Abstract
Seri/Babax spring wheat RIL population was developed to minimize the confounding effect of phenology in the genetic dissection of abiotic stress traits. An existing linkage map (< 500 markers) was updated with 6470 polymorphic Illumina iSelect 90K array and DArTseq SNPs to a genetic map of 5576.5 cM with 1748 non-redundant markers (1165 90K SNPs, 207 DArTseq SNPs, 183 AFLP, 111 DArT array, and 82 SSR) assigned to 31 linkage groups. We conducted QTL mapping for yield and related traits phenotyped in several major wheat growing areas in Egypt, Sudan, Iran, India, and Mexico (nine environments: heat, drought, heat plus drought, and yield potential). QTL analysis identified 39 (LOD 2.5–23.6; PVE 4.8–21.3%), 36 (LOD 2.5–15.4; PVE 2.9–21.4%), 30 (LOD 2.5–13.1; PVE 3.6–26.8%), 39 (LOD 2.7–14.4; PVE 2.6–15.9%), and 22 (LOD 2.8–4.8; PVE 6.8–12.9%) QTLs for grain yield, thousand-grain weight, grain number, days to heading, and plant height, respectively. The present study confirmed QTLs from previous studies and identified novel QTLs. QTL analysis based on high-yielding and low-yielding environmental clusters identified 11 QTLs (LOD 2.6–14.9; PVE 2.7–19.7%). The updated map thereby provides a better genome coverage (3.5-fold) especially on the D genome (4-fold), higher density (1.1-fold), and a good collinearity with the IWGSC RefSeq v1.0 genome, and increased the number of detected QTLs (5-fold) compared with the earlier map. This map serves as a useful genomic resource for genetic analyses of important traits on this wheat population that was widely distributed around the world.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Bread wheat (Triticum aestivum L.) is one of the staple food crops worldwide, contributing ~ 28% of the global grain production and 20% of the calories and protein consumed by human (FAOstat 2018). With a predicted world population of nine billion by 2050, the demand for wheat is expected to increase (Rosegrant and Cline 2003). As the most widely cultivated crop geographically, wheat covers a large variance of climate and soil conditions. Abiotic stresses, such as drought, heat, salinity, and nutrient imbalances reduce wheat yield in many environments (Trethowan and Mujeeb-Kazi 2008). Improvement of grain yield in these target environments is the primary target of wheat research considering climate change (Reynolds et al. 2009).
Grain yield (GY) is a complex quantitative trait that is strongly influenced by interacting genetic and environmental factors (Quarrie et al. 2006). GY can be dissected into the following direct components: thousand-grain weight (TGW) and grain number (GN) per unit area of land. Both TGW and GN are quantitatively inherited and not easy to improve simultaneously through the conventional approaches due to the tradeoff between them (Yano and Sasaki 1997; Griffiths et al. 2015). Quantitative trait loci (QTL) mapping in bi-parental populations has provided an effective approach to dissect quantitative traits into component loci to study their relative effects on a specific trait of interest (Doerge 2002). A large number of QTLs for yield and yield components have been identified to date on almost all wheat chromosomes (Bennett et al. 2012; Golabadi et al. 2011; Kirigwi et al. 2007; Liu et al. 2019b; Maccaferri et al. 2008; Pinto et al. 2010; Tahmasebi et al. 2016). However, the identification of QTL for yield may be confounded by flowering time and plant height (PH), whereby genes of major effects have been shown to mask the identification of minor effects, especially under abiotic stress where the onset of stress treatments will occur at different growth stages depending on the genotypes’ phenological pattern (Reynolds et al. 2009). Therefore, effects of phenology and PH may be restricted in the population to identify QTLs for major traits without the confounding effect of phenology (Lopes et al. 2013; Pinto et al. 2010; Reynolds and Tuberosa 2008).
The narrow range of PH and flowering time of the Seri/Babax recombinant inbred line (RIL) population makes it ideal for genetic and physiological studies. Many QTLs have been identified for agronomic and physiological traits in the Seri/Babax population under water-limiting and heat stress conditions over the past decade (Lopes et al. 2013; Mathews et al. 2008; McIntyre et al. 2010; Pinto et al. 2010, 2016; Tahmasebi et al. 2016). Seri/Babax population is still widely used by researchers around the world for genetic and physiological studies of different traits in various environments, and some of the lines have been valuable in breeding. Genetic linkage map is the foundation for mapping QTLs and subsequent marker-assisted selection or map-based cloning. The existing genetic map of Seri/Babax was primarily based on 475 markers comprising Simple Sequence Repeat (SSR), Amplified Fragment Length Polymorphism (AFLP), and Diversity Arrays Technology (DArT) markers (Lopes et al. 2013). With the rapid development of new genotyping technologies, numerous molecular makers for wheat are available now. Therefore, a new linkage map is necessary for researchers to compare their results with peers. By genotyping the Seri/Babax population with the wheat 90K Illumina iSelect array and DArTseq platform, the objectives of the study were (1) to update the genetic linkage map of the Seri/Babax population by integrating 90K and DArTseq markers to the existing linkage map; (2) to identify QTLs for yield and yield-related traits with phenotypic data from multiple environments, combining data not used earlier and from earlier published data; and (3) to compare the properties of the updated genetic map with the existing linkage map.
Materials and methods
Wheat population and phenotyping
The Seri/Babax population comprising 156 RILs was derived from a reciprocal cross between two elite spring bread wheat, Seri M82 (a released line from the “Veery” cross, KVZ/BUHO//KAL/BB) and Babax (a line derived from the cross “Babax,” BOW/NAC//VEE/3/BJY/COC) (Olivares-Villegas et al. 2007). Seri M82 carries the 1BL.1RS translocation and was characterized by moderate tolerance to drought conditions and high yield potential. Babax is without the 1BL.1RS translocation and is highly tolerant to severe drought (Mathews et al. 2008). Seri M82 and Babax both have the same photoperiod-insensitive allele at Ppd-D1 and spring-type alleles for vernalization (Vrn-B1 and Vrn-D1) loci (Pinto et al. 2010). Hence, this population is characterized by its narrow range of plant height and flowering time (ca. 17 cm, 10–15 days across environments as reported by Lopes et al. (2013)), and is ideal for genetic mapping and physiological studies. The phenotyping was conducted in eight major wheat growing regions from five countries around the world (Fig. 1a), including ten environments (Darab in Iran, Sohag in Egypt, Dongola and Wad Medani in Sudan, Ludhiana and Karnal in India, and yield potential, drought stress, heat stress, and heat plus drought stress in Obregon, Mexico in 2008–2009) reported in an earlier study (Lopes et al. 2013) and six new environments (one from Shandaweel, Egypt in 2008–2009, and five from different stresses in Obregon, Mexico in 2004–2005 and 2005–2006). The sowing and harvest date, temperature, precipitation, total amount of water applied by irrigation, relative humidity, and evapotranspiration during the growing season were reported in an earlier study (Lopes et al. 2013). In Mexico, the experiments were conducted at the Campo Experimental Norman E. Borlaug (CENEB), Cd. Obregon, where the environments were designated as yield potential, drought stress, heat stress, and heat plus drought stress. Yield potential (YP) was applied by normal sowing (late November) with optimum irrigation (total water supply > 700 mm); drought stress (D) was applied by normal planting (late November) with significantly reduced irrigation (total water supply < 300 mm); heat stress (H) was applied by late sowing (late February) with supplementary irrigation (total water supply > 700 mm) to avoid the effect of drought; and the combined heat plus drought stress (HD) was applied by delayed planting date (late February) with reduced irrigation (total water supply < 400 mm). By late sowing, the average maximum temperature during wheat growing cycle was above 32 °C, almost 8 °C higher than that in normal sowing cycles (Liu et al. 2019b; Pinto et al. 2010), a typical method for applying heat stress in Obregon that has been demonstrated to be successful in generating germplasm for heat stress environments, such as ME5 (Reynolds et al. 1994). The experimental design was randomized lattice with two replications. Seeds were sown on raised beds of 2 m length with 2 rows (25 cm between rows) and 80 cm between the beds, with seed rates of 120 kg/ha. The following five agronomic traits were measured: grain yield (GY; t ha−1), thousand-grain weight (TGW, g), grain number (GN; estimated as GY (t ha−1)/TGW (g) × 100 × 1000), days to heading (DTH, i.e., the number of days from emergence when 50% of the spike were emerged), and plant height (PH, cm from the soil surface to the tip of the spike without awns).
Phenotypic data analysis
The adjusted means for each genotype in individual environment were calculated using the MIXED procedure in SAS as described by Lopes et al. (2013). Cluster of environments based on yield performance was generated using the “cluster” package in R (Maechler 2019).
For the individual environment, the broad sense heritability (H2) was estimated as:
where \( {\sigma}_{\mathrm{g}}^2 \) and \( {\sigma}_{\mathrm{e}}^2 \) were genotype and error variance and r was the number of replications.
For the combined analysis of high-yielding and low-yielding clusters, H2 was estimated as:
where \( {\sigma}_{\mathrm{ge}}^2 \) was the genotype by environment interaction variance and l was the number of environment.
Genotyping, map construction, and QTL mapping
A genetic map with 475 markers (120 SSR, 211 AFLP, and 144 DArT) distributed over 29 linkage groups (LGs) had been previously constructed for this population (Lopes et al. 2013). Recently, the Seri/Babax RIL population was genotyped with the wheat 90K Illumina iSelect array and DArTseq platform (data available at https://data.cimmyt.org/dataset.xhtml?persistentId=hdl:11529/10548196). After quality control (removing markers with minor allele frequency < 5%, missing data > 15%, and segregation distortion p < 0.05), additional 5386 90K SNP and 609 DArTseq SNP markers were added to construct the linkage map. A new genetic map was constructed using QTL IciMapping v4.1 (Meng et al. 2015), which combined the map with the previous 475 markers and the new SNP markers. The 90K and DArTseq markers were assigned to chromosomes based on the IWGSC RefSeq v1.0 genome. Within each chromosome, genetic distance was determined using the Kosambi mapping function (Kosambi 2016). Chromosomes were split into LGs and reordered when the gap between two adjacent markers was > 50 cM.
QTL mapping was performed using QTL IciMapping v4.1 software (Meng et al. 2015). Inclusive composite interval mapping of additive (ICIM-ADD) function was selected as mapping method to detect additive QTL. The LOD threshold for QTL detection was determined by using 1000 permutations at p < 0.05. Putative QTLs were declared using a LOD of 2.5. QTLs detected across environments were considered the same if the distance between the peak positions was less than 10 cM. Stable QTLs were declared when QTLs were identified in at least two environments.
Results
Phenotypic variations across environments
The phenotypic traits of the Seri/Babax RIL population showed a wide range of variation across environments (Supplementary Table 1). The highest mean GY was found in Sohag (7.5 t ha−1) and Shandaweel (7.5 t ha−1, H2 = 0.45) in Egypt, while the lowest mean GY was found in Obregon under heat stress in 2006 (1.1 t ha−1, H2 = 0.79; Table 1). Cluster analysis of the 16 environments for GY generated the following two main groups: The high-yielding group (> 5.5 t ha−1, H2 = 0.35) comprising Sohag and Shandaweel in Egypt, Ludhiana in India, Dongola in Sudan, and two yield potential environments in Obregon, Mexico. The low-yielding group (< 4.5 t ha−1, H2 = 0.37) encompassed Karnal in India, Darab in Iran, Wad Medani in Sudan, and all drought, heat, and heat plus drought environments in Obregon, Mexico (Fig. 1b). The correlation analysis for GY was consistent with the results of cluster analysis, with Sohag and Shandaweel in Egypt showing the highest correlation. Most of the environments showed positive correlations with each other, while Wad Medani was negatively correlated with Obregon drought stress in 2004–2005 and 2005–2006 growing seasons (Fig. 1c).
Similar to GY, the highest average TGW (45.4 g, H2 = 0.54) was observed in Sohag and Shandaweel in Egypt, while the lowest TGW (26.6 g, H2 = 0.78) was found in Obregon heat plus drought stress environment (Table 1). The highest GN (18,558, H2 = 0.88) was found in Ludhiana, India, with a large range, and the lowest GN (3405, H2 = 0.48) was observed in Obregon drought stress in 2005–2006. The phenology range was 6 to 18 days across environments. In Darab, Iran, the Seri/Babax population had the longest time—112 days—for days to heading, while, in Obregon heat plus drought environment, the mean day to heading was 53 days. The Seri/Babax population had the highest PH in Sohag and Shandaweel (107.5 cm) and lowest mean PH (71 cm) for the population in Obregon heat plus drought condition. The largest and the smallest range for PH were observed in Wad Medani (a range of 40 cm) and Obregon heat stress in 2009 (a range of 15.5 cm), respectively.
Trait associations across environments
Across the 16 environments, GY was significantly positively correlated with TGW (r range 0.29–0.77), GN (0.48–0.94), DTH (0.32–0.69), and PH (0.24–0.73) in most of the environments with a few exceptions: GY was weakly negatively correlated with TGW under Obregon heat stress (2005, 2006; r = − 0.03), with GN under Obregon drought stress (2005–2006; r = − 0.03), and with DTH in Shandaweel of Egypt (r = − 0.13; Supplementary Fig. 1). TGW and GN were positively correlated with each other in 10 environments (r range 0.19–0.55) except in Shandaweel of Egypt (r = − 0.63), Obregon yield potential (2008–2009; r = − 0.62), drought (2005–2006; r = − 0.36), and heat (2005, 2006; r ranges from − 0.32 to − 0.46) conditions, where negative correlations were observed. TGW was positively correlated with DTH (r range 0.87–0.96) except in Shandaweel of Egypt (r = − 0.33). TGW was positively correlated with PH (r range 0.90–0.97) except in Darab, Iran. GN was positively correlated with DTH (r range 0.14–0.61) and PH (r range 0.90–0.97) in all environments.
Genetic linkage map construction
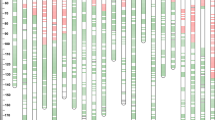
We used 6470 markers comprising SSR (120), AFLP (211), DArT (144), 90K SNP (5386), and DArTseq SNP (609) to construct the genetic map. Discarding redundant markers at the same loci, the final genetic map was 5576.5 cM with 1748 markers (82 SSR, 183 AFLP, 111 DArT, 1165 90K SNPs, and 207 DArTseq SNPs) arranged to 31 LGs (Fig. 2, Supplementary Table 2). The total length of the updated map increased 3.5-fold than that of the previous one (Lopes et al. 2013). The A, B, and D genomes harbored 768 (44%), 648 (37%), and 332 (19%) markers with a total length of 1959.5, 1961.9, and 1655.1 cM, respectively. The current map provided a better coverage of all the 21 chromosomes than the previous one, in which only 11% of the markers were distributed on D genome and no markers were assigned to 3D. On the updated map, the longest LG was 4A (352.5 cM), followed by 6A (351.6 cM) and 1D (350.6 cM). The shortest LG was 7Dc (4.7 cM), followed by 3Ab (7.2 cM). Marker density was lowest in 3D (16.2 cM per marker), followed by 1Ba (14.1 cM per marker), and highest in 7Dc (0.9 cM per marker), followed by 1A (1.6 cM per marker, Table 2). We obtained the physical position of 1059 mapped markers on the linkage map and projected our linkage map to the IWGSC RefSeq v1.0 genome. Results showed that there was a good collinearity between the updated genetic map and the physical map (Fig. 3).
Of these 1748 mapped markers, significant segregation distortion (CHITEST, p < 0.001) was found in 68 (4%) markers, including 28 DArT, 25 AFLP, 9 SSR, and 6 DArTseq markers. Fifty-eight and ten markers showed distortion with alleles biased towards parent Babax and Seri, respectively. The markers that showed segregation distortion towards the Babax parent were mainly distributed on 1Bb (35.6–104.9 cM, 43 markers), while those towards the Seri parent were mainly on 3Aa (142–153 cM, 8 markers).
Clusters of markers from the same marker type on the map were frequently observed (Fig. 2). Of the 1748 mapped markers, 1270 (73%) were adjoined with markers from the same type. Marker intervals with a distance < 0.5 cM between two adjacent markers from the same type were observed in 326, 26, 6, 4, and 1 intervals for 90K, DArTseq, AFLP, DArT, and SSR markers, accounting for 34, 29, 8, 15, and 7% of their total intervals, respectively (Supplementary Fig. 2). The frequency of clustering was primarily related to the number of markers from different types.
We also built a linkage map only using the 90K SNPs to a map length of 5333 cM with 1222 non-redundant 90K markers, while there were many large gaps on the map with 90K SNPs (Supplementary Table 3), indicating the necessity to include different types of markers. As was shown in Fig. 2, DArTseq, DArT, SSR, and AFLP markers could cover most of the large gaps.
QTLs identified in individual environments
A total of 166 QTLs with LOD scores from 2.5 to 23.7 were identified for GY, TGW, GN, DTH, and PH in the 16 environments with the updated genetic map (Supplementary Table 4). The number of QTLs was higher than the earlier map where 31 QTLs were detected for the same traits (Table 2). The largest number of QTLs was identified on chromosome 4A (23), followed by 7Da (22). No QTL was identified on 1Ba, 2Da, 3Ab, 3Ba, 5Db, 7Bb, 7Db, and 7Dc (Supplementary Fig. 3).
QTLs for GY were identified on chromosomes 1A, 1Bb, 1D, 2Db, 3Aa, 3Bb, 4A, 4B, 5A, 5Da, 6A, 6B, 6Da, 7A, and 7Da (LOD 2.6–23.6; PVE 4.8–21.3%; Supplementary Table 4, Supplementary Fig. 3). Seven stable QTLs on chromosomes 3Aa (128–134 cM), 3Bb (228–232 cM), 4A (268 cM, 280–286 cM, 304–308 cM), 6B (206–208 cM), and 7Da (124 cM) were detected in at least two environments (Table 3). The stable QTL on chromosome 4A (304–308 cM, Fig. 4) was detected across the yield potential (2005–2006, 2008–2009), drought (2008–2009), and heat plus drought (2009) conditions in Obregon, Mexico. QTLs for TGW were identified on chromosomes 1A, 1D, 2B, 3Bb, 4A, 4B, 5A, 5B, 5Da, 6A, 6B, 7A, 7Ba, and 7Da (LOD 3–15.4; PVE 3–21.4%). Six stable QTLs on chromosomes 2B (44–46 cM), 3Bb (44–48 cM), 6A (218 cM), 7Ba (26–32 cM), and 7 Da (108–116 cM, 124 cM) were identified (Table 3). The QTL on chromosome 7Da (108–116 cM) was detected in six environments across Karnal in India, Darab in Iran, and Obregon yield potential (2005–2006, 2008–2009) and drought (2005–2006, 2008–2009) conditions.
QTLs for GN were identified on chromosomes 1A, 1Bb, 1D, 2A, 2B, 3Aa, 3Bb, 4A, 4B, 4Db, 5A, 6B, 6Db, 7A, and 7Da (LOD 2.5–13.1; PVE 3.6–26.9%). Three stable QTLs were identified on chromosomes 1Bb (72–80 cM), 4A (308 cM, Fig. 4), and 6B (214 cM) in Obregon, Mexico (Table 3), of which the QTL on 6B (214 cM) was a heat adaptive QTL. QTLs for DTH were located on chromosomes 1Bb, 1D, 2A, 2B, 3D, 4B, 4Da, 5A, 5Da, 6B, 6Db, 7A, 7Ba, and 7Da (LOD 2.7–14.4; PVE 2.6–15.9%). Eight stable QTLs were identified on chromosomes 2A (158–160 cM), 4B (70 cM, 186 cM), 5Da (38 cM), 6B (206 cM), 7Ba (20–22 cM), and 7Da (110–112 cM, 120–124 cM) across environments. QTLs for PH were identified on chromosomes 1D, 2B, 3Aa, 4A, 4B, 4Db, 5A, 6A, 7Ba, and 7Da (LOD 2.8–4.8; PVE 6.9–12.9%). Four stable QTLs on chromosomes 2B (84 cM), 3Aa (166 cM), 4A (268–272 cM), and 4B (192–194 cM) were identified across environments (Table 3).
QTLs identified for high-yielding and low-yielding clusters
A QTL analysis based on the high- and low-yielding clusters identified 32 QTLs (LOD range 2.6–14.9, PVE 2.7–19.7%) for GY, TGW, GN, DTH, and PH, of which 11 QTLs were new and absent in individual environments (Supplementary Table 5). Six QTLs on chromosomes 1A, 1Bb, 2B, 4A, and 4Db were specific for high-yielding cluster; 16 QTLs on chromosomes 1A, 1Bb, 4A, 4B, 4D, 6B, 7A, 7Ba, and 7Da were specific for low-yielding cluster; and five QTLs on chromosomes 1D, 3Aa, 7Ba, and 7Da were common between the two clusters (Fig. 5).
Venn diagram illustrating the common and specific QTLs for high-yielding and low-yielding environment clusters of the Seri/Babax RIL population. Subscript numbers indicate the centi morgan (cM) position on the linkage map, followed by traits. GY, grain yield (t ha−1); TGW, thousand-grain weight (g); GN, grain number; DTH, days to heading (d); PH, plant height (cm)
QTLs for GY were found on chromosomes 1Bb (28 cM, new) and 4A (26 cM) in high-yielding cluster; on 4A (4 cM, 282 cM) and 4B (186 cM) in low-yielding cluster; and the common QTL was located on 1D (26 cM, new). For TGW, the QTLs for low-yielding cluster were identified on chromosomes 1A (242 cM) and 7Ba (26 cM), and the common QTL was identified on 7Da (114–116 cM). For GN, QTLs were located on chromosomes 1A (86 cM, new) and 4A (2 cM, new) in high-yielding cluster and on 1Bb (74 cM), 4A (158 cM, new; 268 cM), 4Db (90 cM), 6B (218 cM), and 7A (60 cM, new) in low-yielding cluster. For DTH, common QTLs were identified on 1D (30–32 cM) and 7Ba (20 cM); high-yielding cluster specific QTL was located on chromosome 4Db (102 cM, new); and low-yielding cluster specific QTLs were located on chromosomes 4B (70 cM), 6B (206 cM), 7A (264 cM), 7Ba (20 cM), and 7Da (114 cM). For PH, QTL in high and low yield cluster was identified on chromosomes 2B (64 cM, new) and 7Ba (130 cM), respectively. The common QTL for PH in both clusters was located on chromosome 3Aa (122 cM, new).
Pleiotropic QTL
Pleiotropic QTLs were observed in this study (Fig. 6). Among the 21 QTL hotspots for multiple traits, nine were independent from DTH and/or PH QTL. Co-location of QTL for GY, TGW, and GN was on chromosome 1D at 194–198 cM, where the favorable allele for GY and TGW was contributed by Babax, while GN was from Seri. Co-locations of QTLs for GY and TGW were observed on chromosomes 3Bb (228–236 cM) where the favorable allele was contributed by Babax and on chromosome 5A (26 cM) the favorable allele was contributed by Seri. Co-location of QTLs for GY and GN was on chromosome 1A (120 cM) where the favorable allele was from Seri and on chromosomes 1Bb (68–92 cM), 4A (56–62 cM, 280–286 cM, 304–308 cM), and 4B (32 cM) favorable alleles were contributed by Babax.
Venn diagram illustrating the common genomic regions for grain yield (GY, t ha−1), thousand-grain weight (TGW, g), grain number (GN), days to heading (DTH, d) and plant height (PH, cm). Subscript numbers indicate the centi morgan (cM) position on the linkage map. For the GY, TGW, and GN QTLs independent of DTH and/or PH, the red up arrows indicate the favorable allele contributed by the same parent and the black down arrows indicate the favorable allele contributed by different parents
Discussion
The rapid advances in genotyping technologies exponentially increase the number of markers available for genetic studies. Genetic maps are very useful tools in the identification of molecular markers closely linked to QTLs of interest, isolation of genes via map-based cloning, comparative mapping, and genome organization studies (Varshney et al. 2007). In the present study, with the addition of the 90K SNP and DArTseq SNP markers, the number of mapped markers (1748) on the genetic map of Seri/Babax increased 3.5-fold compared with the previous map of 475 markers (Lopes et al. 2013). The final size of the linkage map was 5576.5 cM, almost 3.5-fold the length of the previous one (Lopes et al. 2013). The current map provides a better coverage of the 21 chromosomes of wheat than the previous one where linkage group 3D was absent (Lopes et al. 2013; McIntyre et al. 2010). Around 19% of the markers were assigned to D genome, which was higher than the earlier map with 11% of the markers. Though the updated map showed a better genome coverage, gaps with a distance > 50 cM still remained on chromosomes 1B, 2D, 3A, 3B, 4D, 5D, 6D, 7B, and 7D, where small LGs were generated. Gaps of distances > 20 cM were observed on almost all LGs and need to be saturated with more markers for the non-centromeric regions. Of the 1748 mapped markers, 11 markers were assigned to 3D, confirming the difficulty in identifying polymorphic markers for 3D, perhaps due to the large common regions of 3D between the Seri and Babax (McIntyre et al. 2010). In addition, the lower frequency of polymorphic markers in the D genome compared with the AB genome in wheat makes it difficult to have a good coverage of D genome (Allen et al. 2011; Chao et al. 2009). The final map comprised of markers from AFLP, SSR, DArT, 90K, and DArTseq, and clustering of markers from the same marker platform on the map was frequently observed. The frequency of marker clustering was primarily related to the number of markers from different types. For example, clustering of 90K markers was more frequent than the other types, which was not surprising since the number of 90K markers was more abundant than the others. The occurrence of clusters is very common when different kinds of markers are integrated into a single linkage map (Peleg et al. 2008; Semagn et al. 2006). Overall, the updated genetic map provided a higher density (1.1-fold) map with more markers (3.5-fold) and a good collinearity with the IWGSC RefSeq v1.0 genome, representing a step forward in mapping analysis and also in comparing QTLs with other studies using the 90K genetic map.
With the updated integrated genetic map, the number of identified QTLs increased by 5-fold compared with the previous study (Lopes et al. 2013), and 25 QTLs were common between the two studies (Supplementary Table 6). Since the Seri/Babax population was widely used for QTL studies (Mathews et al. 2008; McIntyre et al. 2010; Pinto et al. 2016; Pinto et al. 2010; Tahmasebi et al. 2016), we managed to locate all the reported QTLs on the updated map and found that the QTLs were clustered on several chromosome regions (Supplementary Table 6). For example, on chromosome 1Bb at 70–105 cM, there was a cluster of QTLs for GY, TGW, GN, DTA, PH, NDVI, CT, SPAD, harvest index, rate of senescence, and percentage of greenness lost. On chromosome 2B (24–84 cM), QTLs for GY, TGW, GN, DTH, PH, CT, maturity, grains per spike, leaf rolling, and spikelet compactness were co-located. On chromosome 4A at 265–300 cM, QTLs for GY, GN, and PH in the present study were co-located with QTLs for CT, NDVI, grain-filling rate, and water-soluble carbohydrates. On chromosome 5A (69–139 cM), QTLs for GY, GN, DTH, PH, spikelet per spike, spikelet compactness, flowering time, maturity, and leaf rolling were clustered. On chromosome 6B at 182–216 cM, QTLs for GY, TGW, GN, DTH, grain-filling rate, spikelet per spike, spikelet compactness, grains per spike, and percentage of greenness lost at mid grain-filling stage were frequently identified. Pinto et al. (2010) reported six common QTLs for drought and heat stress on chromosomes 1B-a, 2B-a, 3B-b, 4A-a, 4B-b, and 7A-a in the Seri/Babax population in Obregon, Mexico, environments, while in the present study, the QTL on chromosome 1B-a can be detected in Obregon heat stress in 2006 (71–75 cM on 1Bb in the present study); the QTL on chromosome 2B-a was detected in Wad Medani, Dongola, Darab, and Obregon heat stress in 2009 (39–47 cM on 2B); the QTL on chromosome 3B-b was detected in Obregon yield potential (2005–2006), drought (2004–2005), and heat (2006) conditions (219–235 cM on 3Bb); the QTL on chromosome 4A-a was detected across Obregon heat stress (2006, 2009), Ludhiana, Shandaweel, and Sohag (265–269 cM on 4A); the QTL on chromosome 4B-b was identified in Obregon heat plus drought stress (2009), Darab, Dongola, and Ludhiana (189–195 cM on 4B); and the QTL on 7A-a was detected in Wad Medani and Karnal (265–267 cM on 7A). These results suggested that the experiments and breeding conducted in drought and heat stress environments in Obregon, Mexico, could be used to predict the performance of the germplasm in other parts of the world. Reynolds et al. (1994) also reported that the typical heat stress in Obregon was demonstrated to be successful in generating germplasm for heat-stressed environments, such as ME5 comprising India and Sudan (Hodson and White 2007; Rajaram et al. 1993).
Typically, there is a trade-off between grain weight and grain number, making it difficult to improve yield through increasing grain weight and grain number at the same time (Griffiths et al. 2015; Sukumaran et al. 2018a; Sukumaran et al. 2018b). However, in the present study, TGW and GN were observed to be positively correlated, and both TGW and GN were positively correlated with GY in most environments. Several QTLs co-located for GY and GN were found on chromosomes 1A (120 cM), 1Bb (68–92 cM), 4A (56–62, 280–286, 304–308 cM), and 4B (32 cM), where QTLs for GY and GN shared the favorable allele from the same parent. QTLs co-located for GY and TGW were on chromosomes 3Bb (228–236 cM) and 5A (26 cM), where the same parent contributed the favorable allele for both GY and TGW. Among the QTLs co-located for TGW and GN, most of the favorable alleles for TGW and GN were contributed by different parents, while the QTL on chromosome 6B (194–200 cM) shared the favorable allele from the same parent for TGW and GN, indicating that it may be possible to improve grain yield through selecting complementary TGW and GN QTLs (Groos et al. 2003).
The stable heat adaptive QTL on chromosome 1D (84 cM) for GY could be projected to 484–493 Mb on the IWGSC RefSeq v1.0 genome, where genes were located for disease resistance, heat shock family protein, and early flowering. The QTL on chromosome 2Db (110 cM) for GY was previously identified for photochemical reflectance index (PRI) in a CIMMYT spring wheat population (WAMI) under yield potential condition (Liu et al. 2019a). The QTL on chromosome 3Bb (228–236 cM) for GY and TGW was very close to a locus associated with multiple traits (GY, maturity, and chlorophyll content at vegetative state) in the temperate irrigated environments in spring wheat (Sukumaran et al. 2015). The QTL on chromosome 4A (308 cM) for GY and GN could be mapped to 531–535 Mb in physical map, which contains 34 genes, including heat-shock protein genes. The TGW QTL on chromosome 7Ba (26–32 cM) was located at 3.7–5.9 Mb on the physical map, which harbored 33 genes. The GN QTL on chromosome 1A (86 cM) was associated with ground cover in winter wheat (Gao et al. 2016). The GN QTL on chromosome 6Db (8 cM) was co-located with a QTL for DTH in a synthetic derived RIL population (Liu et al. 2019b).
The Seri/Babax population was initially developed in Obregon, Mexico; so, the lowest range of DTH (8 days) and PH (15–23 cm) was observed in the Obregon environments in this study. When grown in international trials, longer DTH and higher PH with a wider range were observed in most environments. For DTH, the QTL on chromosome 4Da was previously reported for maturity in spring wheat (Sukumaran et al. 2015). The QTL on chromosome 5D (38 cM) was located at a distance (about 60 Mb) from the Vrn-D4 locus (Yoshida et al. 2010). The stable QTL for DTH on chromosome 7Ba (20–22 cM) was co-located with a previously reported QTL for kernel number per spike (Gao et al. 2015). Projecting this QTL to the physical map, it was located at the Vrn-B3 locus (Yan et al. 2006), which could accelerate flowering and bypass the vernalization requirement of its dominant allele if present. This QTL was contributed by the parent Seri and has never been reported before by other studies using Seri/Babax. For PH, the locus on chromosome 4B (192–194 cM) was associated with GY in a CIMMYT spring wheat panel (Sukumaran et al. 2013). This locus can be projected to 3.5–4.7 Mb on the physical map, which was very close to the Rht-B1 gene (Pearce et al. 2011). The PH QTL on chromosome 5A (72 cM) was also associated with Cephalosporium stripe resistance in wheat (Quincke et al. 2011).
This study updated the genetic map of the widely used Seri/Babax RIL population with the 90K and DArTseq SNPs. The updated genetic map has good genome coverage, higher marker density, good collinearity with the IWGSC physical map and increased the number of detected QTLs compared to the earlier studies. A larger number of QTLs were identified with this genetic map across various environments, and some of them were stable in different environments. QTLs that are unique to the current study indicate the importance of an updated genetic map for QTL detection. This updated genetic map is expected to serve as a platform for genetic analyses of important quantitative traits, map-based cloning, and marker-assisted selection studies.
Abbreviations
- QTL:
-
Quantitative trait locus
- RIL:
-
Recombinant inbred line
- SNP:
-
Single-nucleotide polymorphism
- PVE:
-
Phenotypic variation explained by QTL
References
Allen AM, Barker GL, Berry ST, Coghill JA, Gwilliam R, Kirby S, Robinson P, Brenchley RC, D’Amore R, McKenzie N (2011) Transcript-specific, single-nucleotide polymorphism discovery and linkage analysis in hexaploid bread wheat (Triticum aestivum L.). Plant Biotechnol J 9(9):1086–1099
Bennett D, Reynolds M, Mullan D, Izanloo A, Kuchel H, Langridge P, Schnurbusch T (2012) Detection of two major grain yield QTL in bread wheat (Triticum aestivum L.) under heat, drought and high yield potential environments. Theor Appl Genet 125(7):1473–1485
Chao S, Zhang W, Akhunov E, Sherman J, Ma Y, Luo M-C, Dubcovsky J (2009) Analysis of gene-derived SNP marker polymorphism in US wheat (Triticum aestivum L.) cultivars. Mol Breed 23(1):23–33
Doerge RW (2002) Multifactorial genetics: mapping and analysis of quantitative trait loci in experimental populations. Nat Rev Genet 3(1):43
FAOstat (2018) The Statistics Division, Food and Agriculture Organization of the United Nations. http://www.fao.org/statistics/en/. Accessed 26 Aug 19
Gao F, Wen W, Liu J, Rasheed A, Yin G, Xia X, Wu X, He Z (2015) Genome-wide linkage mapping of QTL for yield components, plant height and yield-related physiological traits in the Chinese wheat cross Zhou 8425B/Chinese Spring. Front Plant Sci 6:1099
Gao F, Liu J, Yang L, Wu X, Xiao Y, Xia X, He Z (2016) Genome-wide linkage mapping of QTL for physiological traits in a Chinese wheat population using the 90K SNP array. Euphytica 209(3):789–804. https://doi.org/10.1007/s10681-016-1682-6
Golabadi M, Arzani A, Maibody SM, Tabatabaei BS, Mohammadi S (2011) Identification of microsatellite markers linked with yield components under drought stress at terminal growth stages in durum wheat. Euphytica 177(2):207–221
Griffiths S, Wingen L, Pietragalla J, Garcia G, Hasan A, Miralles D, Calderini DF, Ankleshwaria JB, Waite ML, Simmonds J (2015) Genetic dissection of grain size and grain number trade-offs in CIMMYT wheat germplasm. PLoS One 10(3):e0118847
Groos C, Robert N, Bervas E, Charmet G (2003) Genetic analysis of grain protein-content, grain yield and thousand-kernel weight in bread wheat. Theor Appl Genet 106(6):1032–1040
Hodson D, White J (2007) Use of spatial analyses for global characterization of wheat-based production systems. J Agric Sci (Camb) 145(1):115–125
Kirigwi F, Van Ginkel M, Brown-Guedira G, Gill B, Paulsen GM, Fritz A (2007) Markers associated with a QTL for grain yield in wheat under drought. Mol Breed 20(4):401–413
Kosambi DD (2016) The estimation of map distances from recombination values. In: Ramaswamy R (ed) D.D. Kosambi: Selected Works in Mathematicsand Statistics. Springer, New Delhi, pp 125-130. https://doi.org/10.1007/978-81-322-3676-4_16
Liu C, Pinto F, Cossani CM, Sukumaran S, Reynolds PM (2019a) Spectral reflectance indices as proxies for yield potential and heat stress tolerance in spring wheat: heritability estimates and marker-trait associations. Front Agric Sci Eng. https://doi.org/10.15302/j-fase-2019269
Liu C, Sukumaran S, Claverie E, Sansaloni C, Dreisigacker S, Reynolds M (2019b) Genetic dissection of heat and drought stress QTLs in phenology-controlled synthetic-derived recombinant inbred lines in spring wheat. Mol Breed 39(3):34
Lopes MS, Reynolds MP, McIntyre CL, Mathews KL, Kamali MJ, Mossad M, Feltaous Y, Tahir IS, Chatrath R, Ogbonnaya F (2013) QTL for yield and associated traits in the Seri/Babax population grown across several environments in Mexico, in the West Asia, North Africa, and South Asia regions. Theor Appl Genet 126(4):971–984
Maccaferri M, Sanguineti MC, Corneti S, Ortega JLA, Salem MB, Bort J, DeAmbrogio E, del Moral LFG, Demontis A, El-Ahmed A (2008) Quantitative trait loci for grain yield and adaptation of durum wheat (Triticum durum Desf.) across a wide range of water availability. Genetics 178(1):489–511
Maechler M, Rousseeuw PR, Struyf A, Gonzalez J (2016) Cluster: “Finding Groups in Data”: Cluster Analysis Extended Rousseeuw et al. R package version 2.1.0. https://cran.rproject.org/web/packages/cluster/index.html. Accessed 26 Aug 19
Mathews KL, Malosetti M, Chapman S, McIntyre L, Reynolds M, Shorter R, van Eeuwijk F (2008) Multi-environment QTL mixed models for drought stress adaptation in wheat. Theor Appl Genet 117(7):1077–1091. https://doi.org/10.1007/s00122-008-0846-8
McIntyre CL, Mathews KL, Rattey A, Chapman SC, Drenth J, Ghaderi M, Reynolds M, Shorter R (2010) Molecular detection of genomic regions associated with grain yield and yield-related components in an elite bread wheat cross evaluated under irrigated and rainfed conditions. Theor Appl Genet 120(3):527–541. https://doi.org/10.1007/s00122-009-1173-4
Meng L, Li H, Zhang L, Wang J (2015) QTL IciMapping: integrated software for genetic linkage map construction and quantitative trait locus mapping in biparental populations. Crop J 3(3):269–283. https://doi.org/10.1016/j.cj.2015.01.001
Olivares-Villegas JJ, Reynolds MP, McDonald GK (2007) Drought-adaptive attributes in the Seri/Babax hexaploid wheat population. Funct Plant Biol 34(3):189–203
Pearce S, Saville R, Vaughan SP, Chandler PM, Wilhelm EP, Sparks CA, Al-Kaff N, Korolev A, Boulton MI, Phillips AL, Hedden P, Nicholson P, Thomas SG (2011) Molecular characterization of <em>Rht-1</em> dwarfing genes in hexaploid wheat. Plant Physiol 157(4):1820–1831. https://doi.org/10.1104/pp.111.183657
Peleg Z, Saranga Y, Suprunova T, Ronin Y, Röder MS, Kilian A, Korol AB, Fahima T (2008) High-density genetic map of durum wheat × wild emmer wheat based on SSR and DArT markers. Theor Appl Genet 117(1):103
Pinto RS, Reynolds MP, Mathews KL, McIntyre CL, Olivares-Villegas J-J, Chapman SC (2010) Heat and drought adaptive QTL in a wheat population designed to minimize confounding agronomic effects. Theor Appl Genet 121(6):1001–1021
Pinto RS, Lopes MS, Collins NC, Reynolds MP (2016) Modelling and genetic dissection of staygreen under heat stress. Theor Appl Genet 129(11):2055–2074. https://doi.org/10.1007/s00122-016-2757-4
Quarrie S, Pekic Quarrie S, Radosevic R, Rancic D, Kaminska A, Barnes J, Leverington M, Ceoloni C, Dodig D (2006) Dissecting a wheat QTL for yield present in a range of environments: from the QTL to candidate genes. J Exp Bot 57(11):2627–2637
Quincke MC, Peterson CJ, Zemetra RS, Hansen JL, Chen J, Riera-Lizarazu O, Mundt CC (2011) Quantitative trait loci analysis for resistance to Cephalosporium stripe, a vascular wilt disease of wheat. Theor Appl Genet 122(7):1339–1349
Rajaram S, Van Ginkel M, Fischer R (1993) CIMMYT’s wheat breeding mega-environments (ME). In: Proceedings of the 8th International Wheat Genetic Symposium
Reynolds M, Tuberosa R (2008) Translational research impacting on crop productivity in drought-prone environments. Curr Opin Plant Biol 11(2):171–179. https://doi.org/10.1016/j.pbi.2008.02.005
Reynolds M, Balota M, Delgado M, Amani I, Fischer R (1994) Physiological and morphological traits associated with spring wheat yield under hot, irrigated conditions. Funct Plant Biol 21(6):717–730
Reynolds M, Foulkes MJ, Slafer GA, Berry P, Parry MAJ, Snape JW, Angus WJ (2009) Raising yield potential in wheat. J Exp Bot 60(7):1899–1918. https://doi.org/10.1093/jxb/erp016
Rosegrant MW, Cline SA (2003) Global food security: challenges and policies. Science 302(5652):1917–1919. https://doi.org/10.1126/science.1092958
Semagn K, Bjørnstad Å, Skinnes H, Marøy AG, Tarkegne Y, William M (2006) Distribution of DArT, AFLP, and SSR markers in a genetic linkage map of a doubled-haploid hexaploid wheat population. Genome 49(5):545–555
Sukumaran S, Ellis MH, Lopes MS, Pinto S, Bonnett D, Dreisigacker S, Mullan D, Reynolds MP (2013) Wheat mapping populations available at CIMMYT for yield potential research. Wheat Yield Consortium 782013
Sukumaran S, Dreisigacker S, Lopes M, Chavez P, Reynolds MP (2015) Genome-wide association study for grain yield and related traits in an elite spring wheat population grown in temperate irrigated environments. Theor Appl Genet 128(2):353–363
Sukumaran S, Lopes M, Dreisigacker S, Reynolds M (2018a) Genetic analysis of multi-environmental spring wheat trials identifies genomic regions for locus-specific trade-offs for grain weight and grain number. Theor Appl Genet 131(4):985–998. https://doi.org/10.1007/s00122-017-3037-7
Sukumaran S, Reynolds MP, Sansaloni C (2018b) Genome-wide association analyses identify QTL hotspots for yield and component traits in durum wheat grown under yield potential, drought, and heat stress environments. Front Plant Sci 9:81
Tahmasebi S, Heidari B, Pakniyat H, McIntyre CL (2016) Mapping QTLs associated with agronomic and physiological traits under terminal drought and heat stress conditions in wheat (Triticum aestivum L.). Genome 60(1):26–45
Trethowan RM, Mujeeb-Kazi A (2008) Novel germplasm resources for improving environmental stress tolerance of hexaploid wheat. Crop Sci 48:1255–1265. https://doi.org/10.2135/cropsci2007.08.0477
Varshney RK, Mahendar T, Aggarwal RK, Börner A (2007) Genic molecular markers in plants: development and applications. In: Genomics-assisted crop improvement. Springer, pp 13–29
Yan L, Fu D, Li C, Blechl A, Tranquilli G, Bonafede M, Sanchez A, Valarik M, Yasuda S, Dubcovsky J (2006) The wheat and barley vernalization gene VRN3 is an orthologue of FT. Proc Natl Acad Sci 103(51):19581–19586
Yano M, Sasaki T (1997) Genetic and molecular dissection of quantitative traits in rice. In: Sasaki T, Moore G (eds) Oryza: from molecule to plant. Springer, Netherlands, Dordrecht, pp 145–153. https://doi.org/10.1007/978-94-011-5794-0_14
Yoshida T, Nishida H, Zhu J, Nitcher R, Distelfeld A, Akashi Y, Kato K, Dubcovsky J (2010) Vrn-D4 is a vernalization gene located on the centromeric region of chromosome 5D in hexaploid wheat. Theor Appl Genet 120(3):543–552
Acknowledgments
This work was implemented by CIMMYT as part of IWYP-HUB, made possible by the generous support of the International Wheat Yield Partnership (IWYP). Any opinions, findings, conclusion, or recommendations expressed in this publication are those of the authors and do not necessarily reflect the view of the donors. We also acknowledge Dr. Marc Ellis for providing DArTseq data for the population. Thanks are due to the staff at Wheat Physiology lab at CENEB, Cd. Obregon, México.
Author information
Authors and Affiliations
Contributions
SS designed the current study; ML and MR provided the phenotypic data; CS contributed with the explanation of DArTseq data; SD and MR genotyped the population; CL, SM, and SS created the genetic maps and analyzed the data; CL and SS wrote the manuscript. All authors reviewed and accepted the contents.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Supplementary Figure 1
Phenotypic correlations between grain yield (GY, t ha−1), thousand-grain weight (TGW, g), grain number (GN), days to heading (DTH, d), and plant height (PH, cm) of the Seri/Babax RIL population grown in different environments. (PNG 764 kb)
Supplementary Figure 2
Distribution of markers between two consecutive loci along genetic map on all chromosomes. (PNG 124 kb)
Supplementary Figure 3
An updated genetic map of Seri/Babax RIL population with QTLs identified for grain yield (GY, t ha−1), thousand-grain weight (TGW, g), grain number (GN), days to heading (DTH, d), and plant height (PH, cm) in different environments. Markers in red and green color are distorted markers towards Babax and Seri parents, respectively. (PNG 3306 kb)
Supplementary Table 1
Phenotypic data, including grain yield (GY, t ha−1), thousand-grain weight (TGW, g), grain number (GN), days to heading (DTH, d), and plant height (PH, cm), of the Seri/Babax RIL population grown in 16 environments in five countries. (XLSX 114 kb)
Supplementary Table 2
Linkage map information of the Seri/Babax population. Seri is coded as “0,” Babax is coded as “2,” and missing data is coded as “− 1.” (XLSX 897 kb)
Supplementary Table 3
Linkage map information of the Seri/Babax population using 90K SNPs. Seri is coded as “0,” Babax is coded as “2,” and missing data is coded as “− 1”. (XLSX 611 kb)
Supplementary Table 4
QTLs detected for grain yield (GY, t ha−1), thousand-grain weight (TGW, g), grain number (GN), days to heading (DTH, d), and plant height (PH, cm) of the Seri/Babax RIL population in 16 individual environments. (XLSX 24 kb)
Supplementary Table 5
QTLs detected for grain yield (GY, t ha−1), thousand-grain weight (TGW, g), grain number (GN), days to heading (DTH, d), and plant height (PH, cm) of the Seri/Babax RIL population in high-yielding and low-yielding clusters. (XLSX 12 kb)
Supplementary Table 6
Comparison of QTLs detected in the present study with the previous studies using the Seri/Babax RIL population. (XLSX 73 kb)
Rights and permissions
About this article
Cite this article
Liu, C., Khodaee, M., Lopes, M.S. et al. Multi-environment QTL analysis using an updated genetic map of a widely distributed Seri × Babax spring wheat population. Mol Breeding 39, 134 (2019). https://doi.org/10.1007/s11032-019-1040-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11032-019-1040-1