Abstract
Drought is a major abiotic stress that adversely affects wheat production in many regions of the world. The objective of this study was to identify quantitative trait loci (QTL) controlling grain yield and yield components under reduced moisture. A cross between common wheat cultivars ‘Dharwar Dry’ (drought tolerant) and ‘Sitta’ was the source of one hundred twenty-seven recombinant inbred lines evaluated for two-seasons in a field under differing soil moisture regimes in Ciudad Obregon, Sonora, Mexico. An SSR/EST-STS marker map was constructed and a grain yield QTL on the proximal region of chromosome 4AL was found to have a significant impact on performance under reduced moisture. This region was associated with QTL for grain yield, grain fill rate, spike density, grains m−2, biomass production, biomass production rate, and drought susceptibility index (DSI). Molecular markers associated with these traits explained 20, 33, 15, 23, 30, 26, and 41% of phenotypic variation, respectively on chromosome 4A. Microsatellite locus Xwmc89 was associated with all significant QTL covering a 7.7 centiMorgans (cM) region and generally explained the greatest proportion of phenotypic variation. The alleles associated with enhanced performance under drought stress were contributed by Dharwar Dry. Microsatellite marker wmc89 may be useful for marker assisted selection to enhance drought tolerance.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Wheat (Triticum aestivum L.) production is adversely affected by drought in 50% of the area under production in the developing and 70% in the developed countries (Trethowan and Pfeiffer 2000). Genetic gain for yield in these environments is estimated at about half of the yield under well-watered environments (Rajaram et al. 1996). Apart from poor or erratic rainfall, the developing countries’ wheat growing areas suffer from poor soils, aggressive diseases, and sometimes extreme heat or cold. The Green Revolution technology was slow to spread to these regions and resulted in only modest gains due to the harsh agricultural conditions (Lantican et al. 2002). By 2020, demand for wheat in marginal environments will rise 40% over current levels (Rosegrant et al. 2001), a demand unlikely to be met unless wheat productivity in these environments is increased (Lantican et al. 2002).
It is difficult to make progress for grain yield and yield components under drought as they are complex characters that are influenced by environmental factors and are characterized by low heritabilities and large genotype × environment interactions under drought conditions (Smith et al. 1990). Molecular markers associated with quantitative trait loci (QTL) for drought adaptive traits could greatly enhance progress in breeding for drought tolerance.
Quantitative trait loci for agronomic traits have not been reported for grain yield and yield components under drought-stress in wheat. The objective of this work was to identify QTL associated with yield and other agronomic traits under reduced soil moisture.
Materials and methods
Line development and phenotypic measurements
The cross between cultivars ‘Dharwar Dry’ (-0IND) and ‘Sitta’ (CM77091) was developed at the International Center for Maize and Wheat Improvement (CIMMYT). Dharwar Dry originated from Central India, where wheat is grown mainly on residual moisture while Sitta was developed at CIMMYT and is considered to be moderately tolerant to drought. The F1 seed was planted at Cd. Obregon (27°29′ N, 109°55′ W, 40 m above sea level), Sonora, Mexico, under adequate moisture supply for increase purposes. The F2 seed was space-planted (10–15 cm between plants), while subsequent generations were seeded at 80 kg ha−1. Diseases and pests were controlled. One-hundred plants were advanced from the F2 population, and lines were discarded based on visual assessment for agronomic type by the same breeder in each generation. At the F6, a total of 140 progeny rows were selected, and 10 spikes from each row were harvested and threshed separately. Kernels from these spikes of each selected F2,6 line were planted as head rows at El Batan (19°31′ N, 98°51′ W, 2,275 m above sea level) on sandy-clay soil in central Mexico for advance to the F7 and purification. Uniform rows from each F2,6 were bulked and used as seed for the experiment.
Evaluation
Field evaluation was conducted at the Centro de Investigaciones Agricolas de Noroeste (CIANO) Experimental Station in the Yaqui Valley, Cd. Obregon, Sonora, Mexico during the 1998–1999 and 1999–2000 growing seasons. The lines were evaluated under 1-irrigation (stress) and 3-irrigation regimes during 1998–1999 growing season, and under 1-irrigation (stress) and 2-irrigation regimes during the 1999–2000 growing season. For the 1-irrigation regime, only a pre-seeding irrigation supplying 120 mm of water was applied to the plots while the increased irrigation regimes received 3 and 2 irrigations in the first and the second year, respectively. The 3-irrigation regime was irrigated after planting and later at 2nd node stage (Feek’s growth stage 7) and at anthesis (Feek’s growth stage 10.5). The 2-irrigation regime only received the first two irrigations. Pre-irrigation field soil samples indicated that the soil had a reserve of about 250 mm of moisture. For the 1-irrigation regime, the plots were treated to terminal drought stress because no water was applied after planting.
Green leaf area was assessed from 10 days after anthesis and thereafter weekly for 4 weeks in the LY environment and up to 6 weeks in the HY environment. Plants were scored visually on a scale of 0 (all leaves green and viable) to 9 (all leaves senesced and necrotic). Intermediate values were assigned according to the extent of loss of green color. A green leaf index was calculated as the total green leaf rating attained by a plot summed over all days during the grain-filling period (Cox et al. 1995). The green leaf index (GLI) was calculated as:
where s i is the green leaf rating for date i, t i is the number of days between the heading date of the earliest plot in the trial and date i, s n is the maximum green leaf rating recorded on the final date, and t n is the total number of days of grain filling.
Days to heading and maturity were recorded when 50% of the spikes had emerged from the boot and when 50% of the peduncles had turned yellow, respectively. At maturity, plant height (cm) was measured from the soil surface to the top of the spikes excluding the awns. A sample of 50 tillers was harvested randomly from each plot to measure the yield components. This sample was also used to estimate biomass production (ton ha−1), grains spike−1, kernel weight (mg) and spike density (number m−2). The remaining plants in each plot were harvested with a small plot combine to determine grain yields (ton ha−1). Grain yields were adjusted to 12% moisture after drying the samples at 72°C for 36 h.
A drought susceptibility index (DSI) was calculated using the procedure of Fischer and Maurer (1978) and was expressed by the following relationship: DSI = [1−(Ys1)/(Yp1)]/SI, where Ys1 and Yp1 are the yields of a line under stressed and non-stressed (potential) conditions, respectively. SI is the stress intensity and is estimated as: [1−(Ys2)/(Yp2)], where Ys2 and Yp2 are the mean yield over all genotypes evaluated under stressed and non-stress (potential) conditions, respectively. Grain fill duration (days) was the number of days from days to heading to days to maturity. Grain production rate (kg ha−1 day−1) was determined as the ratio of grain yield and grain-fill duration while biomass production rate (kg ha−1 day−1) was calculated as the ratio of biomass production and days to physiological maturity (Kirigwi et al. 2004).
SAS (SAS Inst. 1990) Proc Mixed procedure was used for analyses. The genetic correlations and their standard errors were estimated using multivariate restricted maximum likelihood (REML)(Holland 2006). The 2-year data from the 1-irrigation environment were analyzed separately and also combined, whereas those from 2- and 3-irrigations analyzed separately because errors were non-homogenous. For the combined data, Year was considered a random effect while lines were considered a fixed effect.
Molecular marker analysis
DNA was isolated from Dharwar Dry and Sitta following the CTAB DNA extraction protocol (Richards et al. 1994). SSR markers were screened for polymorphism between the parents. Polymorphic markers were used to genotype the entire RIL population. Subsequently, 96 SSR markers with an average of four markers per chromosome were used to perform the initial single marker analysis by simple regression (Qgene 3.06). A 210 bp band amplified by SSR marker wmc48 was significantly associated with DSI, grain yield, days to heading, grain fill duration, grain fill rate, biomass and spike density with R 2 values of 0.35, 0.33, 0.11, 0.14, 0.22, 0.26, and 0.08, respectively. No other markers were significantly associated with phenotypic traits. Due to this result, the focus was subsequently placed on further characterization of the chromosomal region surrounding Xwmc48. The first step was to identify the chromosomal location of Xwmc48. Previous reports had placed this marker on the Group 4 chromosomes (Sourdille et al. 2004). DNA from Chinese Spring (CS) and CS aneuploids (nulli-tetras and deletion lines for Group 4 chromosomes) were used as template DNA for amplification with the wmc48 primer pair. It was not possible to physically map Xwmc48 with CS stocks because the 210 bp band was not present in CS (Fig. 3). The wmc48 primer pair was then used to amplify products from the ITMI mapping population data (http://wheat.pw.usda.gov/ggpages/maps/Triticum/Wheat%20 W7984%20 x%20 Opata85%20group%201%20mapping%20, verified 24th February, 2007). The full set of marker data for this population was downloaded from the Graingenes database and used for linkage analysis using Mapmaker 2.0 (Lander et al. 1987). The target fragment was mapped on chromosome 4A (Fig. 1) and this chromosome was subsequently targeted for marker enrichment. Wheat ESTs from each of the deletion bins on chromosome 4A (Qi et al. 2003) were selected and sequences downloaded from the NCBI database (http://www.ncbi.nlm.nih.gov/entrez/query.fcgi, verified 24th February, 2007). These sequences were used to design primers for EST-based STS markers using Mac Vector 6.5.3 and targeting amplicons between 300 bp and 600 bp. An EST-based STS marker (Gene bank designation BE637912) from deletion bin C-4AL12-0.43 mapped 6 cM from Xwmc48. This EST is syntenic to an EST on BAC clone AC027038 of rice chromosome 10. Sequences from wheat ESTs syntenic to rice ESTs on this and adjacent rice BAC clones were used to design additional STS primers. A consensus Group 4A genetic map of wheat from the integrated wheat science database (http://www.shigen.nig.ac.jp/wheat/komugi/maps/) also showed Xwmc48 to be located on chromosome 4A. Microsatellite markers near Xwmc48 on the Komugi map were also used. For STS markers that did not amplify a product, target region amplification polymorphism (TRAP) marker sequences were used as arbitrary sequences together with the targeted EST-based STS sequence (Hu and Vick 2003). Primers producing polymorphism on parents were mapped on the population. Eventurally, 102 SSR- and 34 EST-STS primers polymorphic between Dharwar Dry and Sitta were identified making a total of 136 markers.
A linkage map of wheat chromosome 4A constructed from marker data from the Dharwar Dry × Sitta population using the Mapmaker 2.0 computer program (Lander et al. 1987). Data on the left not in brackets are in cM
The procedures for polymerase chain reactions were performed as described by Röder et al. (1998) in 25 μl volumes using a peltier thermocycler (MJ research). Thermal cycling was programmed at 94°C for 3 min, specific primer temperature 1 min, 72°C for 90 s with 36 cycles. Amplified PCR products were separated on 2.3% GenePure HiRes Agarose (ISC Bioexpress), stained with ethidium bromide, and visualized under ultra violet (UV) light. Non-polymorphic PCR products using electrophoresis with agarose between the parents were separated in a 6% 100 ml denaturing gel run in 1× TBE buffer at 85 W for 2 h and then silver stained according to the protocol of Bassam et al. (1991). PCR products from EST-STS primers were separated using the mutation detection enhancement (MDE®) matrix (Cambrex) according to Martins-Lopes et al. (2001).
Of the 136 markers, only 25 of them mapped on chromosome 4A and were evaluated on 127 lines of the population. The rest of the markers were not pursued further because no association was found between any of them and the traits. Map construction was done using the MAPMAKER 2.0 computer program (Lander et al. 1987). Linkage groups were established using the Two-point/Group command at a LOD score of 3.0. Marker order within linkage groups was established using the ‘Try’ and ‘Ripple’ commands. Marker order on the Komugi map was used for linkage map confirmation (Fig. 1).
The QTL analysis was performed by composite interval mapping (CIM) (Wang et al. 2001–2004). A window size of 10 cM and a maximum of five marker cofactors per model were used. The forward-backward regression model was used to select marker co-factors with a critical P-value of 0.10. Threshold levels were estimated using P < 0.05 for the comparison of 1000 data permutations which is adequate for estimating critical thresholds at α = 0.10, and α = 0.05 (Churchill and Doerge 1994).
Results
During the 1998–1999 and 1999–2000 seasons, rainfall was in the range of 3.1–3.4 mm, therefore, water for wheat growth almost entirely came from the irrigation treatments. The 1-irrigation regime simulated increasing drought stress throughout the growing season. The mean number of days to heading was 79.7 (range: 73−89 days) for the 1-irrigation regime. The genetic correlation between grain yield and days to heading was −0.06 (Std. error 0.071) (Table 1). The 3-irrigation, 2-irrigation and 1-irrigation regimes averaged 6.03, 4.36, and 3.08 ton ha−1of grain, respectively (Table 2). The range for grain yield under the 1-irrigation regime was about twice as large for year 1 compared to year 2. Combined analysis for the 2 years under this regime showed that the year effect was not significant for grain yield and spike density. All entry effects were significant except for plant height (data not shown). Line effect mean squares for DSI for grain yield computed from the mean of 1-irrigation and 2-, and with 3-irrigation regimes were significant (P < 0.0001)(data not shown).
In the single marker analysis, the data combined over years for the 1-irrigation regime revealed significant associations between markers on all traits except days to physiological maturity (Table 3). Dharwar Dry alleles were associated with higher drought tolerance as indicated by DSI results (Table 4). Significant associations for DSI were found at Xgwm601, BE637912, CA487065a, and b, Xwmc48, Xwmc89, and at Xwmc420 using combined data from the 1-irrigation regimes with the individual increased-irrigation regimes. For data from the 1-irrigation regime of year 1, only Xwmc48 was significantly associated with DSI.
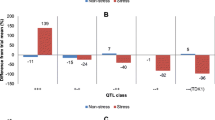
Composite interval mapping was performed on 127 RILs using 25 markers mapped to the region identified by single marker analysis (Fig. 1). Only a few of the QTL identified using single marker analysis were also identified by CIM. These QTL were from data for 1-irrigation regime year 2 (stress) and one QTL from combined data for the 2- and the 3-irrigation regimes (days to heading, data not shown) that was not under water stress. No QTL was identified for combined data from the 1-irrigation regime and neither from 1-irrigation year Results for the from CIM showed that one genomic region on chromosome 4A had a considerable influence on DSI, grain yield, biomass, spike density, grains m−2, grain fill rate, biomass production rate (all from 1-irrigation year 2 data) (Table 5, Fig. 2), and days to heading (combined data for the 2- and 3-irrigation regimes, data not shown). The QTL for grain yield was associated with three markers, BE637912, Xwmc89, and Xwmc420 and accounted for 15, 18 and 20% of the observed phenotypic variation, respectively. Similarly, these markers were associated with QTL for biomass production explaining 29, 32 and 30% of the observed phenotypic variation, respectively.
Coincidence of QTL for grain yield (a), grain fill rate (b), biomass (c), biomass production rate (d), drought susceptibility index (e), and grains m−2 (f). The figures represent data for 1-irrigation year 2 (1999–2000). The DSI figure is from 1-irrigation year 2 and 3-irrigation regime. (Data on Y-axis is in LOD units. Horizontal lines in graph indicate significant LOD level after permutation)
In this study, QTL for spike density was associated with marker Xwmc89 and explained 15% of the phenotypic variation. Similarly, a QTL for grain m−2 was found associated with Xwmc89 and Xwmc420 and explained 16% and 23% of the phenotypic variation, respectively. Both GFR and biomass production rate were significantly associated with marker loci. Marker loci Xwmc601, Xwmc89, and Xwmc420 were significant and explained 9, 28, and 33% of phenotypic variation for GFR. Similarly, a QTL for biomass production rate was found at Xwmc89 with a LOD score of 7.78 and explained 26% of phenotypic variation.
Significant QTL identified for DSI were mainly associated with Xwmc89. Additionally, significant DSI QTL were also found at Xwmc601 and BE637912 (Table 5). DSI QTLs associated with Xgwm601 and at BE637912 accounted for 41% and 12% of phenotypic variation, respectively whereas, at Xwmc89, the proportion of phenotypic variation explained ranged from 13% to 38%. For all QTLs identified under stress, alleles from Dharwar Dry were associated with early heading, increased grain yield, GFR, biomass production and spike density and low DSI values indicative of drought tolerance.
Rice synteny and physical mapping
Wheat EST BE637912 and CA487065 were found to map near the target region. Both BE637912 and CA487065 map to the wheat region syntenic to rice chromosome 10 (BAC clone AC027038). The Graingenes database showed that probe UMW135BE637912, which is derived from BE637912, maps to wheat chromosomes 4A, 6A, 1AL and 2AL. The 4AL fragment maps to bin C-4AL12-0.43. Xgwm601 and Xwmc420 were physically mapped to bins C-4AL12-0.43 and 4AL12-0.43-0.59, respectively (Fig. 3). In addition, the wheat consensus map from the Komugi map showed that markers Xbcd808a-4A and XGermin fall within the region flanked by significant markers in this study. Xbcd808a is a barley (Hordeum vulgare L.) probe from an EST with Genbank designation BE538742. An STS marker designed from this EST was mapped to wheat deletion bin 4AL12-0.43-0.59 but is not polymorphic between Dharwar Dry and Sitta (Fig. 4). A wheat EST (BE406676) with sequence homology to the predicted Germin gene on rice chromosome 3L was mapped to deletion bin C-4AL12-0.43. Xbcd808a-4A and XGermin are 0.77 cM apart on the wheat consensus map, but cover 17 BAC clones spanning 1,790 KB of rice sequence (Fig. 4). The mapping of BE406676 to deletion bin C-4AL12-0.43 provided evidence that the QTL identified in this study is located in the C-4AL12-0.43 bin.
Deletion mapping of Xwmc48, Xgwm601, Xwmc420 and Xbcd808a (Gene bank accession BE538742 (wheat syntenic EST BE499340)) using Chinese Spring (CS) aneuploids and deletion lines of chromosome Group 4A. Xwmc48 band that maps on chromosome 4A and amplified in Dharwar Dry (DD) did not amplify in CS lines. Xbcd808a was not polymorphic between Dharwar Dry and Sitta (ST)(arrow). However, it had polymorphisms for deletion lines N4A and dt4AS relative to the other aneuploids and the diagnostic fragment was missing. Deletion line dt4AL had a distinct polymorphism for the diagnostic fragment
Discussion
Cultivar Dharwar Dry was selected for its outstanding tolerance to drought by CIMMYT scientists in India where wheat is usually grown under residual moisture in the rice-wheat rotation system. The growing conditions under residual moisture are characterized by receding moisture throughout the growing season and therefore, increasing water stress. In this experiment, alleles from this cultivar contributed to better performance under increasing water stress in the mapping population. Cultivar Sitta was developed at CIMMYT, Mexico, and has high yield potential and moderate drought stress tolerance. The level of polymorphism between Dharwar Dry and Sitta was lower than expected, given the origins of the cultivars. The Dharwar Dry/Sitta cross consists of tolerant to moderately tolerant parents rather than tolerant by susceptible and thus the parents may contain common alleles for tolerance. Since the pedigree of Dharwar Dry is unknown, it is also possible that it is derived, in part, from CIMMYT germplasm.
The population was not developed by standard methods of producing recombinant inbred lines, as selection was practiced throughout the development of the population. Each RIL, however, does trace to a unique F2 plant. The consistency of marker order and distances with previously published maps is also good evidence for the validity of the population. Selected populations are often useful for these types of studies as undue influence of plants with extreme phenological characteristics can be mitigated. The remaining variability represents the useful variation available to the breeder for practical plant improvement. Additionally, procurement of good phenotypic data for traits such as drought tolerance is far more difficult to develop compared to the creation of genetic maps. The presence of a high quality set of phenotypic data far outweighed any slight potential disadvantages associated with population structure.
Our efforts to increase marker density around Xwmc48 revealed low levels of polymorphism. This result was not completely unexpected as homoeologous group 4 of wheat is known to have lower levels of polymorphism (Huang et al. 2002). Xwmc48 was found to be located in a highly recombinogenic region of chromosome 4AL in deletion bin 4AL12-0.43 (Mickelson-Young et al. 1995).
This experiment identified a major QTL for grain yield and yield components, spike density, grain m−2 and DSI under increasing drought stress on chromosome 4AL. Traits that are correlated, as in the case of yield and yield components, are likely to have QTL mapping to similar locations (Kato et al. 2000). Similar correspondence of QTL for grain yield and correlated traits has been reported in maize (Zea mays L.)(Austin and Lee 1996), rice (Xiao et al. 1996), and barley (Bezant et al. 1997). The coincidence of QTL in one region of chromosome 4A may point to the involvement of several genes for performance under stress located together or a major gene located upstream in the drought response pathway that influences several yield traits. An earliness per se gene (eps4L) (Laurie et al. 1995) and dehydrin (Dhn6) (Choi et al. 2000) have been reported on this chromosome in barley. Dehydrin expression has been associated with increased drought tolerance in wheat cultivars ‘Tam105’, ‘Gene’ and ‘Connie’ (Lopez et al. 2003) but is not a likely explanation for our results based on expression experiments with Dharwar Dry and Sitta (data not shown). The QTL identified in this work may correspond to the QTL for yield reported by Araki et al. (1999) and Börner et al. (2002) on chromosome 4 although their work was carried out under adequate moisture. In barley, chromosome 4H was also found to carry important genes for adaptation to water-stressed environments (Handley et al. 1994).
In the CIM analysis, significance for markers in the target region of chromosome 4A was limited to the second year, indicating a significant genotype by environment effect. The inability to detect this QTL in the first year could have been due to the relatively small number of replicates used in the trial. In this scenario, QTL with small effects would not be detected at the threshold levels set for QTL declaration (Shah et al. 1999). The single marker analysis for year 1 does provide some evidence that the Dharwar Dry alleles in this region possess the ability to influence drought tolerance across environmental conditions.
In this study, the syntenic relationship between rice and wheat was used to attempt to increase marker density and obtain insight into the basis of tolerance to drought. Both BE637912 and CA487065 have been mapped onto BAC clone OSJNBa0071K18. This BAC clone has 35 genes, including an early nodulin gene (Enod), a response regulator gene, and a mitochondrial chaperone-60 protein gene. Genes in the neighboring upstream and downstream BAC clones include a stress inducible protein and a putative flowering-time gene, respectively.
The use of syntenic relationships can, however, be complicated. A consensus group 4A genetic map of wheat from the integrated wheat science database (Komugi) was used for comparison with the map from this study (Fig. 4). Microsatellite markers Xgwm601, Xwmc48, Xwmc89 and Xwmc420 mapped in the same order and at similar distances on the two maps. The consensus map also showed two markers, Xbcd808a-4A and XGermin, fall within the QTL region in this study. Xbcd808a is a barley probe from an EST (Genebank designation BE538742) that codes for an alpha tubulin. A marker based on STS primers designed from this EST mapped to wheat deletion bin 4AL12–0.43–0.59. Germin is a hydrogen peroxide-generating oxalate oxidase with manganese superoxide dismutase (MnSOD) activity and is important for both abiotic and biotic stress resistance (Druka et al. 2002). A wheat EST (BE406676) with sequence homology to the predicted Germin gene on rice chromosome 3L was mapped to deletion bin C-4AL12-0.43. Since the wheat EST homologous to the germin gene maps to C-4AL12-0.43 and is distal to the most significant markers, the QTL in this study is likely to be in the most distal region of C-4AL12-0.43. It should be noted that BE637912 and CA487065 both map to this region of wheat chromosome 4AL but are syntenic to a region on rice chromosome 10L, while Xbcd808a and XGermin map to the same region of wheat 4AL but are syntenic to a region on rice chromosome 3L. The presence of markers based on these genes in our target region indicates that large scale synteny between rice and wheat in this region is not fully preserved.
In summary, Dharwar Dry chromosome 4A appears to carry important genes for production under drought stress. The present results and earlier reports of QTL for earliness per se, water use efficiency, dehydrins (Dhn6), heat shock proteins and early salt induced (Esi) genes in homoelogous Triticeae group 4 chromosomes highlight the importance of this chromosome group for stress tolerance. The QTL is near breakpoint FL 0.43, but further saturation of the region is needed to determine how far the QTL is from the breakpoint. In this study, Xwmc89 was associated with all significant QTL and, in most cases, explained the greatest proportion of phenotypic variation. This marker could therefore be used for marker assisted selection for drought tolerance. Further work to verify the effect of this region in various genetic backgrounds and environments is needed.
References
Araki E, Miura H, Sawada S (1999) Identification of genetic loci affecting amylase content and agronomic traits on chromosome 4A of wheat. Theor Appl Genet 98:977–984
Austin DF, Lee M (1996) Comparative mapping in F2:3 and F6:7 generations of quantitative trait loci for grain yield and yield components in maize. Theor Appl Genet 92:817–826
Bassam BJ, Caetano-Anolles G, Gresshoff PM (1991) Fast and sensitive silver staining of DNA in polyacrylamide gels. Anal Biochem 196:80–83
Bezant J, Laurie D, Pratchett N, Chojecki J, Kearsey M (1997) Mapping QTLs controlling yield and yield components in a spring barley (Hordeum vulgare L.) cross using marker regression. Mol Breed 3:29–38
Börner A, Schumann E, Fürste A, Cöster H (2002) Mapping of quantitative trait loci determining agronomic important characters in hexaploid wheat (Triticum aestivum L.). Theor Appl Genet 105:921–936
Choi H, Hong J, Ha J, Kang J, Kim S (2000) ABFs, a family of ABA-responsive element binding factors. J Biol Chem 275:1723–1730
Churchill GA, Doerge RW (1994) Empirical threshold values for quantitative trait mapping. Genetics 138:963–971
Cox TS, Sears RG, Bequette RK, Martin TJ (1995) Germplasm enhancement in winter wheat x Triticum tauchii backcross populations. Crop Sci 35:913–919
Druka A, Kudrna D, Gamini Kannangara C, von Wettstein D, Kleinhofs A (2002) Physical and genetic mapping of barley (Hordeum vulgare) germin-like cDNAsPNAS 99:850–855
Fischer RA, Maurer R (1978) Drought resistance in spring wheat cultivars. I. Grain yield responses. Aust J Agric Res 29:897–912
Handley LL, Nevo E, Raven JA, Martínez-Carrasco R, Scrimgeour CM, Pakniyat H, Forster BP (1994) Chromosome 4 controls potential water use efficiency (δ13) in barley. J Exp Bot 45:1661–1663
Holland JB (2006) Estimating genotypic correlations and their standard errors using multivariate restricted maximum likelihood estimation with SAS Proc MIXED. Crop Sci 46:642–654
Hu J, Vick BA (2003) Target region amplification polymorphism: a novel marker technique for plant genotyping. Plant Mol Biol 21:289–294
Huang XQ, Börner A, Röder MS, Ganal MW (2002) Assessing genetic diversity of wheat (Triticum aestivum L.) germplasm using microsatellite markers. Theor Appl Genet 105:699–707
Kato K, Miura H, Swada S (2000) Mapping QTLs controlling grain yield and its components on chromosome 5A of wheat. Theor Appl Genet 101:1114–1121
Kirigwi FM, van Ginkel M, Trethowan R, Sears RG, Rajaram S, Paulsen GM (2004) Evaluation of selection strategies for wheat adaptation across water regimes. Euphytica 135:361–371
Lander ES, Green P, Abrahamson J, Barlow A, Daly MJ, Lincoln SE, Newburg L (1987) MAPMAKER: an interactive computer package for constructing primary genetic linkage maps of experimental and natural populations. Genomics 1:174–181
Lantican MA, Pingali PL, Rajaram S (2002) Are marginal environments catching up? In: Ekboir J (ed) CIMMYT world wheat overview and outlook 2000–2001. Developing no-till packages for small-scale farmers. CIMMYT, Mexico, DF
Laurie DA, Pratchett N, Bezant JH, Snape JW (1995) RFLP mapping of five major genes and eight quantitative trait loci controlling flowering time in a winter × spring barley (Hordeum vulgare L.) cross. Genome 38:575–585
Lopez CG, Banowetz GM, Peterson CJ, Kronstad WE (2003) Dehydrin expression and drought tolerance in seven wheat cultivars. Crop Sci 43:577–582
Martins-Lopes P, Zhang H, Koebner R (2001) Detection of single nucleotide mutations in wheat using single strand conformation polymorphism gels. Plant Mol Biol Rep 19:159–162
Mickelson-Young L, Endo TR, Gill BS (1995) A cytogenetic ladder-map of the wheat homologous group-4 chromosomes. Theor Appl Genet 90:1007–1011
Qi L, Echalier B, Friebe B, Gill BS (2003) Molecular characterization of a set of wheat deletion stocks for use in chromosome bin mapping of ESTs. Funct Integr Genomics 3:39–55
Rajaram S, Braun H-J, van Ginkel M (1996) CIMMYT’s approach to breed for drought tolerance. Euphytica 92:147–153
Richards E, Reichardt M, Rogers S (1994) Preparation of genomic DNA from plant tissue. In: Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K (eds) Current protocols in molecular biology, Vol. 1. John Wiley and Sons, New York, pp 231–237
Röder MS, Korzun V, Wendehake K, Plaschke J, Tixier HM, Leroy P, Ganal MW (1998) A microsatellite map of wheat. Genetics 149:2007–2023
Rosegrant MW, Paisner MS, Meijer S, Witcover J (2001) Global food projections to 2020: emerging trends and alternative futures. International food policy research institute (IFPRI), Washington, DC
SAS Institute (1990) SAS Users Guide. Statistics (Version 6.0). SAS Inst, Cary, NC
Shah MM, Gill KS, Baenziger PS, Yen Y, Kaeppler SM, Ariyarathme HM (1999) Molecular mapping of loci for agronomic traits on chromosome 3A of bread wheat. Crop Sci 39:1728–1732
Smith ME, Coffman WR, Baker TC (1990) Environmental effects on selection under high and low-input conditions. In: Kang MS (ed) Genotype-by-environment interaction and plant breeding. Louisiana State University, Baton Rouge, LA, USA, pp 261–272
Sourdille P, Singh S, Cadalen T, Brown-Guedira GL, Gay G, Qi L, Gill BS, Dufour P, Murigneux A, Bernard M (2004) Microsatellite-based deletion bin system for the establishment of genetic-physical map relationships in wheat (Triticum aestivum L.). Funct Integr Genomics 4:12–25
Trethowan RM, Pfeiffer WH (2000) Challenges and future strategies in breeding wheat for adaptation to drought stressed environments: A CIMMYT wheat program perspective. In: Ribaut JM, Poland D (eds) Molecular approaches for the genetic improvement of cereals for stable production in water-limited environments. A strategic planning workshop held at CIMMYT El Batan, Mexico, 21–25 June 1999. CIMMYT, Mexico DF, pp 45–48
Wang S, Basten CJ, Zeng ZB (2001–2004) Windows QTL Cartographer 2.0. Department of Statistics, North Carolina State University, Raleigh, NC
Xiao J, Li J, Yuan L, Tanksley SD (1996) Identification of QTLs affecting traits of agronomic importance in a recombinant inbred population derived from a subspecific rice cross. Theor Appl Genet 92:230–244
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Kirigwi, F.M., Van Ginkel, M., Brown-Guedira, G. et al. Markers associated with a QTL for grain yield in wheat under drought. Mol Breeding 20, 401–413 (2007). https://doi.org/10.1007/s11032-007-9100-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11032-007-9100-3