Abstract
Background
Recent investigations suggested that deregulated levels of Circular RNAs (circRNAs) could be associated with type 2 diabetes mellitus (T2DM) pathogenesis. Accordingly, this study aimed to determine the expression levels of circulating CircHIPK3, CDR1as and their correlation with biochemical parameters in patients with T2DM, pre-diabetes and control subjects.
Methods and results
The expression of circRNAs in peripheral blood was determined using QRT-PCR in 70 patients with T2DM, 60 pre-diabetes and in 69 age and sex matched healthy controls. Moreover, bioinformatics tools were applied to explore and predict the potential interactions between circRNAs and other non-coding RNAs (ncRNAs). Our analysis revealed that the expression level of CircHIPK3 was significantly elevated in T2DM patients compared to healthy participants (P < 0.001) and pre-diabetes subjects (P = 0.018). In addition, ROC analysis suggested that at the cutoff value of 0.24 and the sensitivity and specificity of 50% and 88.4%, respectively, CircHIPK3 could distinguish between T2DM patients and control subjects. Furthermore, it was observed that the expression level of CDR1as is higher in pre-diabetic individuals than healthy individuals (P = 0.004). Finally, Spearman correlation analysis showed that there was a significant correlation between CircHIPK3 and CDR1as expression levels and clinical and anthropometrical parameters such as BMI, systolic and diastolic blood pressure, HbA1c and fasting blood glucose (P < 0.005).
Conclusions
The data of this study provided evidence that the expression levels of CircHIPK3, CDR1as increased in T2DM and pre-diabetes subjects, respectively.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Diabetes mellitus (DM) is a major global health problem resulting from the interplay of genetic, environmental and epigenetic factors [1]. The data from the International Diabetes Federation (IDF) revealed that, nearly 463 million adults were living with diabetes in 2019 worldwide, and this number is expected to increase to 700 million worldwide by 2045 [2].
It has been demonstrated that the human genome can be transcribed widely into a large number of non-coding RNAs (ncRNAs), which are closely related to the incidence and development of human disorders such as type 2 diabetes mellitus (T2DM) [3]. One of the most recently discovered type of ncRNAs is circular RNAs (circRNAs) [4]. CircRNAs are single-stranded molecules deferentially generated by the non-sequential back-splicing of pre-mRNAs or lariat introns and exon-skipping events [5, 6]. Because of the lack of 5′ or 3′ ends, circRNA possesses marked stability in body fluids and they could act as an ideal class of blood-based biomarkers [7]. It has been reported that circRNAs play essential roles in a wide range of biological processes that are included: regulation of gene splicing/transcription, microRNA (miRNA) and protein sponging, and modulation of protein translation and protein function [8, 9].
Several lines of evidence suggested that deregulated levels of circRNAs could be involved in human diseases [10, 11]. In this regards, previous studies shown that circRNAs are involved in β-cell function and T2DM related complications [12]. Huanyu Xu et al. found that the over-expression of CDR1as could improve insulin secretion by sponging miR-7 [13]. CircHIPK3 found to regulate the expression of several key β-cell genes involved in insulin signaling pathway [14]. This circRNAs could also affect hyperglycemia and insulin resistance by sponging some miRNAs such as miR-192-5p and modulation of FOXO1 expression [15]. Furthermore, it has been reported that CircHIPK3 is highly abundant in diabetic neuropathic pain patients [16] and its silencing could alleviate retinal vascular disorders [16].
To date, only few studies have investigated the circulating level of CircHIPK3, and CDR1as in T2DM patients. Accordingly, in this study, we determined and compared the expression levels of CircHIPK3, and CDR1as in peripheral blood samples of T2DM patients, pre-diabetes and control subjects. Moreover, we evaluated the correlation of these circRNAs with biochemical and clinical parameters.
Material and methods
Study subjects
A total of 199 age and sex-matched subjects including 70 patients with T2DM, 60 pre-diabetes subjects, and 69 healthy volunteers were included in this case–control study. The control group was chosen among the healthy volunteers who had a fasting blood sugar (FBS) level of < 100 mg/dL or hemoglobin A1c (HbA1c) < 5.7% and no history of diabetes. Patients were diagnosed according to the Standards of the American Diabetes Association criteria for the diagnosis of diabetes. Accordingly, cases have either of the following criteria were diagnosed as having T2DM: FBS level of 126 mg/dL or higher; 2-h plasma glucose (2-h PG) level of 200 mg/dL or higher or HbA1c level of 6.5% or higher. Moreover, patients with FBS 100–125 mg/dL, 2-h PG 140–199 mg/dL or an HbA1c level 5.7–6.4% diagnosed as pre-diabetes. Subjects with liver or kidney dysfunction, inflammatory diseases, malignancies, autoimmune diseases and any endocrine diseases were excluded.
The study was approved by the Yasuj University of Medical Sciences Ethics Committee (IR.YUMS.REC.1397.153) and performed in compliance with the Helsinki declaration. Moreover, the informed consent was obtained from all subjects before enrolling in the study.
Anthropometrical and biochemical measurements
The demographic characteristics of the study population were collected by an interviewer-administered questionnaire and from medical records. Anthropometric parameters were measured by a nutritionist using standardized techniques. Estimation of body mass index (BMI) was done by dividing body weight in kilograms by height in square meters. Biochemical parameters including, FBS, HbA1c%, total cholesterol (TC), triglycerides (TG), high density lipoprotein cholesterol (HDL-c) and low density lipoprotein cholesterol (LDL-c) levels were measured using the routine laboratory methods. In addition, serum insulin levels were measured by enzyme-linked immune-sorbent assay. Insulin resistance was assessed by the use of homeostasis model assessment of insulin resistance (HOMA-IR) equation.
Total RNA extraction and cDNA synthesis
Whole blood samples were collected from participants after an overnight fasting in EDTA-containing tubes. Total RNA was isolated from the peripheral blood using a miRNeasy Mini kit (Qiagen, Valencia, CA, USA) according to the manufacturer’s instructions. The purity and concentration of the total RNA were evaluated using a NanoDrop Lite spectrophotometer (Termo Scientifc). Subsequently, total RNA digested with RNase R to remove linear RNAs and enrich circRNAs. RNA was reverse-transcribed into cDNA using Primescript RT reagent kit (TaKaRa, Japan) and primed with random primers.
Quantitative reverse transcription-polymerase chain reaction (qRT-PCR)
For quantification, real-time PCR analysis was performed using the Applied Biosystems StepOnePlus Real-Time PCR System and SYBR Premix Ex TaqTM II (Takara, Japan). qRT-PCR was done using forward primer 5′-TGGAGACTGGGGGAAGATGA-3′ and reverse primer 5′-CACACTAACTGGCTGAGGGG-3′ for CircHIPK3, and forward primer 5′-TCTGCTCGTCTTCCAACATC-3′ and reverse primer 5′-AGATCAGCACACTGGAGACG-3′ for CDR1as. All the melting curves contain single peaks, indicating specific PCR amplification. Moreover, the PCR product size was tested by agarose gel electrophoresis. Furthermore, PCR efficiency was evaluated by LinRegPCR software. Finally, the relative expression of circRNAs was calculated using the 2−ΔCt method and normalized using the GAPDH as the internal control.
Functional in-silico analysis
CircRNAs plays important regulatory roles in T2DM pathogenesis by interacting with other ncRNAs such as miRNAs (miRNA sponging). Moreover, they have several binding sites for RNA-binding proteins (RBPs). Therefore, to identify the miRNAs that bind to circRNAs, we performed a circRNA-miRNA interaction network prediction on circinteractome database [available on: https://circinteractome.nia.nih.gov/] [17]. In addition, the circRNA-RBP interaction was predicted with CircFunBase database (available on: http://bis.zju.edu.cn/CircFunBase/) [18]. Cytoscape (available on: http://www.cytoscape.org/) was applied to build a circRNA–miRNA–RBP interaction network.
Statistical analysis
All statistical analysis was performed by SPSS Statistical Software Package (version 20.0). The assessment of normality performed by the Kolmogorov–Smirnov test. The comparisons of the circRNA expression levels between groups calculated by one-way analysis of variance (ANOVA) for the normally distributed data or Kruskal–Wallis test for the nonparametric data. Pearson’s and Spearman’s correlation analysis carried out to determine the correlation between the expression levels of circRNAs and parametric and nonparametric variables, respectively. P value < 0.05 was considered to be statistically significant.
A receiver operating characteristic (ROC) curve provided by MedCalc software was used to assess the feasibility of using circRNAs as a diagnostic marker for the T2DM. The best sensitivity/specificity pair was selected based on the maximum Youden Index.
Results
Characteristics of the study population
The clinical and biochemical characteristics of study subjects (n = 199) are presented in Table 1. There was no significant difference between age (P = 0.562) and gender (P = 0.562) among the three studied groups. Our results showed that T2DM patients had significantly higher values for FBS (P = 0.000), HbA1c (P = 0.000), insulin (P = 0.011), HOMA-IR (P = 0.000), systolic blood pressure (P = 0.000), diastolic blood pressure (P = 0.015), BMI (P = 0.00), total cholesterol (P = 0.019) and LDL-c (P = 0.043) levels than other groups. We did not observe significant differences between groups for TG (P = 0.46), HDL-c (P = 0.33) levels.
Comparison of circulating circRNA levels between groups
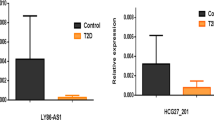
Our analysis revealed that the expression level of CircHIPK3 was significantly elevated in T2DM patients compared to healthy participants (P < 0.001). In addition, the circulating level of this circRNA was significantly increased in T2DM patients than that in pre-diabetes subjects (P = 0.0180 (Fig. 1A). For further analysis, the ROC curve was applied to evaluate the diagnostic values of CircHIPK3. ROC analysis showed that at the cutoff value of 0.24 and the sensitivity and specificity of 50% and 88.4%, respectively, CircHIPK3 was able to distinguish between T2DM patients and control subjects (AUC = 0.665, 95% CI 0.589–0.752, P = 0.001) (Fig. 2A). Moreover, CircHIPK3 could distinguish T2DM patients from the pre-diabetes by a sensitivity and specificity of 50% and 75% respectively at a cutoff of 0.25 (AUC = 0.613, 95% CI 0.524–0.697, P = 0.023) (Fig. 2B).
It was observed that the expression level of CDR1as is higher in the pre-diabetic individuals than healthy individuals (P = 0.0040 (Fig. 1B). We did not observe a significant difference for this circRNA between other groups. Furthermore, ROC analysis revealed that at a cutoff of 0.47, 86.9% sensitivity and 36.7% specificity (AUC = 0.605, 95% CI 0.516–0.690, P = 0.041), CDR1as could distinguish pre-diabetes patients from the healthy controls (Fig. 2C).
Correlation of circRNA levels with biochemical and anthropometrical parameters
Spearman correlation analysis was applied to determine the correlation of circRNAs expression levels and clinical parameters using 2−ΔCt values as input data. The results revealed that there was a significant positive correlation between the CircHIPK3 level with BMI (R = 0.216, P = 0.002), systolic blood pressure (R = 0.224, P = 0.001), FBS (R = 0.22, P = 0.002), HbA1c (R = 0.168, P = 0.018), and HOMA-IR (R = 0.154, P = 0.03) (Fig. 3A–E). In addition, a significant correlation was found between CDR1as and FBS (R = 0.180, P = 0.011) (Fig. 3F). We did not observe a significant correlation between the CircHIPK3 and CDR1as circulating levels with each other and another parameter (Table 2).
Prediction of circRNAs interaction with miRNAs and proteins
To identify the miRNAs that bind to circRNAs, we performed a circRNA-miRNA interaction network prediction on circinteractome database. Forty-two miRNA binding sites matching to CircHIPK3 (hsa_circ_0000284) were identified by Circinteractome (Fig. 4). Moreover, this database provided 24 miRNA binding site residues in CDR1as (hsa_circ_0001946) (Fig. 4). In addition, the circRNA-RBP interaction was predicted with CircFunBase database. Obtained data revealed a total of 13 and 8 RBPs sites matching to CircHIPK3 and CDR1as, respectively (Fig. 5).
Discussion
It has been reported that deregulated expression levels of circRNAs are involved in several human diseases. However, little is known about the role of circRNAs in the T2DM [14]. Accordingly, the present study compared the expression profile of CircHIPK3 and CDR1as in the peripheral blood samples of pre-diabetes, T2DM and control subjects. Our findings revealed that the expression level of CircHIPK3 elevated in T2DM patients compared to healthy participants and pre-diabetes subjects. There is no previous report on the evaluation of CircHIPK3 level in the peripheral blood samples of diabetic patients. However, the association of the CircHIPK3 expression level with diabetes complications evaluated in several studies. In this regard, it has been shown that CircHIPK3 level is elevated in patients with diabetic retinopathy [19], diabetic neuropathic pain [16], cardiovascular disease [20], carotid acute and myocardial infarction [21]. There is some reason related to the elevated expression level of circHIPK3 under diabetic conditions in vivo and in vitro. Shan et al. revealed that high-glucose treatment could upregulate CircHIPK3 expression in HRVECs cells. Moreover, they reported that diabetes mellitus–related pathological factors, such as oxidative stress and inflammatory stimulus, could significantly upregulate CircHIPK3 expression [19].
Multiple lines of evidence suggested that CircHIPK3 could be involved in the pathogenesis of T2DM by different mechanisms. It has been reported that CircHIPK3 impairs retinal vascular function in diabetes by sponging miR-30a-3p and regulation of the expression of its target gene including VEGFC, FZD4 and WNT2 [19]. Interestingly, Wang.L et al. shown that CircHIPK3 silencing reduced diabetic neuropathic pain by inhibiting neuro-inflammatory factors Including IL-1b, IL-6, IL-12, and TNF-α by interaction with miR-124 [16]. In addition, it has been revealed that silencing of CircHIPK3 could ameliorate the damage to renal function by acting as a sponge for miR-185 [22].
In this study, we observed a positive correlation between the CircHIPK3 expression with FBS, HbA1c, blood pressure and BMI. In agreement with these findings, Yi Sun et al. showed that BMI and CircHIPK3 expression levels are correlated in cardiovascular disease [11]. In addition, it has been reported that CircHIPK3 could promote hyperglycemia and insulin homeostasis by sponging with miR-192-5p and increasing FOXO1 [15]. Interestingly, a recent study suggested that miR-192-5p is a target for ATP1B1, which reduces renal tubular reabsorption via Na + / K + -ATPase and eventually reduce blood pressure [23].
In the current investigation, we reported that the expression of CDR1as in the peripheral blood of pre-diabetes was higher than control. To the best of our knowledge, this is the first report evaluating the expression levels of CDR1as in diabetic patients. However, there are some reports about the dysregulated levels of this circRNAs in human disease such as myocardial infarction and cardiovascular patients [20, 24]. In-vitro studies suggested that CDR1as could be involve in the development of T2DM by different mechanisms [25, 26]. Huanyu Xu et al. Reported that CDR1as may regulate insulin transcription and secretion in the pancreatic islets by interacting with miR-7 [13]. Moreover, we observed a positive correlation between expression of CDR1as with FBS. In line with this finding, Zhang et al. reported that the expression levels of ZFAS1 and CDR1as in whole blood samples of myocardial infarction patients were positively correlated with FBS [24].
Experimental analysis reported that circRNAs could function as a “molecular sponge” for their related miRNAs and thereby modulates their inhibitory effects on the expression of target genes [12]. Therefore, to further investigation, we performed a circRNA-miRNA interaction network prediction to identify the miRNAs that bind to circRNAs. Annotation and function prediction revealed that CircHIPK3 could interact with a total of 42 miRNAs. It has been reported that some of these could be involve in T2DM pathogenesis. For example, hsa-miR-1179, hsa-mir-149, hsa-mir-382 and hsa-mir-377 are involved in proliferative diabetic retinopathy [27], impaired insulin synthesis and secretion in beta cells [28], diabetic nephropathy [29, 30] respectively. Moreover, we found that a total of 24 miRNAs have an interaction with CDR1as. Among them, hsa-miR-21 plays an important role in the pathogenesis of diabetes and its complications [31, 32].
CircRNAs also could participate in sponging of RBPs and thereby affect the fate of their target mRNAs and protein [12]. Our results revealed that CircHIPK3 and CDR1as could interact with several common RBPs including AGO1, AGO2, IGF2BP2 and FUS. It has been reported that AGO1 involves in the pathways of angiogenesis, adipose tissue function, and insulin sensitivity [33]. Furthermore, hepatic AGO2 could regulate energy expenditure during obesity and its inactivation protects against obesity-related glucose intolerance in mice [34]. In addition, it has been shown that the IGF2BP2 genotypes is associated with impaired insulin levels, beta cell function, and insulin resistance, and increases the risk of T2DM. It is also involved in adipogenesis and pancreatic development [35, 36].
Conclusion
In the current study, we determined and compared the expression levels of CircHIPK3, and CDR1as in peripheral blood samples of T2DM patients, pre-diabetes and control subjects. Our analysis revealed that the expression level of CircHIPK3 elevated in T2DM patients compared to healthy participants and pre-diabetes subjects. In addition, it was observed that the expression level of CDR1as is higher in pre-diabetic individuals than healthy individuals. The data of this study provided evidence that the expression of CircHIPK3, CDR1as increased in T2DM and pre-diabetes subjects, respectively. However, further in vitro and in vivo investigations are needed to confirm our results.
References
Tremblay J, Hamet P (2019) Environmental and genetic contributions to diabetes. Metabolism 100:153952
Saeedi P, Salpea P, Karuranga S, Petersohn I, Malanda B, Gregg EW et al (2020) Mortality attributable to diabetes in 20–79 years old adults, 2019 estimates: results from the International Diabetes Federation Diabetes Atlas. Diabetes Res Clin Pract 162:108086
Maass PG, Luft FC, Bähring S (2014) Long non-coding RNA in health and disease. J Mol Med 92(4):337–346
Cheng Y, Zhao P, Wu S, Yang T, Chen Y, Zhang X et al (2018) Cisplatin and curcumin co-loaded nano-liposomes for the treatment of hepatocellular carcinoma. Int J Pharm 545(1–2):261–273
Chen L-L (2016) The biogenesis and emerging roles of circular RNAs. Nat Rev Mol Cell Biol 17(4):205
Awan AR, Manfredo A, Pleiss JA (2013) Lariat sequencing in a unicellular yeast identifies regulated alternative splicing of exons that are evolutionarily conserved with humans. Proc Natl Acad Sci 110(31):12762–12767
Sanger HL, Klotz G, Riesner D, Gross HJ, Kleinschmidt AK (1976) Viroids are single-stranded covalently closed circular RNA molecules existing as highly base-paired rod-like structures. Proc Natl Acad Sci 73(11):3852–3856
Li Z, Huang C, Bao C, Chen L, Lin M, Wang X et al (2015) Exon-intron circular RNAs regulate transcription in the nucleus. Nat Struct Mol Biol 22(3):256
Hentze MW, Preiss T (2013) Circular RNAs: splicing’s enigma variations. EMBO J 32(7):923–925
Haque S, Harries LW (2017) Circular RNAs (circRNAs) in health and disease. Genes 8(12):353
Ghosal S, Das S, Sen R, Basak P, Chakrabarti J (2013) Circ2Traits: a comprehensive database for circular RNA potentially associated with disease and traits. Front Genet 4:283
Ghasemi H, Sabati Z, Ghaedi H, Salehi Z, Alipoor B (2019) Circular RNAs in β-cell function and type 2 diabetes-related complications: a potential diagnostic and therapeutic approach. Mol Biol Rep 46:5631–5643
Xu H, Guo S, Li W, Yu P (2015) The circular RNA Cdr1as, via miR-7 and its targets, regulates insulin transcription and secretion in islet cells. Sci Rep 5(1):1–12
Stoll L, Sobel J, Rodriguez-Trejo A, Guay C, Lee K, Venø MT et al (2018) Circular RNAs as novel regulators of β-cell functions in normal and disease conditions. Mol Metab 9:69–83
Cai H, Jiang Z, Yang X, Lin J, Cai Q, Li X (2020) Circular RNA HIPK3 contributes to hyperglycemia and insulin homeostasis by sponging miR-192-5p and upregulating transcription factor forkhead box O1. Endocr J. https://doi.org/10.1507/endocrj.EJ19-0271
Wang L, Luo T, Bao Z, Li Y, Bu W (2018) Intrathecal circHIPK3 shRNA alleviates neuropathic pain in diabetic rats. Biochem Biophys Res Commun 505(3):644–650
Dudekula DB, Panda AC, Grammatikakis I, De S, Abdelmohsen K, Gorospe M (2016) CircInteractome: a web tool for exploring circular RNAs and their interacting proteins and microRNAs. RNA Biol 13(1):34–42
Meng X, Hu D, Zhang P, Chen Q, Chen M (2019) CircFunBase: a database for functional circular RNAs. Database. https://doi.org/10.1093/database/baz003
Shan K, Liu C, Liu B-H, Chen X, Dong R, Liu X et al (2017) Circular noncoding RNA HIPK3 mediates retinal vascular dysfunction in diabetes mellitus. Circulation 136(17):1629–1642
Sun Y, Chen R, Lin S, Xie X, Ye H, Zheng F et al (2019) Association of circular RNAs and environmental risk factors with coronary heart disease. BMC Cardiovasc Disord 19(1):223
Bazan HA, Hatfield SA, Brug A, Brooks AJ, Lightell DJ Jr, Woods TC (2017) Carotid plaque rupture is accompanied by an increase in the ratio of serum circR-284 to miR-221 levels. Circ: Cardiovasc Genet 10(4):e001720
Liu R, Zhang M, Ge Y (2020) Circular RNA HIPK3 exacerbates diabetic nephropathy and promotes proliferation by sponging miR-185. Gene 765:145065
Baker MA, Wang F, Liu Y, Kriegel AJ, Geurts AM, Usa K et al (2019) MiR-192-5p in the kidney protects against the development of hypertension. Hypertension 73(2):399–406
Zhang Y, Sun L, Xuan L, Pan Z, Li K, Liu S et al (2016) Reciprocal changes of circulating long non-coding RNAs ZFAS1 and CDR1AS predict acute myocardial infarction. Sci Rep 6:22384
Li P, Yang X, Yuan W, Yang C, Zhang X, Han J et al (2018) CircRNA-Cdr1as exerts anti-oncogenic functions in bladder cancer by sponging MicroRNA-135a. Cell Physiol Biochem 46(4):1606–1616
Hansen TB, Jensen TI, Clausen BH, Bramsen JB, Finsen B, Damgaard CK et al (2013) Natural RNA circles function as efficient microRNA sponges. Nature 495(7441):384–388
Qing S, Yuan S, Yun C, Hui H, Mao P, Wen F et al (2014) Serum miRNA biomarkers serve as a fingerprint for proliferative diabetic retinopathy. Cell Physiol Biochem 34(5):1733–1740
Sun Q, Yang Q, Xu H, Xue J, Chen C, Yang X et al (2019) miR-149 negative regulation of mafA is involved in the arsenite-induced dysfunction of insulin synthesis and secretion in pancreatic beta cells. Toxicol Sci 167(1):116–125
Wang S, Wen X, Han XR, Wang YJ, Shen M, Fan SH et al (2018) Repression of micro RNA-382 inhibits glomerular mesangial cell proliferation and extracellular matrix accumulation via FoxO1 in mice with diabetic nephropathy. Cell Prolif 51(5):e12462
Chen G, Lu L, Liu C, Shan L, Yuan D (2015) MicroRNA-377 suppresses cell proliferation and invasion by inhibiting TIAM1 expression in hepatocellular carcinoma. PLoS ONE 10(3):e0117714
Sekar D, Venugopal B, Sekar P, Ramalingam K (2016) Role of microRNA 21 in diabetes and associated/related diseases. Gene 582(1):14–18
Mazzeo A, Beltramo E, Lopatina T, Gai C, Trento M, Porta M (2018) Molecular and functional characterization of circulating extracellular vesicles from diabetic patients with and without retinopathy and healthy subjects. Exp Eye Res 176:69–77
Tang X, Miao Y, Luo Y, Sriram K, Qi Z, Lin F-M et al (2020) Suppression of endothelial AGO1 promotes adipose tissue browning and improves metabolic dysfunction. Circulation 142:365–379
Zhang C, Seo J, Murakami K, Salem ES, Bernhard E, Borra VJ et al (2018) Hepatic Ago2-mediated RNA silencing controls energy metabolism linked to AMPK activation and obesity-associated pathophysiology. Nat Commun 9(1):1–15
Huang Q, Yin J-Y, Dai X-P, Pei Q, Dong M, Zhou Z-G et al (2010) IGF2BP2 variations influence repaglinide response and risk of type 2 diabetes in Chinese population. Acta Pharmacol Sin 31(6):709–717
Chistiakov DA, Nikitin AG, Smetanina SA, Bel’chikova LN, Suplotova LA, Shestakova MV et al (2012) The rs11705701 G> A polymorphism of IGF2BP2 is associated with IGF2BP2 mRNA and protein levels in the visceral adipose tissue-a link to type 2 diabetes susceptibility. Rev Diabetic Stud: RDS 9(2–3):112
Funding
This work was financially supported by a Grant (960416) from the Deputy of Research, Yasuj University of Medical Sciences.
Author information
Authors and Affiliations
Contributions
BA planned the studies; BA and AM conducted analysis and interpretation of all experiments; FR and BK conducted all experiments. BA and GS wrote the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflicts of interest.
Ethical approval
The study was approved by the Yasuj University of Medical Sciences Ethics Committee (IR.YUMS.REC.1397.153) and performed in compliance with the 1964 Helsinki declaration. Moreover, the informed consent was obtained from all subjects before enrolling in the study.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Rezaeinejad, F., Mirzaei, A., Khalvati, B. et al. Circulating expression levels of CircHIPK3 and CDR1as circular-RNAs in type 2 diabetes patients. Mol Biol Rep 49, 131–138 (2022). https://doi.org/10.1007/s11033-021-06850-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-021-06850-8