Abstract
Dasypyrum villosum (Dv), a wild relative of wheat, is an important and useful gene resource for wheat improvement. A large number of wheat-Dv aneuploid lines harboring whole or fragments of Dv chromosomes have been developed. However, the lack of sufficient molecular markers hindered accurate identification of Dv chromatin, especially when the introgressed fragments are small. Development of molecular markers covering the whole Dv genome and evenly distributed on different chromosome regions is not only useful for the detection of the introgressed alien chromatin in wheat background, but also provides evidence of the syntenic relationship between homoeologous chromosomes. In the present study, in order to develop high density and evenly distributed molecular markers on individual Dv chromosomes, genomic DNA of Dv leaves was sequenced and assembled. Sequence assemblies of all wheat chromosomes were first used to identify exon–exon junctions and localize introns in Dv. Intron length polymorphisms suitable for designing Dv primers flanking introns were evaluated, and a total of 1624 intron targeting (IT) markers was designed. By using the Chinese Spring, the Triticum durum-Dv amphiploid and the Dv sequenced DNA libraries, 841 IT molecular markers specific for Dv chromosomes were developed, with maximum efficiency up to 51.79%. We assigned the 841 IT markers to seven Dv chromosomes (1V–7V) using seven wheat–Dv chromosome addition and substitution lines: 135 to 1V, 175 to 2V, 120 to 3V, 89 to 4V, 140 to 5V, 71 to 6V, and 111 to 7V, respectively. Using T. aestivum-Dv telosomic and whole arm translocation lines, they were further located on the short or long chromosome arms. These specific markers for individual chromosomes of Dv provided efficient tools for the characterization of structural variation involving the individual chromosome of Dv, as well as for the selection of useful genes located on individual Dv chromosome in breeding programs.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Dasypyrum villosum (L.) (Candargy, Dv; 2n = 14, VV) (L.) Schur [syn. Haynaldia villosa (L.) Schur], a wild relative of wheat, has been used to introgress genes that confer resistances to powdery mildew, rusts, take-all, eyespot, wheat spindle streak mosaic virus (WSSMV), tolerances to drought and frost, good tiller ability, and high grain protein content (Blanco et al. 1987; Murray et al. 1994; Chen et al. 1995; Grądzielewska 2006; De Pace et al. 2011). A large amount of common wheat-Dv cytogenetic materials, including amphiploid, disomic and ditelosomic addition lines, and disomic substitution lines, as well as translocation lines that display the expression of those useful genes have been developed (Grądzielewska 2006; De Pace et al. 2011). For example, the translocation line T6VS·6AL has both powdery mildew (Pm21 on 6VS) and stripe rust (Yr26 on 1B introduced from Triticum turgidum) resistance. The introduction of 6VS (and corresponding absence of 6AS) has no significant negative effect on yield or quality traits (Chen et al. 1995; Li et al. 2005b, 2007), and T6VS•6AL lines are widely used as parents in wheat breeding programs. More genes have been transferred into wheat by the development of alien translocation lines, including wheat yellow mosaic (WYM) resistance gene Wss1 (Zhang et al. 2005; Zhao et al. 2013), stem rust resistance gene Sr52 (Qi et al. 2011), grain softness genes (Zhang et al. 2010, 2012), seed storage protein genes (Zhao et al. 2010; Vaccino et al. 2010; Zhang et al. 2014), yield-related genes (Zhang et al. 2015), the powdery mildew resistance gene Pm55 (Zhang et al. 2016b) and the cereal cyst nematode resistance gene CreV (Zhang et al. 2016a). Such wheat lines represent excellent resources, and greatly enrich the genetic basis of wheat.
To improve the efficiency of gene transfer from Dv to wheat, it is necessary and important to develop molecular markers linked to useful genes for accelerating wheat plant breeding by marker-assisted selection (MAS). The development of a large number of molecular markers specific for Dv will help to accurately trace the introduced alien chromatin carrying useful alien genes. Although it is a diploid genome, the outcrossing breeding system of Dv makes its genome very complex. Several different types of Dv chromosome-specific molecular markers have been reported, including random amplified polymorphic DNAs (RAPD) (Qi et al. 1996; Li et al. 2005a), restriction fragment length polymorphism (RFLP) (Qi et al. 1996; Li et al. 2005a), simple sequence repeat (SSR) (Zhang et al. 2006), sequence characterized amplified region (SCAR) (Liu et al. 1999; Cao et al. 2006; Chen et al. 2006), conserved-intron scanning primers (CISPs) (He et al. 2013), and expressed sequence tags (EST-PCR) (Wang et al. 2007; Cao et al. 2009; Chen and Chen 2010; Zhao et al. 2012; Zhang et al. 2012; Chen et al. 2013; He et al. 2013; Zhao et al. 2014; Zhang et al. 2015; Bie et al. 2015; Zhang et al. 2016a). However, the number of molecular markers specific for Dv chromosomes is still very limited. Therefore, there is an urgent need to mine more molecular markers, especially those having a wider genomic distribution and amenable to high throughput genotyping, in order to identify alien introgression and translocation lines and also aid molecular genetics and breeding studies involving Dv.
Introns are an attractive source of polymorphism for marker development because insertions/deletions and base substitutions are more common within intron than within exon sequences (Kimura 1983). Intron length polymorphism has been considered as a convenient and reliable source of informative markers with high interspecies transferability (Poczai et al. 2013). Based on the orthologous gene conservation between rice and wheat (Ishikawa et al. 2007), a set of PCR-based landmark unique gene (PLUG) primer pairs were designed; subsequent analysis showed that PLUG markers are suitable not only for detecting the polymorphisms among wheat A, B and D genomes (Ishikawa et al. 2009), but also for identifying the homology between wheat and alien chromosomes. Thus, they are useful in MAS, comparative genomics, alien chromosome tracing, taxonomic studies and genotyping (Poczai et al. 2010; Li et al. 2013; Hu et al. 2012; Zhan et al. 2013).
Next-generation high-throughput DNA sequencing techniques (NGS) have opened opportunities for sequencing an entire plant genome at low cost, allowing fast characterization of genome structure at high resolution and detailed comparative study across species (Egan et al. 2012). All wheat chromosomes have been sequenced using NGS and their gene contents has been determined (International Wheat Genome Sequencing Consortium 2014). These new genomic resources have opened an avenue for efficient high-throughput marker development, high-density mapping, gene cloning, comparative genomics, functional studies and phylogenetic analyses.
In this paper, the Dv genome was sequenced and assembled to develop more specific gene markers. Based on blasting against the genome sequences of Triticum aestivum cv Chinese Spring (Poczai et al. 2013), intron length polymorphisms were identified and chromosome-specific molecular markers for Dv were successfully developed and allocated on specific Dv chromosome arms.
Materials and methods
Plant materials
Triticum durum-Dv amphiploid (AABBVV), T. aestivum-Dv disomic addition lines [DA1V, 3V–7V, in a Chinese Spring (CS) background] and T. aestivum -Dv disomic substitution line DS2V (in a Xiangmai1 background), T. aestivum -Dv translocation lines involving different Dv chromosome arms, including T1VL·1DS(CS), T1VS·1DL(CS), T2VL·2DS(CS), T2VS·2DL(CS), T4VL·4DS(CS), T4VS·4DL(CS), T5VS·5DL(CS), T6VL·6AS(CS) and T6VS·6AL(Yangmai 5) were developed by the Cytogenetics Institute, Nanjing Agricultural University (hereafter CINAU). The Dv (accession No. 91C43) was introduced from Cambridge Botanical Garden (Cambridge, UK).
NGS and assembling of Dv
About 24 μg genomic DNA from young leaves of Dv was used to create the eight shotgun DNA-seq libraries with 626–636 bp and 311–321 bp inserted sizes, respectively. The libraries were sequenced in a single lane of Illumina HiSeq 2500 platform. De novo assembly of the Illumina paired-end reads was performed using the software SOAPdenovo (version 2.0) using k-mer size of 31, which provided the assembly with the best sequence coverage and N50 size to generate Dv genome scaffolds.
Sequence resources for primer design
DNA sequences used in this work included the Dv assembly (SRP077844), assemblies and annotated genes of 21 chromosomes of CS (T. aestivum Ensembl plants, Release 30, http://plants.ensembl.org/index.html).
Homology and alignment analysis
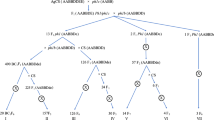
The flowchart for designing the Dv chromosome-specific markers is shown in Fig. S1. In the first step, we chose a set of annotated genes on the subgenomic D genome of CS to calculate the exon–exon junction sizes in the genomic sequences of individual homoeologous groups of wheat chromosomes (WHG1–WHG7) and 1V–7V chromosomes of Dv, respectively. In the second step, all genes were compared with the genomic sequences of individual homoeologous groups of wheat chromosomes (WHG1–WHG7) and chromosomes 1V–7V of Dv using a local Blastn program, respectively. All genes matching individual homoeologous groups of wheat chromosomes (WHG1 to WHG7) and appropriate Dv 1V–7V assemblies and possessing at least one predicted exon–exon junction were selected. Intron sizes of the corresponding genes were then calculated and compared against each other. Genes whose intron sizes in Dv 1V–7V chromosomes differed by at least 10% from those in corresponding allele of individual wheat homoeologous groups (WHG1–WHG7) were chosen for marker design, respectively. The primer pairs were designed in the exons flanking the targeted introns.
Intron targeting primer design
Intron targeting (IT) primers were designed in the exon regions flanking a targeted intron using the online software Primer 3 (version 4.0, http://frodo.wi.mit.edu/primer3//primer3/). The designed primers were expected to amplify the genomic DNA both in common wheat and Dv. The PCR setting for these primers is: melting temperature 55–65 °C (optimum: 60 °C), primer length 18–25 bp (optimum: 20), the estimated sizes of the amplified fragments were approximately 50 bp more than the targeted introns. All primers were synthesized by Shanghai Invitrogen Biotechnology Company (Shanghai, China).
DNA extraction and PCR
Genomic DNA was extracted from 2 g fresh leaves of plants at three-leaf stage with the SDS-phenol-chloroform method according to Sharp et al. (1989) and Devos et al. (1992) and purified to eliminate RNA, amylase and other unwanted compounds. The DNA purity and concentration was assessed with microplate reader (M200, TECan, Männedorf, Switzerland). Each DNA sample was finally diluted to approximately 50 ng/μl and stored at −20 °C until use.
PCR amplification was carried out in a 10-μl reaction volume containing 40 ng genomic DNA, 2 μM each of the primer pairs, 2.5 mM each dNTPs, 2.5 mM MgCl2, 1 × PCR buffer (10 mM Tris-HCl, pH 8.5, 50 mM KCl), and 0.5 U Taq DNA polymerase with a PTC-200 thermal cycler (Bio-Rad, Hercules, CA). The amplification was conducted at 94 °C for 3 min, followed by 35 cycles at 94 °C for 30 s, annealing of different primers at 50, 55, or 60 °C for 50 s at a ramp rate of 0.5 °C/s, 72 °C for 1 min 10 s, and a final extension at 72 °C for 10 min. PCR products were resolved in 8% non-denaturing polyacrylamide gels (Acr: Bis = 19: 1 or 39: 1) and the band patterns were visualized by silver staining (Bassam and Gresshoff 2007).
Results
Amplification patterns of different IT markers
In our previous study, we developed IT markers specific for chromosome arms 4VS and 6VS of Dv by chromosome sorting and NGS. In the present paper, we focused on the remaining arms of Dv chromosomes. A total of 1624 primer pairs was designed, including 265 for 1V, 332 for 2V, 211 for 3V, 151 for 4VL, 328 for 5V, 138 for 6VL and 199 for 7V (Table 1). They were used for amplification in CS, Dv and T. durum-Dv amphiploid. The results indicated that the majority (97.84%) of these primer pairs were suitable for producing PCR amplicons in all the three materials. Only 841 primer pairs (51.79%) amplified specific polymorphic fragments that were common in Dv and T. durum-Dv amphiploid, indicating they were specific for the genome of Dv. We also found that 35 primer pairs had no amplicons in any of the three materials.
Allocation of IT markers to specific Dv chromosomes
To allocate specific markers on Dv chromosomes, the 841 primer pairs were used for amplification in different wheat-Dv addition/substitution lines involving different Dv chromosomes. If a primer pair amplified a distinct PCR product in Dv, T. durum-Dv amphiploid and an alien chromosome line involving one of the Dv chromosomes, while not in T. durum and CS, it might serve as a specific marker for this alien chromosome. To summarize, we assigned 135, 175, 120, 89, 140, 71, and 111 IT specific markers to 1V, 2V, 3V, 4V, 5V, 6V and 7V, respectively (Tables S1, S2).
Some of the amplification results for the 21 primer pairs are shown in Fig. 1. The representative three types of IT markers corresponding to the sequence information in subgenome AA, BB, DD and VV, the sequences of forward and reverse primers, and the intron sizes are given in Tables 2 and 3, respectively. For example, as shown in Fig. 1a, primer CINAU954 produced three bands with different sizes in Chinese Spring (AABBDD), presumably on AA, DD and BB, respectively, according to the predicted amplified size of the flanking intron. The primers also produced a band in Dv (VV) that has different size (500 bp) from those in CS, indicating this band is V genome specific. When amplified in T. durum-Dv amphiploid (AABBVV), CS-Dv addition/substitution lines (DA1V, DA3V–7V and DS2V), the primer pair produced the same band in DA1V and T. durum-Dv amphiploid as that in Dv (VV); thus, we assigned this 500 bp band to chromosome 1V.
Bands in non-denaturing 8% polyacrylamide gels electrophoresis evidencing PCR products of intron targeting (IT) markers specific for individual chromosome of Dv. Lanes: M Marker, 1 Chinese Spring (CS), 2 Dv, 3 T. aestivum-Dv amphiploid, 4 DA1V, 5 DS2V, 6 DA3V, 7 DA4V, 8 DA5V, 9 DA6V, 10 DA7V. a The amplification of representative Type I IT markers (CINAU954, CINAU1068, CINAU1205, CINAU1331, CINAU1454, CINAU1532, CINAU1566) specific for Dv chromosome of 1V, 2V, 3V, 4V, 5V, 6V and 7V from top to bottom, respectively. b The amplification of representative Type II IT markers (CINAU862, CINAU1010, CINAU1162, CINAU1325, CINAU1459, CINAU1542, CINAU1661) specific for Dv chromosome of 1V, 2V, 3V, 4V, 5V, 6V and 7V from top to bottom, respectively. c The amplification of representative Type III IT markers (CINAU893, CINAU1067, CINAU1161, CINAU1295, CINAU1474, CINAU1517, CINAU1608) specific for Dv chromosome of 1V, 2V, 3V, 4V, 5V, 6V and 7V from top to bottom, respectively. Arrows Amplified bands specific for Dv individual chromosomes
We classified the developed IT markers into different types based on the observed two, three or four band patterns on the polyacrylamide gel. Type I markers evidenced four bands, three of which each contained an amplicon from one of the orthologous introns in the three chromosomes of the same homoeologous group of CS (WHG1 to WHG7), and the fourth band containing an amplicon from the orthologous intron in the Dv-chromosome syntenic to that wheat homoeologous group. For example, CINAU954 is 1V specific, and amplified fragments specific for 1V-, 1A- 1B- and 1D- simultaneously (Fig. 1). These markers are co-dominant, and are useful to simultaneously trace the alien Dv chromosome and its wheat homoeologous group chromosomes.
Type II markers evidenced three bands: one band containing amplicons of the same size from orthologous introns (Tables 3, S3, S4) in any of two chromosomes of the same wheat homoeologous group, the second band containing the amplicon from the third chromosome of that homoeologous group, and the third band containing an amplicon from the orthologous intron in the Dv-chromosome syntenic to that wheat homoeologous group. For example, CINAU1010 is specific for 2V and amplified fragments specific for 2V-, and any two of wheat chromosomes 2A- 2B- or 2D- (Fig. 1).
Type III markers evidenced two bands: one band containing amplicons of the same size from introns (Tables 3, S3, S4) on three homoeologous chromosomes of the same wheat homoeologous group, and the second band containing an amplicon from a Dv-chromosome corresponding to that wheat homoeologous group. For example, CINAU1161 is specific for 3V and amplified a specific fragments from 3V-, and another fragment from CS chromosomes (possibly one of the 3A or 3B or 3D), or two CS chromosomes of (3A and 3D, 3D and 3B, or 3A and 3D); or all three chromosomes (3A, 3D and 3B) (Fig. 1).
Allocation of the IT markers to specific arms of Dv chromosomes
To further map these specific IT markers on specific arms of individual Dv chromosomes, all the IT markers specific for Dv chromosomes were used for amplification in different alien translocation lines. Using chromosome 2V as an example, if the amplification of a primer pair specific for 2V generated a common polymorphic band in Dv, T. durum-Dv amphiploid, translocation line T2VS·2DL (or T2VL·2DS), but not in CS, it is considered as a specific marker for chromosome arm 2VS (or 2VL)(Fig. 2, Tables S3, S4). Using those criteria, we assigned 47, 57, 44, 3, 51 and 51 IT specific markers to chromosome arms 1VS, 2VS, 3VS, 4VS, 5VS and 7VS, respectively. Similarly, 88, 118, 76, 86, 89, 71, and 60 IT specific markers were assigned to chromosome arms 1VL, 2VL, 3VL, 4VL, 5VL, 6VL and 7VL, respectively (Tables S1, S2).
Bands in non-denaturing 8% polyacrylamide gel electrophoresis evidencing PCR products of IT markers specific for individual chromosome arms of Dv. Lanes: M Marker; 1 CS; 2 Dv; 3 T.aestivum-Dv amphiploid; 4, 5 different T.aestivum-Dv translocations. a Lanes: 4 T1VL·1DL, 5 T1VS·1DS. The amplification of different representative IT markers (CINAU944, CINAU939, CINAU967 from left to right panels, respectively) specific for chromosome arm 1VS. b Lanes: 4 T2VL·2DS, 5 T2VS·2DL. The amplification of different representative IT markers (CINAU1011, CINAU1015, CINAU1013 from left to right panels, respectively) specific for chromosome arm 2VL. c Lanes: 4 T4VL·4DS, 5 T4VS·4DL. The amplification of different representative IT markers (CINAU1295, CINAU1325, CINAU1319 from left to right panels, respectively) specific for chromosome arm 4VL. d Lanes: 4 DA5V, 5 T5VS·5DL. The amplification of different representative IT markers (CINAU1404, CINAU1363, CINAU1454 from left to right panels, respectively) specific for chromosome arm 5VL. e Lanes: 4 T6VL·6AS, 5 T6VS·6AL. The amplification of different representative IT markers (CINAU1517, CINAU1541, CINAU1532 from left to right panels, respectively) specific for chromosome arm 6VL
Discussion
In recent years, the great accumulation of genetic markers and a large number of DNA sequences have made the study of comparative genomics in the grass family more feasible. It is found, in most cases, that the linear order (colinearity) of genetic markers is well conserved and is retained at the molecular level (microcolinearity) (Gale and Devos 1998). Sponteneous substitution usually occurs among chromosomes belonging to the same homoeologous groups; thus, the identified T. aestivum-Dv substitution lines combined with isozyme analysis have been used successfully for assigning the added V genome chromosomes to specific homoeologous groups (Liu et al. 1995). RFLP and microsatellite molecular markers further confirmed the homoeology relationship between Dv and wheat chromosomes (Qi et al. 1999; Zhang et al. 2006). Zhao et al. (2014) developed EST-based molecular markers and proved that chromosome 4V is highly homoeologous to wheat homoeologous group 4 chromosomes.
However, the above types of molecular markers developed for Dv chromosomes have some disadvantages. RFLPs are dependent on radioactive probes and are also time-consuming. SSRs have low transferability among wild relatives and their application in wheat breeding is limited by the lack of locus specificity (Mullan et al. 2005). Out of 276 SSR markers, 148 could amplify polymorphic amplicons between Dv and CS, but only 7 (wmc49 on 1BS, wmc25 on 2BS, gdm36 on 3DS, gdm145 on 4AL, wmc233 on 5DS, wmc256 on 6AL and gwm344 on 7BL) amplified specific polymorphic DNA fragments from chromosome 1V to 7V, respectively (Zhang et al. 2006). Wheat EST bin maps have been explored as a source for development of specific markers for alien chromosomes, but the polymorphic rate is very low (Qi et al. 2007). For example, Zhao et al. (2014) designed a total of 607 EST-based primer pairs, but only 58 (polymorphic rate being 9.23%) could amplify bands specific for chromosome 4V. A total of 240 EST-based STS primer pairs was designed, and only 13 (polymorphic rate being 5.42%) were specific for chromosomes of Dv (Cao et al. 2009). In the present study, we developed 841 chromosome-specific IT markers for Dv based on NGS technology, and the polymorphic rate was as high as 51.79%, indicating this is a highly efficient approach for developing chromosome-specific markers with higher success rate, specificity, stability and lower cost.
The IT markers discovered in this study are based on the sequence conservation of orthologous genes. All annotated genes from subgenomic D genome of CS were compared with genomic sequences of Dv using a local Blastn program. We found that most genes located on homoeologous group 1 to 7 wheat chromosomes could hit the genes from the corresponding homoeologous 1 to 7 Dv chromosomes, respectively. Using different alien translocation lines previously identified by FISH and molecular markers in our laboratory, these specific IT markers were mapped to specific arms of individual Dv chromosomes. The 1VL (88), 3VL (76), 5VL (89), 6VL (71) and 7VL (60) IT markers were derived from the long arm of the wheat homoeologous group 1, 3, 5, 6, and 7, respectively (Table S2). The IT markers for the 1VS (47), 3VS (44), 5VS (51) and 7VS (51) were derived from the wheat 1S, 3S, 5S and 7S arm, respectively.
However, we also observed some exceptions. For chromosome 2VL, all 118 IT markers were derived from the long arm of wheat group 2, while for 57 IT markers for 2VS, 56 were derived from short arm of wheat group 2, and one (CINAU975) was from the long arm of wheat group 2. For chromosome 4V, all 86 4VL IT markers were derived from the long arm of wheat group 4, and three 4VS IT markers (CINAU1275, CINAU1290 and CINAU1349) were also derived from the wheat 4L. This confirmed that Dv and T. aestivum had a close phylogenetic relationship, and suggested that their collinear regions were highly conserved, reinforcing the homoeologous relationship between Dv and wheat chromosomes (Table S5) (Qi et al. 1999; Zhang et al. 2006). The four exceptions indicated there present complex syntenic relationships between the wheat and Dv genomes, which may be the results of chromosomal rearrangements that most likely occurred during the karyotype evolution of Dv.
The IT markers developed in this study will dramatically increase the density of the Dv physical and cytological map to detect structural changes in Dv chromosomes or arms. Furthermore, some of the IT markers are co-dominant and hence will be very helpful in identifying chromosome changes both for the alien chromosomes and wheat chromosomes in a large population. In breeding program, they can be used to distinguish the homozygous and heterozygous individuals by MAS.
References
Bassam BJ, Gresshoff PM (2007) Silver staining DNA in polyacrylamide gels. Nat Protoc 2:2649–2654
Bie TD, Zhao RH, Jiang ZN, Gao DR, Zhang BQ, He HG (2015) Efficient marker-assisted screening of structural changes involving Haynaldia villosa chromosome 6V using a double-distal-marker strategy. Mol Breeding 35:34
Blanco A, Simeone R, Resta P (1987) The addition of Dasypyrum villosum (L.) Candargy chromosomes to durum wheat (Triticum durum Desf.) Theor Appl Genet 74:328–333
Cao AZ, Wang XE, Chen YP, Zou XW, Chen PD (2006) A sequence-specific PCR marker linked with Pm21 distinguishes chromosomes 6AS, 6BS, 6DS of Triticum aestivum and 6VS of Haynaldia villosa. Plant Breed 125:201–205
Cao YP, Cao AZ, Wang XE, Chen PD (2009) Screening and application of EST-based PCR markers specific to individual chromosomes of Haynaldia villosa. Acta Agro Sin 35:1–10
Chen PD, Qi LL, Zhou B, Zhang SZ, Liu DJ (1995) Development and molecular cytogenetic analysis of wheat-Haynaldia villosa 6VS/6AL translocation lines specifying resistance to powdery mildew. Theor Appl Genet 91:1125–1128
Chen PD, You CF, Hu Y, Chen SW, Zhou B, Cao AZ, Wang X (2013) Radiation-induced translocations with reduced Haynaldia villosa chromatin at the Pm21 locus for powdery mildew resistance in wheat. Mol Breeding 31:477–484
Chen SW, Chen PD (2010) Developmant of the specific EST markers for the 6V chromosome short arm of Haynaldia villosa and location of their deletion. J Triticeae Crops 30:789–795
Chen YP, Wang HZ, Cao AZ, Wang CM, Chen PD (2006) Cloning of a resistance gene analog from wheat and development of a co-dominant PCR marker for Pm21. J Int Plant Biol 48:715–721
De Pace C, Vaccino P, Cionini G, Pasquini M, Bizzarri M, Qualset CO (2011) Dasypyrum. In: Kole C (ed) Wild crop relatives: genomic and breeding resources, vol 1. Springer, Heidelberg, pp 185–292
Devos KM, Atkinson MD, Chinoy CN, Liu CJ, Gale MD (1992) RFLP-based genetic map of the homoeologous group 3 chromosomes of wheat and rye. Theor Appl Genet 83:931–939
Egan AN, Schlueter J, Spooner DM (2012) Applications of next-generation sequencing in plant biology. Am J Bot 99:175–185
Gale MD, Devos KM (1998) Comparative genetics in the grasses. Proc Natl Acad Sci USA 95:1971–1974
Grądzielewska A (2006) The genus Dasypyrum—part 2. Dasypyrum villosum—a wild species used in wheat improvement. Euphytica 152:441–454
He HG, Zhu SY, Sun WH, Gao DR, Bie TD (2013) Efficient development of Haynaldia villosa chromosome 6VS-specific DNA markers using a CISP-IS strategy. Plant Breed 132:290–294
Hu LJ, Liu C, Zeng ZX, Li GR, Song XJ, Yang ZJ (2012) Genomic rearrangement between wheat and Thinopyrum elongatum revealed by mapped functional molecular markers. Genes Genom 34:67–75
International Wheat Genome Sequencing, Consortium (2014) A chromosome-based draft sequence of the hexaploid bread wheat (Triticum aestivum) genome. Science 345:6194
Ishikawa G, Nakamura T, Ashida T, Saito M, Nasuda S, Endo TR, Wu J, Matsumoto T (2009) Localization of anchor loci representing five hundred annotated rice genes to wheat chromosomes using PLUG markers. Theor Appl Genet 118:499–514
Ishikawa G, Yonemaru J, Saito M, Nakamura T (2007) PCR-based landmark unique gene (PLUG) markers effectively assign homoeologous wheat genes to a, B and D genomes. BMC Genomics 8:135
Kimura M (1983) Rare variant alleles in the light of the neutral theory. Mol Biol Evolution 1(1):84–93
Li GP, Chen PD, Zhang SZ, Wang X, He ZH, Zhang Y, Zhao H, Huang HY, Zhou XC (2007) Effects of the 6VS.6AL translocation on agronomic traits and dough properties of wheat. Euphytica 155:305–313
Li H, Chen X, Shi AN, Kong FJ, Leath S, Murphy J, Jia X (2005a) Characterization of RAPD markers and RFLP marker linked to powdery mildew resistant gene derived from different H. villosa. Sci Agric Sin 38:439–445
Li H, Chen X, Xin ZY, Ma YZ, Xu HJ, Chen XY, Jia X (2005b) Development and identification of wheat-Haynaldia villosa T6DL.6VS chromosome translocation lines conferring resistance to powdery mildew. Plant Breed 124:203–205
Li J, Endo TR, Saito M, Ishikawa G, Nakamura T, Nasuda S (2013) Homoeologous relationship of rye chromosome arms as detected with wheat PLUG markers. Chromosoma 122:555–564
Liu DJ, Chen PD, Raupp WJ (1995) Determination of homoeologous groups of Haynaldia villosa chromosomes. Proceedings of the 8th international wheat genetics symposium, vol 1. China Agricultural Scientech, Beijing, China, pp 181–185
Liu Z, Sun Q, Ni Z, Yang T (1999) Development of SCAR markers linked to the Pm21 gene conferring resistance to powdery mildew in common wheat. Plant Breed 118:215–219
Mullan DJ, Platteter A, Teakle NL, Appels R, Colmer TD, Anderson JM, Francki MG (2005) EST-derived SSR markers from defined regions of the wheat genome to identify Lophopyrum elongatum specific loci. Genome 48:811–822
Murray TD, Delapena RC, Yildirim A, Jones SS (1994) A new source of resistance to Pseudocercosporella-Herpotrichoides, cause of eyespot disease of wheat, located on chromosome-4V of Dasypyrum-villosum. Plant Breed 113:281–286
Poczai P, Cernak I, Gorji AM, Nagy S, Taller J, Polgar Z (2010) Development of intron targeting (IT) markers for potato and cross-species amplification in Solanum nigrum (Solanaceae). Am J Bot 97:e142–e145
Poczai P, Varga I, Laos M, Cseh A, Bell N, Valkonen JP, Hyvonen J (2013) Advances in plant gene-targeted and functional markers: a review. Plant Methods 9:6
Qi L, Cao M, Chen P, Li W, Liu D (1996) Identification, mapping, and application of polymorphic DNA associated with resistance gene Pm21 of wheat. Genome 39:191–197
Qi L, Friebe B, Zhang P, Gill BS (2007) Homoeologous recombination, chromosome engineering and crop improvement. Chromosom Res 15:3–19
Qi LL, Chen PD, Liu DJ, Gill BS (1999) Homoeologous relationships of Haynaldia villosa chromosomes with those of Triticum aestivum as revealed by RFLP analysis. Genes Genet Syst 74:77–82
Qi LL, Pumphrey MO, Friebe B, Zhang P, Qian C, Bowden RL, Rouse MN, Jin Y, Gill BS (2011) A novel Robertsonian translocation event leads to transfer of a stem rust resistance gene (Sr52) effective against race Ug99 from Dasypyrum villosum into bread wheat. Theor Appl Genet 123:159–167
Sharp PJ, Chao S, Desai S, Gale MD (1989) The isolation, characterization and application in the Triticeae of a set of wheat RFLP probes identifying each homoeologous chromosome arm. Theor Appl Genet 78:342–348
Vaccino P, Banfi R, Corbellini M, De Pace C (2010) Improving the wheat genetic diversity for end-use grain quality by chromatin introgression from the wheat wild relative Dasypyrum villosum. Crop Sci 50:528–540
Wang CM, Feng YG, Zhuang LF, Cao YP, Qi ZJ, Bie TD, Cao AZ, Chen PD (2007) Screening of chromosome-specific markers for chromosome 1R of Secale cereale, 1V of Haynaldia villosa and 1Rk# 1 of Roegneria kamoji. Acta Agron Sin 33:1741–1747
Zhan HX, Li GR, Chang ZJ, Zhang XJ, Li X, Yang ZJ (2013) Molecular identification of a new wheat-Thinopyrum intermedium cryptic translocation line for resistance to powdery mildew. IJBBB 4:376–378
Zhang QP, Li Q, Wang X, Wang HY, Lang SP, Wang YN, Wang SL, Chen PD, Liu DJ (2005) Development and characterization of a Triticum aestivum-Haynaldia villosa translocation line T4VS.4DL conferring resistance to wheat spindle streak mosaic virus. Euphytica 145:317–320
Zhang R, Hou F, Feng Y, Zhang W, Zhang M, Chen P (2015) Characterization of a Triticum aestivum-Dasypyrum villosum T2VS.2DL translocation line expressing a longer spike and more kernels traits. Theor Appl Genet 128:2415–2425
Zhang RQ, Wang XE, Chen PD (2012) Molecular and cytogenetic characterization of a small alien-segment translocation line carrying the softness genes of Haynaldia villosa. Genome 55(9):639–646
Zhang RQ, Cao YP, Wang XE, Feng YG, Chen PD (2010) Development and characterization of a Triticum aestivum-H. villosa T5VS·5DL translocation line with soft grain texture. J Cereal Sci 51:220–225
Zhang RQ, Feng YG, Li HF, Yuan HX, Dai JL, Cao AZ, Xing LP, Li HL (2016a) Cereal cyst nematode resistance gene CreV effective against Heterodera Filipjevi transferred from chromosome 6VL of Dasypyrum villosum to bread wheat. Mol Breeding 36:122
Zhang RQ, Sun BX, Chen J, Cao AZ, Xing LP, Feng YG, Lan CX, Chen PD (2016b) Pm55, a developmental-stage and tissue-specific powdery mildew resistance gene introgressed from Dasypyrum villosum into common wheat. Theore Appl Genet 129:1975–1984
Zhang RQ, Zhang MY, Wang XE, Chen PD (2014) Introduction of chromosome segment carrying the seed storage protein genes from chromosome 1V of Dasypyrum villosum showed positive effect on bread-making quality of common wheat. Theore Appl Genet 127:523–533
Zhang W, Gao AL, Zhou B, Chen PD (2006) Screening and applying wheat microsatellite markers to trace individual Haynaldia villosa chromosomes. Acta Genet Sin 33:236–243
Zhao R, Wang H, Xiao J, Bie T, Cheng S, Jia Q, Yuan C, Zhang R, Cao A, Chen P, Wang X (2013) Induction of 4VS chromosome recombinants using the CS ph1b mutant and mapping of the wheat yellow mosaic virus resistance gene from Haynaldia villosa. Theor Appl Genet 126:2921–2930
Zhao RH, Wang HY, Jia Q, Xiao J, Yuan CX, Zhang YJ, Hu QS, Wang XE (2014) Development of EST-PCR markers for the chromosome 4V of Haynaldia villosa and their application in identification of 4V chromosome structural aberrants. J Integr Agr 13:282–289
Zhao WC, Dong J, Gao X, Chen Q, Li XY, Shi YG, Chen LG (2012) Development and application of EST-STS markers for 1V chromosome arms of Haynaldia villosa. J Triticeae Crops 32:393–397
Zhao WC, Qi LL, Gao XA, Zhang GS, Dong JA, Chen QJ, Friebe B, Gill BS (2010) Development and characterization of two new Triticum aestivum-Dasypyrum villosum Robertsonian translocation lines T1DS.1V#3L and T1DL.1V#3S and their effect on grain quality. Euphytica 175:343–350
Acknowledgements
This research was supported by the National Key Research and Development Program (2016YFD0102001), the National Natural Science Foundation of China (Grant No. 31571653, 31201204, 31501305), the National High Technology Research Program (‘863’ Program) of China (Grant No. 2011AA10010201), ‘948’ Project of Ministry of Agriculture (2015-Z41), the Fundamental Research Funds for the Central Universities (KYZ201403; KJ2013003), International Cooperation and Exchange of the National Natural Science Foundation of China (Grant No. 31661143005), the Technology Support Program of Jiangsu Province (Grant no. BE2015352-2), the Jiangsu Agricultural Science and Technology Innovation Fund (CX151001), the Jiangsu “333 high-level personnel training project”, the Program of Introducing Talents of Discipline to Universities (No. B08025), A Project Funded by the Priority Academic Program Development of Jiangsu Higher Education Institutions (PAPD) and Six Talent Peaks project in Jiangsu Province.
Author’s contribution
W.X.E., W.H.Y. and X.J. designed experimental plan. Z.X.D., W.X. and Y.C.X. performed all experiments. X.J., W.Y.F., C.A.Z. and X.L.P. carried out sequence analysis and designed IT markers. C.P.D. contributed to the development of the germplasms used in the experiments. Z.S.Z. managed the materials in the field. W.H.Y., W.X.E. and Z.X.D. wrote the manuscript. All authors have read and approved the final manuscript.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Key message
The V-genome specific IT markers were mined to facilitate the identification and utilization of Dv chromatin.
Electronic supplementary material
Fig. S1
Flowchart of designing the specific markers for Dv chromosome (DOC 30 kb)
Table S1
IT markers corresponding to the sequence information in subgenome AA, BB, DD (Triticum aestivum Ensembl plants, Release 30) and VV(SRP077844) (XLSX 98 kb)
Table S2
The sequences of forward and reverse primers and the size of intron of the IT markers (XLSX 90 kb)
Table S3
Catalog of some of the Type I, II, and III markers displaying nucleotide sequence homoeology in sequence scaffolds of different chromosome arms of the wheat sub-genome A, B, D, and V genomes (DOCX 17 kb)
Table S4
Primer sequences for Type I, II, and III IT markers used in the amplification of different alien translocation lines (DOCX 17 kb)
Table S5
Comparative analysis of Dv genome with Chinese Spring subgenomic D genome (DOCX 15 kb)
Rights and permissions
About this article
Cite this article
Zhang, X., Wei, X., Xiao, J. et al. Whole genome development of intron targeting (IT) markers specific for Dasypyrum villosum chromosomes based on next-generation sequencing technology. Mol Breeding 37, 115 (2017). https://doi.org/10.1007/s11032-017-0710-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11032-017-0710-0