:19–29
Costa AF, Humpire-mamani G, Juci A, et al (2012) An Efficient Algorithm for Fractal Analysis of Textures. In: 25th SIBGRAPI Conf. Graph. Patterns Images. IEEE, Ouro Preto, Brazil, pp 39–46
Cover T, Hart P (1967) Nearest neighbor pattern classification. IEEE Trans Inf Theory 13:21–27
Das R (2010) A comparison of multiple classification methods for diagnosis of Parkinson disease. Expert Syst Appl 37:1568–1572
Denoeux T (1995) A k-nearest neighbor classification rule based on Dempster-Shafer theory. IEEE Trans Syst Man Cybern 25:804–813
Derrac J, Chiclana F, García S, Herrera F (2016) Evolutionary fuzzy k -nearest neighbors algorithm using interval-valued fuzzy sets. Inf Sci (Ny) 329:144–163
Emblem KE, Nedregaard B, Hald JK et al (2009) Automatic glioma characterization from dynamic susceptibility contrast imaging: brain tumor segmentation using knowledge-based fuzzy clustering. J Magn Reson Imaging 30:1–10
Fawcett T (2004) ROC graphs: notes and practical considerations for researchers. Mach Learn 31:1–38
Georgiadis P, Cavouras D, Kalatzis I et al (2008) Improving brain tumor characterization on MRI by probabilistic neural networks and non-linear transformation of textural features. Comput Methods Prog Biomed 89:24–32
Gibbs P, Turnbull LW (2003) Textural analysis of contrast-enhanced MR images of the breast. Magn Reson Med 50:92–98
Guo P-F, Bhattacharya P, Kharma N (2010) Advances in detecting Parkinson’s disease. In: Int. Conf. Med. Biometrics. pp 306–314
Gupta N, Bhatele P, Khanna P (2019) Glioma detection on brain MRIs using texture and morphological features with ensemble learning. Biomed Sign Proc Control 47:115–125
Guyon I, Weston J, Barnhill S, Vapnik V (2002) Gene selection for cancer classification using support vector machines. Mach Learn 46:389–422
Haralick RM, Shanmugam K, Dinstein I (1973) Textural features for image classification. IEEE Trans Syst Man Cybern SMC-3:610–621
Harvard Medical School. http://med.harvard.edu/AANLIB/. Accessed 2 Apr 2016
Hemanth JD, Anitha J (2019) Modified genetic algorithm approaches for classification. Appl Soft Comput J 75:21–28
Hemanth DJ, Vijila CKS, Selvakumar AI, Anitha J (2011) Performance Enhanced Hybrid Kohonen-Hopfield Neural Network for Abnormal Brain Image Classification. In: Signal Process. Image Process. Pattern Recognit. Springer, pp 356–365
Herlidou-Meme S, Constans J, Carsin B et al (2003) MRI texture analysis on texture test objects, normal brain and intracranial tumors. Magn Reson Imaging 21:989–993
Hong L, Wan Y, Jain A (1998) Fingerprint image enhancement: algorithm and performance evaluation. IEEE Trans Pattern Anal Mach Intell 20:777–789
Hu X, Xie C (2005) Improving fuzzy k-NN by using genetic algorithm. J Comput Inf Syst 1:203–213
Hui LY, Muftah M, Das T et al (2012) Classification of MR tumor images based on Gabor wavelet analysis. J Med Biol Eng 32:22–28
Iftekharuddin KM, Zheng J, Islam MA, Ogg RJ (2009) Fractal-based brain tumor detection in multimodal MRI. Appl Math Comput 207:23–41
Kaur T, Saini B, Gupta S (2017) Quantitative metric for MR brain tumor grade classification using sample space density measure of analytic intrinsic mode function representation. IET Image Process 11:620–632
Kaur T, Saini BS, Gupta S (2018) A joint intensity and edge magnitude-based multilevel thresholding algorithm for the automatic segmentation of pathological MR brain images. Neural Comput Appl 30:1317–1340
Kaur T, Saini BS, Gupta S (2018) A novel feature selection method for brain tumor MR image classification based on the fisher criterion and parameter-free bat optimization. Neural Comput Appl 29:193–206
Keller JM, Gray MR (1985) A fuzzy K-nearest neighbor algorithm. IEEE Trans Syst Man Cybern SMC-15:580–585
Kucnehva LI (1995) An intuitionistic fuzzy k-nearest neighbors rule.
Lahmiri S (2017) Glioma detection based on multi-fractal features of segmented brain MRI by particle swarm optimization techniques. Biomed Sign Proc Control 31:148–155
Lee S-H (2015) Feature selection based on the center of gravity of BSWFMs using NEWFM. Eng Appl Artif Intell 45:482–487
Lee MC, Nelson SJ (2008) Supervised pattern recognition for the prediction of contrast-enhancement appearance in brain tumors from multivariate magnetic resonance imaging and spectroscopy. Artif Intell Med 43:61–74
Leng L, Zhang J, Xu J, et al (2010) Dynamic weighted discrimination power analysis in DCT domain for face and palmprint recognition. In: Inf. Commun. Technol. Converg. (ICTC), 2010 Int. Conf. pp 467–471
Leng L, Zhang J, Xu J, et al (2010) Dynamic weighted discrimination power analysis: A novel approach for face and palmprint recognition in DCT domain. In: Int. J. Phys. Sci. pp 467–471
Leng L, Zhang J, Chen G, et al (2011) Two-directional two-dimensional random projection and its variations for face and palmprint recognition. In: Int. Conf. Comput. Sci. Its Appl. pp 458–470
Leng L, Zhang S, Bi X, Khan MK (2012) Two-dimensional cancelable biometric scheme. In: 2012 Int. Conf. Wavelet Anal. Pattern Recognit. pp 164–169
Leng L, Li M, Teoh ABJ 2013) Conjugate 2D palmhash code for secure palm-print-vein verification. In: Image Signal Process. (CISP), 2013 6th Int. Congr. pp 1705–1710
Leng L, Li M, Kim C, Bi X (2017) Dual-source discrimination power analysis for multi-instance contactless palmprint recognition. Multimed Tools Appl 76:333–354
Li Z, Tang J (2015) Unsupervised feature selection via nonnegative spectral analysis and redundancy control. IEEE Trans Image Process 24:5343–5355
Li D-C, Liu C-W, Hu SC (2011) A fuzzy-based data transformation for feature extraction to increase classification performance with small medical data sets. Artif Intell Med 52:45–52
Little MA, McSharry PE, Hunter EJ et al (2009) Suitability of dysphonia measurements for telemonitoring of Parkinson’s disease. IEEE Trans Biomed Eng 56:1015–1022
Liu DY, Chen HL, Yang B et al (2012) Design of an enhanced fuzzy k-nearest neighbor classifier based computer aided diagnostic system for thyroid disease. J Med Syst 36:3243–3254
Liu Y, Nie L, Han L, et al (2015) Action2Activity: Recognizing Complex Activities from Sensor Data. In: IJCAI. pp 1617–1623
Lu S, Qiu X, Shi J et al (2017) A pathological brain detection system based on extreme learning machine optimized by bat algorithm. CNS Neurol Disord Targets (Formerly Curr Drug Targets-CNS Neurol Disord) 16:23–29
Luukka P (2011) Feature selection using fuzzy entropy measures with similarity classifier. Expert Syst Appl 38:4600–4607
Mahdavi M, Fesanghary M, Damangir E (2007) An improved harmony search algorithm for solving optimization problems. Appl Math Comput 188:1567–1579
Mahmoud-Ghoneim D, Toussaint G, Constans J-M, de Certaines JD (2003) Three dimensional texture analysis in MRI: a preliminary evaluation in gliomas. Magn Reson Imaging 21:983–987
Materka A, Strzelecki M (1998) Texture analysis methods--a review. Tech. Univ. lodz, Inst. Electron. COST B11 report, Brussels
Meng X-B, Gao XZ, Liu Y, Zhang H (2015) A novel bat algorithm with habitat selection and Doppler effect in echoes for optimization. Expert Syst Appl 42:6350–6364
Menze BH, Jakab A, Bauer S et al (2014) The multimodal brain tumor image segmentation benchmark (BRATS). IEEE Trans Med Imaging 34:1993–2024
Nayak DR, Dash R, Majhi B et al (2016) Brain MR image classification using two-dimensional discrete wavelet transform and AdaBoost with random forests. Neurocomputing 177:188–197
Nayak DR, Dash R, Majhi B (2018) Discrete ripplet-II transform and modified PSO based improved evolutionary extreme learning machine for pathological brain detection. Neurocomputing 282:232–247
Ozcift A, Gulten A (2011) Classifier ensemble construction with rotation forest to improve medical diagnosis performance of machine learning algorithms. Comput Methods Prog Biomed 104:443–451
Psorakis I, Damoulas T, Girolami MA (2010) Multiclass relevance vector machines: sparsity and accuracy. IEEE Trans Neural Netw 21:1588–1598
Rakotomamonjy A (2003) Variable selection using SVM-based criteria. J Mach Learn Res 3:1357–1370
Ramana Murthy G, Senthil Arumugam M, Loo CK (2009) Hybrid particle swarm optimization algorithm with fine tuning operators. Int J Bio-Inspired Comput 1:14–31
Rhee F-H, Hwang C (2003) An interval type-2 fuzzy K-nearest neighbor. In: fuzzy Syst. 2003. FUZZ’03. 12th IEEE Int. Conf. Pp 802–807
Sachdeva J, Kumar V, Gupta I et al (2012) A novel content-based active contour model for brain tumor segmentation. Magn Reson Imaging 30:694–715
Sachdeva J, Kumar V, Gupta I et al (2013) Segmentation, feature extraction, and multiclass brain tumor classification. J Digit Imaging 26:1141–1150
Sachdeva J, Kumar V, Gupta I et al (2016) A package-SFERCB-“segmentation, feature extraction, reduction and classification analysis by both SVM and ANN for brain tumors”. Appl Soft Comput 47:151–167
Sakar CO, Kursun O (2010) Telediagnosis of Parkinson’s disease using measurements of dysphonia. J Med Syst 34:591–599
Shahbaba B, Neal R (2009) Nonlinear models using Dirichlet process mixtures. J Mach Learn Res 10:1829–1850
Shrivastava P, Shukla A, Vepakomma P et al (2017) A survey of nature-inspired algorithms for feature selection to identify Parkinson’s disease. Comput Methods Prog Biomed 139:171–179
Skogen K, Schulz A, Dormagen JB et al (2016) Diagnostic performance of texture analysis on MRI in grading cerebral gliomas. Eur J Radiol 85:824–829
Spadoto AA, Guido RC, Carnevali FL, et al (2011) Improving Parkinson’s disease identification through evolutionary-based feature selection. In: 2011 Annu. Int. Conf. IEEE Eng. Med. Biol. Soc. pp 7857–7860
Subashini MM, Sahoo SK, Sunil V, Easwaran S (2016) A non-invasive methodology for the grade identification of astrocytoma using image processing and artificial intelligence techniques. Expert Syst Appl 43:186–196
Subashini MM, Sahoo SK, Sunil V, Easwaran S (2016) A non-invasive methodology for the grade identification of astrocytoma using image processing and artificial intelligence techniques. Expert Syst Appl 43:186–196
Tang X (1998) Texture information in run-length matrices. IEEE Trans Image Process 7:1602–1609
Tencer L, Reznakova M, Cheriet M (2012) A new framework for online sketch-based image retrieval in web environment. In: Inf. Sci. Signal Process. their Appl. Spec. Sess. IEEE, Montreal, QC, pp 1430–1431
Tien Bui D, Nguyen QP, Hoang ND, Klempe H (2016) A novel fuzzy K-nearest neighbor inference model with differential evolution for spatial prediction of rainfall-induced shallow landslides in a tropical hilly area using GIS. Landslides 14:1–17
Vidya KS, Ng EY, Acharya UR et al (2015) Computer-aided diagnosis of myocardial infarction using ultrasound images with DWT, GLCM and HOS methods: a comparative study. Comput Biol Med 62:86–93. https://doi.org/10.1016/j.compbiomed.2015.03.033
Wagner F, Gryanik A, Schulz-Wendtland R et al (2012) 3D characterization of texture: evaluation for the potential application in mammographic mass diagnosis. Biomed Eng (NY) 57:490–493
Wang S, Kim S, Chawla S et al (2010) Differentiation between glioblastomas and solitary brain metastases using diffusion tensor imaging. Neuroimage 44:653–660
Wang S, Zhang Y, Dong Z et al (2015) Feed-forward neural network optimized by hybridization of PSO and ABC for abnormal brain detection. Int J Imaging Syst Technol 25:153–164
Xu Y, van Beek EJR, Hwanjo Y et al (2006) Computer-aided classification of interstitial lung diseases via MDCT: 3D adaptive multiple feature method (3D AMFM). Acad Radiol 13:969–978
Yang X-S (2010) A new metaheuristic bat-inspired algorithm. In: Nat. inspired Coop. Strateg. Optim. (NICSO 2010). Springer, pp 65–74
Yang M-S, Chen C-H (1998) On the edited fuzzy K-nearest neighbor rule. IEEE Trans Syst Man, Cybern Part B 28:461–466
Yang X-S, Deb S (2009) Cuckoo search via Levy flights. In: Nat. Biol. Inspired Comput. 2009. NaBIC 2009. World Congr. pp 210–214
Yang X-S, Deb S (2010) Engineering optimisation by cuckoo search. Int J Math Model Numer Optim 1:330–343
Yang G, Zhang Y, Yang J et al (2016) Automated classification of brain images using wavelet-energy and biogeography-based optimization. Multimed Tools Appl 75:15601–15617
Yilmaz S, Kucuksille EU (2015) A new modification approach on bat algorithm for solving optimization problems. Appl Soft Comput 28:259–275
Zacharaki EI, Wang S, Chawla S, Soo D (2009) Classification of brain tumor type and grade using MRI texture and shape in a machine learning scheme. Magn Reson Med 62:1609–1618
Zhang Y, Wang S, Ji G, Dong Z (2013) An MR brain images classifier system via particle swarm optimization and kernel support vector machine. Sci World J 2013
Zhang Y-D, Jiang Y, Zhu W et al (2018) Exploring a smart pathological brain detection method on pseudo Zernike moment. Multimed Tools Appl 77:22589–22604
Zollner FG, Emblem KE, Schad LR (2012) SVM-based glioma grading: optimization by feature reduction analysis. J Med Phys 22:205–214
Funding
This research received no specific grant from any funding agency in the public, commercial, or not-for-profit sectors.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
‘None Declared’.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Kaur, T., Saini, B.S. & Gupta, S. An adaptive fuzzy K-nearest neighbor approach for MR brain tumor image classification using parameter free bat optimization algorithm. Multimed Tools Appl 78, 21853–21890 (2019). https://doi.org/10.1007/s11042-019-7498-3
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11042-019-7498-3