Abstract
As an ecologically and economically important seaweed, Saccharina japonica has developed strategies to utilize HCO3-. In general, seaweeds have at least three mechanisms for HCO3- acquisition, one of which is the use of extracellular carbonic anhydrases (CA) to convert HCO3- into CO2 for utilization. However, it is unknown which CA in S. japonica performs this function. In order to find the extracellular CA in S. japonica that exerts utilization of HCO3-, in this study, the cloning, characterization, and subcellular localization of Sjβ-CA are described. This enzyme has a full-length cDNA of 1397 bp with a 170-bp 5’-untranslated region (UTR), a 282-bp 3’-UTR, and a 945-bp open reading frame encoding a protein precursor consisting of 314 amino acids which contains a predicted 28-residue signal peptide. Enzyme activity assays showed that the recombinant Sjβ-CA in Escherichia coli possessed CO2 hydration and dehydration activities, thus identifying this gene Sjβ-CA in function. Immunogold electron microscopic observations with the prepared anti-Sjβ-CA polyclonal antibody illustrated that Sjβ-CA was located in periplasmic space of the kelp gametophyte cells. A positive correlation between the gene transcription and the level of exogenous HCO3- utilization was also established. These findings provide a molecular and cellular basis for understanding the mechanism of inorganic carbon absorption of this important kelp.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The economically important brown seaweed Saccharina japonica (Areschoug) C. E. Lane, C. Mayes, Druehl et G. W. Saunders (Lane et al. 2006), previously known as Laminaria japonica, is a kelp native to the cold temperate coasts from Okhotsk Sea southwards to Japan Sea of the northwestern Pacific Ocean (Tseng 1981; Zhang et al. 2015). It inhabits naturally in the sublittoral zone, where the concentration of dissolved CO2 in water, usually used by algae and plants as a substrate for photosynthesis, can get to 12 μM (Millero 2013), which is very similar to that in air. However, the diffusion coefficient for CO2 in water is four orders of magnitude lower than that in air (Maberly and Gontero 2017). This diffusional resistance to CO2 supply can potentially lead to restricted photosynthetic rates and slow growth. In contrast, the annual productivity of S. japonica grown naturally in Ehime, Japan, for example, has been reported up to 1.0 to 1.4 kg C m−2 (Suzuki et al. 2006). These rates of biomass are comparable to that of tropical rainforests, which are believed to have the highest terrestrial productivity as suggested by Suzuki et al. (2006). It is the reason that S. japonica has developed a strategy to remove HCO3-, which accounts for 85.86% of the total dissolved inorganic carbon (DIC) (Millero 2013), in addition to CO2 from seawater during the evolutionary process, as examined in a large number of brown seaweeds (Surif and Raven 1989; Larsson and Axelsson 1999; Klenell et al. 2004; García-Sánchez et al. 2016). The utilization of HCO3- by S. japonica as an inorganic carbon source for photosynthesis has well been corroborated by Ji et al. (1980) using carbon isotope labeling method.
As summarized by Prins and Elzenga (1989), at least three main mechanisms have been proposed for HCO3- acquisition by seaweeds. Of these mechanisms, the extracellular dehydration of HCO3- to CO2, which is catalyzed by external or periplasmic carbonic anhydrase (CA, EC 4.2.1.1), has been documented in the majority of brown seaweeds as reviewed by Johnston (1991), Badger (2003), and Bi et al. (2019b). The resulting CO2 is then taken into the seaweed cell by diffusion either through plasma membranes (Missner et al. 2008) or via plasmalemma-located aquaporins (Uehlein et al. 2017). In S. japonica, Yue et al. (2001) found that either 4′, 4′-diisothiocyanatosilbene-2, 2-disulfonic acid (DIDS) or 4-acetamido-4′-isothiocyano-2, 2′-stibene-disulfonate (SITS), the inhibitors of plasmalemma-located anion exchange (AE) proteins, had little inhibitory effects on HCO3- acquisition, thus implying that the kelp diploid sporophytes are unable to take directly up HCO3- through AE proteins. When treated with acetazolamide (AZ), an inhibitor of periplasmic CA, Yue et al. (2001) estimated that approximately 75% inorganic carbon acquisition by S. japonica was inhibited. These experimental data suggest that this kelp could take up exogenous HCO3- via periplasmic CA proteins in the same way as many species of brown seaweeds (Bi et al. 2019b). With this mechanism for HCO3- acquisition, S. japonica is capable of possessing high photosynthetic rates which is even higher than sugarcane and other C4 plants (Gao & McKinley 1994).
CA is a metalloenzyme that catalyzes the reversible interconversion of CO2 and HCO3- (Badger 2003; Bi et al. 2019b). It is widely distributed throughout nature, from eukaryotes such as animals (Thiry et al. 2008), plants (Rudenko et al. 2021), and algae (Moroney et al. 2011), to prokaryotes such as archaea and bacteria (Smith and Ferry 2000). The known CA proteins are grouped into eight distinct families, namely α, β, γ, δ, ζ, η, θ, and ι, which are phylogenetically unrelated and possess little to no sequence or structural similarity (Langella et al. 2022). In the haploid gametophytes of S. japonica, a periplasmic CA, i.e. Sjα-CA2, has been characterized and its subcellular localization has also been determined by immuno-electron microscopy (Bi et al. 2021c). Although the transcription of Sjα-CA2 has been reported to be induced by elevated HCO3- levels in the medium (Bi et al. 2021c), the expected positive correlation between Sjα-CA2 transcripts and HCO3- utilization has not yet been conducted. It is speculated that another periplasmic CA could be attributed to the dehydration of exogenous HCO3- as documented by Yue et al. (2001) and Bi et al. (2021b).
In the present study, after searching the transcriptome database of S. japonica (Wang et al. 2023), one 1 041-bp contig coding for a peptide with a conserved β-CA domain was screened, and it was then cloned from the gametophytes of S. japonica. On the basis of in silico analysis, the open reading frame (ORF) of this gene without a putative signal peptide-corresponding cDNA was cloned and then expressed in Escherichia coli for functional identification and polyclonal antibody preparation. Subcellular localization of the gene product was determined using an immuno-electron microscope technique with the prepared antibody. Based on the detected gene transcripts by quantitative real-time PCR (qRT-PCR) and seawater physicochemical parameters, a positive correlation between the gene transcripts and utilized levels of exogenous HCO3- was established. This is another report for the periplasmic CA of S. japonica. The findings provide a molecular and cellular basis for understanding the mechanism of inorganic carbon absorption of this important kelp.
Materials and methods
Gametophytes and culture conditions
Saccharina japonica gametophyte clones germinated from zoospores were isolated according to cell size under a microscope (Zhou and Wu 1998). They were cultured under the vegetative growth conditions of 30 μmol photons m−2 s−1 at 17 ± 1 °C with a photoperiod of 12 h:12 h (light/dark) as described previously (Zhou and Wu 1998). PES medium (Starr and Zeikus 1993) was replaced once every 2 weeks.
To analyze the transcript levels of target genes at different concentrations of CO2 and NaHCO3 by qRT-PCR, the gametophytes collected by centrifugation at 1 500 × g for 5 min were stirred with a magnetic stirrer at 100 rpm. Four hours later, the dissociated fragments from the aggregated gametophytes were transferred into fresh PES medium for 3 − 4 days to allow them to recover from the mechanical damage. Approximately 15 g of fresh weight gametophytes were cultured separately in conical flasks containing 800 mL of PES medium either agitated with filtered air (low CO2) or 3% CO2 (high CO2) at 200 mL min−1 or supplemented with NaHCO3 at a final concentration of 0.018 M in a shaker at 85 rpm under the previously described growth conditions. All the treatments for the culture of female or male gametophytes were simultaneously repeated three times. After incubation for 0, 8, 16, 24, 32, 40, and 48 h, samples were harvested with the same centrifugation for RNA isolation, and the supernatant was for seawater chemistry assay as described as follows.
Nucleic acid extraction and cDNA synthesis
The collected samples were washed three times with sterilized seawater, and then they were ground into a powder in liquid nitrogen with a mortar and pestle. Genomic DNA was extracted from female and male gametophytes separately according to the modified cetyltrimethyl ammonium bromide (CTAB) method as described previously (Hu and Zhou 2001).
Total RNA was extracted using TRIzol reagent (Invitrogen, USA) from the female and male gametophytes separately. The quality and quantity of the isolated RNA were determined by measuring the absorbance at 260/280 nm (A260/A280) and 260/230 nm (A260/A230). RNA samples only with an A260/A280 ratio between 1.8 and 2.0 and an A260/A230 ratio greater than 2.0 were used for subsequent experiments. Agarose gel electrophoresis (1%) was used to evaluate the integrity of the extracted DNA and RNA samples. Complementary DNA used for 3′ rapid amplification of cDNA ends (RACE) was synthesized using a SMART RACE cDNA kit (Clontech, USA) according to the manufacturer’s protocol.
Complementary DNA and genomic DNA cloning of Sjβ-CA
Through searching the transcriptome database of S. japonica obtained by Wang et al. (2013), one 1 041-bp contig was annotated as a β-CA gene. According to the contig sequence, the forward and reverse primers of CAV (Supplementary Table 1) were designed online using the Primer3web server v. 4.1.0 (https://bioinfo.ut.ee/primer3/) for this contig calibration. Twenty five-microliter reaction volume contained 1 μL the synthesized cDNA, 12.5 μL 2 × Pfu PCR MasterMix (Tiangen Biotech, China), 0.5 μL each forward and reverse primers (10 μM) of CAV and 10.5 μL distilled deionized (dd) H2O. Polymerase chain reaction (PCR) was performed in a gradient Mastercycler (Eppendorf, Germany) programmed as follows: 1 cycle of 5 min pre-denaturation at 95 °C, then 35 cycles including 45 s denaturation at 95 °C, 45 s annealing at 63 °C, and 90 s extension at 72 °C, and followed by 1 cycle of 10 min extension at 72 °C.
The amplified product was recovered using an agarose gel purification and extraction kit (Aidlab, China) and was ligated to pMD19-T vector (TaKaRa, Japan) by T4 DNA ligase. The constructed vector was subsequently transformed into E. coli DH5α competent cells (TaKaRa) by a heat shock method (Hanahan 1983). Then the liquid transformed bacteria were evenly spread on solid Luria–Bertani (LB) medium containing 100 μg mL−1 ampicillin, 20 μg mL−1 5-bromo-4-chloro-3-indol β-D-galactopyranoside (X-gal), and 50 μg mL−1 isopropyl-β-D-1-thiogalactoside (IPTG) for blue-white screening. White or positive clones were selected for verification by PCR with the general primers RV-M and M13-20 (TaKaRa) and sent to Sangon Biotech (Shanghai, China) for sequencing analysis using an automated DNA sequencer (ABI Prism 3730, USA).
According to the verified contig sequence, two primers, 3′GSP1 and 3′GSP2 (Supplementary Table 1), were designed for the 3′-RACE PCR reactions. Of these primers, 3′GSP2 was used in the second round of nested PCR reactions. The obtained 3′-RACE product was recovered and ligated to pMD19-T (TaKaRa), and the constructed vector was transformed into E. coli DH5α competent cells (TaKaRa) as described above. Afterwards, the positive clones were sent to Sangon Biotech (Shanghai) for sequencing analysis. In combination with the sequenced 3′-RACE product and the verified contig fragment, full-length cDNA of the target gene was assembled by DNAMAN 5.2.9 software (Lynnon BioSoft, USA). The cDNA sequence was confirmed by PCR amplification using re-designed primers (data not shown).
On the basis of the verified full-length cDNA sequence of target gene, the forward and reverse primers of CA-1 through CA-5 (Supplementary Table 1) were designed for DNA cloning by PCR with the extracted genomic DNA as template. Twenty five-μL reaction volume contained 1 μL genomic DNA, 12.5 μL 2 × Pfu PCR MasterMix (Tiangen Biotech), 0.5 μL each forward and reverse primers and 10.5 μL ddH2O. PCR was programmed as pre-denaturation at 94 °C for 3 min, and then pre-denaturation at 94 °C for 45 s, annealing at the designed temperature (Supplementary Table 1) for 45 s, and extension at 72 °C for 2 min for 35 cycles, and finally extension at 72 °C for 10 min. When the target products were obtained and sequenced, DNAMAN 5.2.9 software (Lynnon BioSoft) was also used to assemble the obtained fragments into the DNA sequence of target gene. The gene structure was brought to light by comparing its corresponding cDNA using Splign program embedded in NCBI website (https://www.ncbi.nlm.nih.gov/sutils/splign/splign.cgi).
Bioinformatics and phylogeny analysis of Sjβ-CA
ORF Finder (https://www.ncbi.nlm.nih.gov/orffinder/) was used to predict open reading frame (ORF) of Sjβ-CA. The deduced amino acids from this gene were translated by Primer Premier 5 software (Premier Biosoft, USA). Isoelectric point (pI), molecular mass, transmembrane region, hydrophobicity, signal peptide, transit peptide, structure domain, functional site, and secondary structure of this putative Sjβ-CA were predicted and shown using the listed websites or servers in Supplementary Table 2.
With Sjβ-CA as a query sequence, β-CA protein sequences were retrieved from NCBI by protein Blast (https://blast.ncbi.nlm.nih.gov/Blast.cgi), and phylogenetic inference was constructed using MEGA 11 program (Tamura et al. 2021) by Maximum Likelihood (ML) and neighbor-joining (NJ) algorithms based on the β-CA amino acid sequences. Homologous sequences of β-CA proteins from different organisms were aligned by Clustal_X software (Thompson et al. 1997) with default parameter settings.
Construction of prokaryotic expression vector
The cDNA corresponding to the mature Sjβ-CA (mSjβ-CA) that resulted from the removal of a putative signal peptide was amplified with the the forward and reverse primers of heCA (Supplementary Table 1). The PCR reaction and program was the same to DNA cloning of Sjβ-CA, except for the template, primers and annealing temperature (Supplementary Table 1). The target product was separated by agarose gel electrophoresis and then recovered using the aforementioned purification and extraction kit (Aidlab). The recovered DNA fragments were ligated to pMD19-T vector (TaKaRa). The resultant constructs were transformed to E. coli DH5α competent cells (TaKaRa) and sequenced as previously described. The transformants harboring the correct orientation and reading frame of Sjβ-CA were used for isolation of cloning plasmids. The isolated plasmids and the empty vector pET-28a were separately digested by restriction endonucleases BamHI and HindIII. The linearized target products were ligated to generate the prokaryotic expression plasmid pET28a-SjβCA. The resulting constructs were transformed to E. coli DH5α competent cells (TaKaRa) for proliferation and sequencing analysis, and then transformed to E. coli BL21 (DE3) pLysS competent cells (Biocolor BioScience, Shanghai, China) for heterologous expression of SjβCA.
Expression and detection of recombinant mSjβ-CA
For expression analysis of recombinant mSjβ-CA, the transformed E. coli harboring the construct pET28a-SjβCA was cultured in liquid LB medium supplemented with 50 μg mL−1 kanamycin (Kan) on a 108 rpm orbital shaker at either 37 °C or 30 °C. When A600 of the culture reached 0.6, IPTG was added at a final concentration of 1 mM. The culture was shaken for another 0, 2, 3, and 4 h at the same temperature, and then harvested by centrifugation at 3 145 × g for 5 min at 4 °C. The collected samples were washed and re-suspended in phosphate-buffered saline (PBS) (0.137 M NaCl, 2.7 mM KCl, 10 mM Na2HPO4, and 2 mM KH2PO4). The re-suspended mixture was frozen and thawed repeatedly with liquid nitrogen for 3 times, followed by sonication with Scientz-IID Ultrasonic Homogenizer (China) at 4 °C for the lysis of bacterial cells. The supernatant and pellet fractions were separated by centrifugation at 20 379 × g for 10 min at 4 °C. Proteins in both fractions, of the pellet which was added with the supernatant equal volume of 1 × PBS, were quantified by a ND-2000c spectrophotometer (NanoDrop) using Bradford’s method (Bradford 1976). The expressed proteins were analyzed by denaturing sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) according to Laemmli (1970).
Western blotting analysis
After electrophoresis by 12% SDS-PAGE, proteins on the gel were electronically transferred onto a nitrocellulose membrane for Western blotting analysis according to Ye et al. (2014). Protein blots on the nitrocellulose membrane were blocked with 5% skim milk powders in Tris-buffered saline Tween-20 buffer (TBST) (0.137 M NaCl, 2.7 mM KCl, 0.025 M Tris, and 500 μL Tween 20 at pH 7.4). Recombinant mSjβ-CA heterologously expressed in E. coli and native Sjβ-CA in S. japonica gametophytes were immuno-blotted with the commercially supplied anti-6 × His tag polyclonal antibody (Youke Biotech, China) and the purified anti-Sjβ-CA polyclonal antibody, respectively, as the primary one. After incubation in a suitable dilution of primary antibodies for 1 h at room temperature, the nitrocellulose membrane was washed in TBST several times and incubated with the secondary antibody, anti-rabbit IgG labeled by horseradish peroxidase (Youke Biotech) diluted 1:1 200 in TBST at room temperature for 1 h and washed again. The color reaction was visualized with diaminobenzidine following the manufacturer’s instructions (Tiangen Biotech). The Western blotting analysis was performed with gradient dilutions of primary antibodies until the hybridization signals were strong with less background.
Purification of recombinant mSjβ-CA and preparation of polyclonal antibody
To mitigate the impact of protein mis-folding on antibody preparation and enzyme activity, recombinant mSjβ-CA expressed in the supernatant was extracted as described above from the transformed E. coli carrying the construct pET28a-SjβCA incubated for 3 h at 30 °C. After that, the extracts were loaded onto Bio-Scale Mini Profinity IMAC Cartridges nickel columns (Bio-Rad, USA) for affinity chromatography purification of the recombinant mSjβ-CA using the fused 6 × His tag. The recombinant mSjβ-CA was eluted with different designed concentrations, i.e. 5, 10, 50, 100, 150, 200, and 250 mM, of imidazole, following the manufacturer’s protocol (Bio-Rad). The eluted fraction containing target product was collected after checking by the aforementioned SDS-PAGE.
The affinity-purified recombinant mSjβ-CA was dialyzed in a 20 mM Tis-HCl (pH 8.0) solution for 12 h. Afterwards, the purified and dialyzed recombinant mSjβ-CA was employed by Youke Biotech to immunize New Zealand white rabbits. After four immunizations, antiserum was collected from the immunized rabbits by centrifugation at 10 379 × g for 30 min to prepare the anti-Sjβ-CA polyclonal antibody. The antibody was purified using a CNBr-activated Sephrose 4B antigen affinity chromatography column (GE Healthcare, Sweden) following the manufacturer’s protocols. Antibody titer was determined by indirect ELISA detection. Specificity or quality of the purified anti-Sjβ-CA polyclonal antibody was checked by Western blotting analysis using the purified recombinant mSjβ-CA as well as crude proteins extracted from S. japonica gametophytes using RIPA lysis and extraction buffer (Thermo Scientific, USA) containing 1 mM phenylmethanesulfonyl fluoride as a protease inhibitor.
Enzyme activity assay of recombinant mSjβ-CA
While obtaining the purified recombinant mSjβ-CA, its CO2 hydration activity was assayed according to Wilbur and Anderson (1948). The reaction was initiated by adding 3 mL of a saturated aqueous CO2 solution into ice-cold barbiturate buffer (pH 8.4) containing about 0.3 mg purified recombinant mSjβ-CA. One unit of enzyme activity was defined as U = (t0-t)/t, where ‘t0’ and ‘t’ denote the time taken for one unit decrease in pH in the absence and presence of recombinant mSjβ-CA, respectively.
The HCO3- dehydration activity of recombinant mSjβ-CA was assessed according to Kikutani et al. (2016) with a slight modification. One milliliter of the purified recombinant mSjβ-CA (ca. 0.3 mg mL−1) was added to 4 mL of pre-cooled 50 mM 2-morpholinoethanesulfonic acid-NaOH buffer (pH 5.5). When the pH meter reading was stable, 2 mL of pre-cooled 50 mM NaHCO3 solution was added to initiate the reaction. One unit of enzyme activity was defined as the same as CO2 hydration, in which ‘t0’ and ‘t’ represented the time required for the non-enzyme and recombinant mSjβ-CA catalytic reactions, respectively, to increase the pH from 5.7 to 6.0.
Both enzyme activity assays of recombinant mSjβ-CA were performed at 4 °C for three times. Thus, the specific activity was expressed as ‘U mg−1 protein’ as mean ± standard deviation (SD).
Immunoprecipitation
After extraction of total proteins from S. japonica gametophytes using the RIPA lysis and extraction buffer (Thermo Scientific), immunoprecipitation (IP) was performed with the purified anti-Sjβ-CA polyclonal antibody according to Liu et al. (2022). Briefly, 10 μL of the purified polyclonal antibody and 150 μL of crude extracts containing native Sjβ-CA were mixed with 200 μL of IP lysis and wash buffer in the Pierce Classic IP Kit (Thermo Scientific) for incubation overnight at 4°C to form immune complexes. At the same time, 20 μL protein A/G resin slurry was pipetted into a spin column and centrifuged at 100 × g for 1 min. Subsequently, the antibody:antigen complexes were loaded to the spin column, and the non-target proteins were eluted and discarded according to the IP Kit manufacturer’s instructions (Thermo Scientific).
Approximately 50 μL of 2 × non-reducing lane marker sample buffer containing 20 mM dithiothreitol was added to the column and incubated at 100 °C for 10 min. The prepared samples were then fractioned by the earlier described SDS-PAGE. The gel was stained with Coomassie Brilliant Blue R250 and the target band gel was excised for mass spectrometry (MS) analysis.
In-gel digestion and MS analysis
Prior to MS analysis, the band gel was cut into pieces and treated according to the described procedures by Kussmann and Roepstorff (2000). The further details of operation process refer to Liu et al. (2022). The processed and lyophilized samples were sent to Bioprofile Technol (Shanghai, China) for MS analysis using an Easy-nLC 1200 system (Thermo Scientific) and a Q Exactive Plus Orbitrap mass spectrometer (Thermo Scientific).
Raw tandem mass spectra were visualized and processed with the Xcalibur 3.0 package (Thermo Scientific). Peptide identification was performed by either correlating the acquired experimental MS/MS spectra with theoretical spectra or searching them against protein sequence databases, for example, from the NCBI website, following the guidelines laid out by Nesvizhskii (2007). Only the best scoring peptide to spectrum match for each MS/MS spectrum was considered to be the potential peptide identification as suggested by Nesvizhskii (2007).
Immunogold electron microscopy
The subcellular localization of Sjβ-CA in S. japonica gametophytes was investigated using immunogold electron microscopy as described by Ye et al. (2014). Freshly harvested S. japonica gametophytes were fixed with 8% (w/v) paraformaldehyde and 6% glutaraldehyde in sterilized seawater, and then subjected to post-fixation with 0.5% osmium tetroxide. The post-fixed samples were dehydrated in ethanol/acetone series from 30 to 100% ethanol and then were infiltrated and embedded in Epoxy resin Epon 812 (Zhongjingkeyi Technol., China) as described by Ouyang et al. (2012). Seventy-nm thick thin-sections were made using a LKB Ultrotome 4802 Ultracut microtome (Leica, Germany).
The ultrathin sections used for immuno-electron microscopic observations were collected on 200-mesh nickel grids (Zhongjingkeyi Technol) with a noncarbonated Formvar-supporting film. The nickel grids carrying ultrathin sections were etched with 1% (w/v) sodium metaperiodate resolved in PBS (Bendayan and Zollinger 1983). Subsequently, the sections were preincubated with 50 mM glycine for 30 min (Miller and Howell 2006) and blocked with 5% (w/v) bovine serum albumin (BSA) in PBS for 20 min. Finally, these treated sections were incubated with the purified anti-Sjβ-CA antibody at 4 °C for 48 h or with the primary antibody absent as a control. The optimal working titer of the primary antibody was based on serial dilutions of the antibody from 1:1000 to 1:3600 as described by Miller and Howell (2006). After washing in PBS and pre-incubating for another 20 min with 1% (w/v) BSA, the sections were incubated with the secondary antibody, antirabbit IgG conjugated to 10 nm gold particles (Sigma, Germany) at room temperature for 1 h. Following sequential washes in PBS and water, and dehydration in the air, the sections were stained with 3% uranyl acetate-lead citrate, and observed in a Tecnai G2 Sprit BioTWIN transmission electron microscope (FEI, USA) at 80 kV.
The labeling density was defined as the number of gold particles per area unit (μm2) as described by Bernal et al. (2007), and the area was estimated using Adobe Photoshop software (ver. 3.0). Following counting the gold particles in periplasmic space or the other areas by subtracting the periplasmic space in each micrograph, the percentage of particles versus the total ones was calculated. A total of 11 images were used for the calculation.
Monitoring of carbonate system parameters of the PES medium
The gametophytes of S. japonica in this experiment were cultivated in PES medium (Starr and Zeikus 1993) with the aforementioned different inorganic carbon sources. Once the gametophytes were harvested as described above every 8 h until 48 h, the centrifuged culture medium was filtrated with 0.2 μm Whatman Polycap TC filter capsule (GE Healthcare Life Sciences, USA) and then used for monitoring of carbonate system parameters. The medium pH was detected using a FiveEasy Plus FE28 pH meter (Mettler-Toledo, China). Total alkalinity of the culture solution was estimated by a T860 automatic potentiometric titrator (Hanon Advanced Technol., China). Dissolved inorganic carbon (DIC) of the medium was measured by TOC-L analyzer (Shimadzu, Japan), and salinity was measured by LS-10 T salinometer (Mingrui, China). Carbonate parameters were calculated from the DIC, salinity and pH measurements of the culture solution corresponding to each treatment according to the formula of Stumm and Morgan (1996). Three parallels were set up for each sample and the results were expressed as mean ± SD.
Quantitative RT-PCR analysis of Sjβ-CA
Freshly collected S. japonica male and female gametophytes were employed for total RNA extraction using the aforementioned approach. First-strand cDNA was synthesized using the Reverse Transcribed Kit II (TaKaRa). Quantitative RT-PCR was analyzed in a Bio-Rad CFX96 Touch Real-time PCR detection system (Bio-Rad) using SYBR RT-PCR kit (TaKaRa) as described previously (Ye et al. 2014). The 18S rRNA gene of S. japonica (GenBank Accession No. EU293553.1) was used as internal reference, and all primers used for qRT-PCR analysis are listed in Supplementary Table 1. Three sets of replicate experiments were performed for each sample. The relative transcriptional levels of Sjβ-CA were presented as mean ± SD using the 2−ΔΔCT method (Livak and Schmittgen 2001).
Statistical analyses
The statistical analysis of the subcellular distribution of Sjβ-CA was carried out using the two-tailed Student’s t-test. A two-way ANOVA was employed to estimate the gene transcription of Sjβ-CA among different time and inorganic carbon sources. Variance of individual carbonate system parameter among different culture time was tested using a one-way ANOVA. All these statistical analyses were performed using SPSS 26.0 software (IBM Corp., USA). A significance level of 5% was set for all tests.
Results
Gene cloning of Sjβ-CA from Saccharina japonica
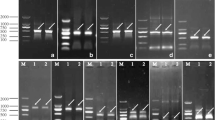
With the primer CAV (Supplementary Table 1), the screened contig was verified by PCR amplification (Lane 1 in Fig. 1). After two-round PCR amplification by RACE approach, a 3′-end product of target gene was amplified, and it was 526 bp long (Lane 2 in Fig. 1). After assembly using DNAMAN software and amplification with re-designed primers (data not shown), a full-length cDNA of target gene was obtained. It was 1 397 bp in length with a 170-bp 5′-UTR and a 282-bp 3′-UTR. The 3′-UTR had a consensus sequence AATAA close to stop codon and the downstream sequence from here on was enriched with GT, thus suggesting that this cloned cDNA would be a full-length one. Using ORF Finder, open reading frame (ORF) of this cloned cDNA was predicted online to be 945 bp long, and it encoded a precursor composed of 314 amino acids with a predicted molecular mass of 34.525 kDa (Supplementary Table 2).
Electrophoresis of amplified products for cDNA and DNA cloning of Sjβ-CA (A) and schematic structure of this gene (B) Lane 1: the amplified products of 3′-RACE; Lane 2: the full-length cDNA of Sjβ-CA; Lane 3: with H2O instead of genomic DNA as a control; Lanes from 4 through 8: the amplified products with primers CA-5, CA-4, CA-3, CA-2, and CA-1, respectively; Lanes M and M1: DL-2000 marker and Marker IV, respectively, of standard DNA (Tiangen Biotech, Beijing, China); Exon and intron in Image B are shown in black boxes and lines, respectively, and UTRs are denoted by gray lines
Based on the assembled full-length cDNA of Sjβ-CA, its DNA sequence was amplified with the designed primers from CA-1 through CA-5 (Supplementary Table 1). After cloning (Fig. 1, Lanes from 4 through 8), sequencing, assembling by DNAMAN 5.2.9 software, and validation with re-designed primers (data not shown), its DNA sequence was obtained. It was 8 810 bp in length, and the ORF of Sjβ-CA was separated by six introns which were 3 311 bp, 213 bp, 1 214 bp, 1 028 bp, 642 bp, and 1 005 bp long, respectively, from 5′-UTR on (Fig. 1, Image B). Both cDNA and DNA sequences of Sjβ-CA were deposited in GenBank under the accession Nos: ARM53418.1 and KY041784.1, respectively.
Characterization and of Sjβ-CA
By a homologous search through BlastP, it got to know that Sjβ-CA, the product of Sjβ-CA, just had a percent identity of 43.63%, 40.96%, and 35.78% with its known function homologs from the red alga Porphyridium purpureum (Mitsuhashi et al. 2000), the green alga Chlamydomonas reinhardtii (Ynalvez et al. 2008), and the higher plant Pisum sativum (Majeau and Coleman 1991), respectively. Nevertheless, multiple sequence alignment (Fig. 2) illustrated that there was a highly conserved Pro_CA domain in these β-CA sequences. As predicted by SMART, this characteristic domain (SM00947) was situated from 109A to 272D of Sjβ-CA (Fig. 2), which was embraced in a predicted β-CA domain (SSF53056) located between 78N and 280 K by InterPro analysis (Supplementary Table 2). In these aforementioned domains, the strictly conserved residues 121C, 123D, 177H, and 180C were suggested to be zinc-binding sites (Fig. 2), while 112Q, 114P, 124S, 125R, 137G, 140F, 162Y, 167L, and 261Y were to be active sites (Mitsuhashi et al. 2000). In addition, 64.97% of Sjβ-CA sequence was modeled by Phyre v. 2.0 with 100.0% confidence by the single highest scoring template, X-ray structure of the β-CA from P. purpureum R-1 (c1ddzA_) (Supplementary Table 2). From these pieces of information it was inferred that Sjβ-CA might function as a β-CA in S. japonica gametophytes.
Multisequence alignment of amino acid sequences of β-CA proteins from the selected species. The putative signal peptides are underlined. Amino acids with high identities more than 75% are shaded in gray, and the conserved ones are shaded in black. Zinc-binding sites and active site clefts are predicted online by InterPro and denoted by upper triangles (Δ) and asterisks (*), respectively. The GenBank accession numbers of these selected β-CA proteins are as follows: CBN77745.1 (Ectocarpus siliculosus), BAA12981.1 (Porphyridium purpureum), ABS87675.1 (Chlamydomonas reinhardtii), AAA33652.1 (Pisum sativum), and ARM53418.1 (Saccharina japonica)
A signal peptide was predicted to be present in the deduced protein of Sjβ-CA by SignalP 5.0, Phobius, PredictProtein, and Protein Prowler v. 1.2 (Supplementary Table 2). Although the signal peptide length was predicted to be inconsistent, most of the predicted results suggested that the cleavage site might be between 28Thr and 29Gly (Fig. 2). Once this signal peptide was cleaved off (Choo et al. 2005; Emanuelsson et al. 2007), the mature Sjβ-CA (mSjβ-CA) was consisted of 286 amino acids, and the predicted molecular mass of mSjβ-CA was thereby reduced to 31.59 kDa. As predicted by TargetP-2.0 and BaCelLo (Supplementary Table 2), mSjβ-CA would enter the secretory pathway under the guidance of its signal peptide, thus meaning that the mature protein could function after secreting to the periplasmic space or specific organelles (Choo et al. 2005) of S. japonica gametophytes.
Phylogeny of algal β-CA proteins
Phylogenetic analysis showed that 96 homologs of β-CAs were significantly clustered into three clades with bootstrap support of 99%, 91%, and 100%, respectively (Fig. 3). Saccharina japonica β-CA was grouped with the majority of green algal β-CA proteins to constitute Clade I. The remaining β-CA proteins of green algae including C. reinhardtii CAH4, CAH5, and CAH6 were grouped with a few of cyanophytic or cyanobacterial β-CA proteins into Clade III, while Clade II predominantly contained the β-CA proteins of Streptophyta (Fig. 3). Clade III stood at the root of this constructed ML phylogenetic tree, indicating that this clade was close to ancestral β-CA.
Maximum Likelihood phylogenetic tree inferred from the deduced amino acid sequences of CA genes from several species. The evolutionary history was inferred by using the Maximum Likelihood method and LG model. The tree with the highest log likelihood (− 16,233.51) is shown. The percentage of trees in which the associated taxa clustered together is shown next to the branches. Initial tree(s) for the heuristic search were obtained automatically by applying Neighbor-Join and BioNJ algorithms to a matrix of pairwise distances estimated using the JTT model, and then selecting the topology with superior log likelihood value. A discrete Gamma distribution was used to model evolutionary rate differences among sites (5 categories (+ G, parameter = 2.0648)). The rate variation model allowed for some sites to be evolutionarily invariable ([+ I], 1.73% sites). The tree is drawn to scale, with branch lengths measured in the number of substitutions per site. This analysis involved 96 amino acid sequences. All positions with less than 95% site coverage were eliminated, i.e., fewer than 5% alignment gaps, missing data, and ambiguous bases were allowed at any position (partial deletion option). There were a total of 173 positions in the final dataset. Evolutionary analyses were conducted in MEGA11
Clade I was divided further into three sub-branches (Fig. 3). The model green alga Chlamydomonas reinhardtii CAH7 and CAH8 were grouped in one sub-branch only consisting of green algal β-CA proteins (94% bootstrap proportion), while S. japonica β-CA was clustered with its homologs from Ochrophyta, Cryptophyta, Haptophyta, and Alveolata to constitute another sub-branch (99% bootstrap proportion, Fig. 3). The red algal sub-branch (Porphyra umbilicalis and Porphyridium purpureum) was between these two mentioned sub-branches (Fig. 3). The sub-branch where Sjβ-CA situated contained a red algal β-CA (GenBank accession No. QWE79112.1) from the extant species Neopyropia yezoensis, thus showing the phylogeny between this sub-branch and the red algal one. In combination with a 96% bootstrap support for these two sub-branches in the NJ phylogenetic inference (Supplementary Fig. 2), the β-CA gene of S. japonica was proposed to evolve from an ancestral red alga.
It was surprised to find that a cyanobacterium Nostoc linckia z16 β-CA (GenBank accession No. PHK25687.1) stood at the root of this sub-branch where Sjβ-CA situated (Fig. 3). In addition, several β-CA proteins from Cyanophyta were positioned in Clades II and III (Fig. 3), implying that these cyanophytes played a possible role in the origin and evolution of algal β-CA genes as put forward by Hewett-Emmett (2000).
Prokaryotic expression and purification of Sjβ-CA
Using primers heCA-F and heCA-R (Supplementary Table 2), a product of 870 bp in size was amplified and then subcloned to generate the vector pMD19T-SjβCA. After digestion by the combined restriction endonucleases BamHI and HindIII, the 870-bp target product (Supplementary Fig. 1, Lane 2) was ligated to the digested pET-28a (Supplementary Fig. 1, Lane 4) to generate the recombinant plasmid pET28a-SjβCA. Then the construct pET28a-SjβCA was verified by the following restriction endonuclease cleavage (Lane 1, 3 and 4 in Supplementary Fig. 1) and sequencing.
Compared to the strain carrying empty plasmid alone as a negative control (Lanes 4, 5, and 9 in Supplementary Fig. 3), cell lysate of the transformant with the construct pET28a-SjβCA presented a dark-color protein band of about 36 kD (Lanes 2 and 3 in Supplementary Fig. 3). The size of this band seemed close to that which was composed of both the target protein (31.59 kD) and a 6 × His tag plus a peptide (3.84 kD in total) as coded by multiple cloning site sequences upstream of Sjβ-CA in the construct pET28a-SjβCA. This band was hence expected to be the recombinant mSjβ-CA.
Western blotting analysis results showed that comparing to the control (Lane 13 in Supplementary Fig. 3), only one signal was present in the line transformed with the construct pET28a-SjβCA (Lane 12 in Supplementary Fig. 3). This signal was the same as one while co-developing with the purified recombinant mSjβ-CA (Lane 14 in Supplementary Fig. 3). In combination with the aforementioned result of molecular mass, these immunoblotting profiles denoted that the recombinant protein might be mSjβ-CA fused with a 6 × His tag.
From the electrophoresis profile (Lane 7, and 8 in Supplementary Fig. 3), it was inferred that the 36-kD band mainly emerged in the insoluble fractions. After decreasing the culture temperature to 30 °C, SDS-PAGE profiles of the cell lysate of transformed line showed that mSjβ-CA could be expressed in both forms of the supernatant and inclusion bodies (Lanes 15 through 22 in Supplementary Fig. 3). In comparison, induction culture for 3 h at 30 °C was more favorable for the expression of mSjβ-CA in the supernatant of E. coli as illustrated by Supplementary Fig. 3 (Lanes 19 vs. 20).
After purification by affinity chromatography with different concentrations of imidazole, electrophoresis profiles of the eluted products (Lane 27 in Supplementary Fig. 3) showed a clear band at approximately 36 kD while eluting with the concentration of 200 mM imidazole. The target protein was thereby purified with this concentration of imidazole for the use of enzyme activity assay and antibody preparation.
Identification of Sjβ-CA by enzyme activity detection
CO2 hydration activity of the purified recombinant mSjβ-CA was detected with the established in vitro reaction system. It spent 98.83 ± 6.306 s (n = 6) for the purified recombinant mSjβ-CA in reducing the reaction system pH by 1 unit. By contrast, it took 151.00 ± 1.549 s (n = 6) when this recombinant protein was absent. Comparison of these two results suggested that this recombinant mSjβ-CA was able to accelerate CO2 hydration, and the specific activity for mSjβ-CA was estimated to be 1.53 ± 0.154 U⋅mg−1 protein (n = 6, Fig. 4).
Similar to the above CO2 hydration assay, the in vitro reaction system of dehydration of HCO3- was established. The purified recombinant mSjβ-CA took 18.33 ± 0.577 s (n = 6) to raise the pH of reaction system by 0.3 unit, but it took 35.67 ± 2.309 s (n = 6) while this recombinant protein was not supplied. The specific activity of HCO3- dehydration for mSjβ-CA was thereby estimated to be 3.05 ± 0.226 U mg−1 protein (n = 6, Fig. 4).
These biochemical data provided direct evidence that the recombinant mSjβ-CA could catalyze the reversible conversion of CO2 and bicarbonate (CO2 + H2O ⇔ HCO3- + H+), thus functionally identifying this gene Sjβ-CA cloned from S. japonica gametophytes.
Preparation and specificity verification of anti-Sjβ-CA polyclonal antibody
After immunization, antiserum was successfully collected from the immunized rabbits and purified. The valence of the purified anti-Sjβ-CA polyclonal antibody was 1:80 000 by an indirect ELISA check, thus suggesting that it could meet the requirements of subsequent experiments.
Western blotting analyses of the purified polyclonal antibody with crude proteins (upper Image A in Fig. 5) extracted from S. japonica female or male gametophytes showed that two distinguishable blotting signals appeared precisely at the same sites as the theoretical molecular mass of precursor (34.525 kD) and mature protein (31.59 kD) of Sjβ-CA (lower Image A in Fig. 5).
Mass spectrometry (MS) spectrum of a digested polypeptide (B) of the immunoprecipitated protein (A) from Saccharina japonica gametophytes with the purified anti-Sjβ-CA polyclonal antibody, and location of this detected polypeptide in the deduced amino acids (C) encoded by Sjβ-CA. The underlined residues in (C) indicate peptide sequences as detected by MS, and the red ones show the amino acid sequence corresponding to the detected one in (B). Lane M: Item #SM0671 (lot specific) PageRuler™ Prestained Protein Ladder (MBI Fermentas, Rockford, Canada); and Lane M1: Item #TSP021 Trelief® Prestained Protein Ladder (Tsingke Biotech, Beijing, China)
To further illustrate that these two protein bands were presented by the blotted target proteins, native Sjβ-CA was isolated from the crude extracts of S. japonica by IP approach with the purified anti-Sjβ-CA polyclonal antibody. After SDS-PAGE analysis these two bands were excised and digested for MS analysis. Of the larger but lighter protein, a total 10 peptide fragments were detected and they constituted 4 longer peptides as found in Image C of Fig. 5 due to overlapping among them. They were composed of 57 amino acids, which were completely matched the corresponding amino acid sequence of Sjβ-CA (Image C in Fig. 5), although the detected amino acids only accounted for 18.15% (i.e. 57/314) of the total ones. With respect to the smaller but darker one, a total of 49 peptide fragments were detected. Although they constituted 4 longer peptides as well as shown in Supplementary Fig. 3, they consisted of 233 amino acids, which accounted for 81.47% (i.e. 233/286) of the total ones. The detected amino acids also matched well the deduced ones of mSjβ-CA in sequence. In combination with the immunoblotting profiles, the MS analysis confirmed that the immunoprecipitated protein was the native Sjβ-CA but took on in two forms, i.e. precursor and mature protein, in S. japonica gametophytes.
In addition, the MS analysis could explain why the two immunoblotting signals were different from each other in size. In the smaller or mature protein, the first detected peptide fragment from its N terminus on was GFVLTAGGSGATTALGK (Supplementary Fig. 4). However, upstream of the corresponding position of this detected peptide fragment in the larger protein or precursor, other 28 residues were present. Moreover, one peptide fragment, AQTRAALR (Images B and C in Fig. 5), was detected by MS from these 28 residues in the larger rather than smaller protein. The last amino acid of the 28-residue peptide was Thr, and it was coincidentally neighbor of the first residue, Gly, in the first detected peptide of the smaller one by MS. This was consistent with the predicted digestion site between 28Thr and 29Gly (Fig. 2) by SignalP 5.0 as well by PredictProtein and Protein Prowler v. 1.2 (Supplementary Table 2). From these pieces of information, it was inferred that these 28 residues could not be detected by MS in the smaller one being as a signal peptide for removal after targeting to destination.
It was necessary to denote that this detected peptide fragment AQTRAALR was not considered for the heterologous expression of mSjβ-CA since it was located at the predicted signal peptide. After IP experiment, AQTRAALR (Images B and C in Fig. 5) was detected by MS, thus showing the specificity of this prepared anti-Sjβ-CA polyclonal antibody which could be reacted immunologically with both mature protein and precursor of Sjβ-CA. Based on the strength of immunoblotting signals (Image A in Fig. 5), the mature protein was expressed more abundantly than its precursor in S. japonica gametophytes. As a consequence, the numbers of detected peptide fragments in the mature protein (i.e. 49) far surpassed that in the precursor (i.e. 10) as described earlier. It thus suggested that mSjβ-CA would be the predominant form that functioned in the kelp gametophytes, both female and male (Image A in Fig. 5).
Subcellular localization of Sjβ-CA using immunoelectron microscopy
After determining the optimal antibody concentration by Western blotting analysis, this concentration of purified anti-Sjβ-CA polyclonal antibody could be employed in the subcellular localization using immunoelectron microscopy. In the immunocolloidal gold electron micrographs of S. japonica gametophyte cells (Fig. 6), it was found that 97.01% (i.e. 267/275, n = 11) of colloidal gold particles were spread in cell wall or periplasmic space with 2.91% (i.e. 8/275, n = 11) in other areas. This immunocytological evidence supported that the mSjβ-CA could be secreted to the periplasmic space of S. japonica gametophyte cells as predicted by TargetP-2.0, BaCelLo, and CELLO v.2.5 rather than to chloroplasts by WoLF PSORT (Supplementary Table 2). This was also reflected by the density of immunocolloidal gold particles and its statistical analysis. The density in the periplasmic space was estimated to be 13.46 ± 8.784 gold particles μm−2 (n = 11) averagely, which was significantly (P = 0.006 < 0.05, t10 = 4.187) higher than that in the other compartments (1.88 ± 2.644 gold particles μm−2, n = 11).
Transmission electron micrographs showing the immunogold labeling distribution of Sjβ-CA in the gametophyte cells of Saccharina japonica. (A) Ultrastructure micrograph of a gametophyte cell of S. japonica; (B), (C), and (D) are the enlarged images corresponding to the marked areas 1, 2, and 3, respectively, in Image A; Gold particles are denoted by white or black arrows; CW, cell wall; Ch: chloroplast
These immunocolloidal gold electron micrographs thereby provided direct evidence that Sjβ-CA was located in the periplasmic space of S. japonica gametophyte cells. Taken together with the in vitro detection of enzyme activity (Fig. 5), it was speculated that Sjβ-CA might be involved in the conversion of environmental HCO3- to CO2, thus facilitating the rapid entry of CO2 into cells for the efficient utilization of HCO3- from seawater.
Relationship between Sjβ-CA transcription and the consumed HCO3 - in PES medium
To investigate into the possible role played by the periplasmic Sjβ-CA in inorganic carbon utilization, S. japonica gametophytes were incubated under various concentrations of inorganic carbon supplemented with CO2 or NaHCO3. During the cultivation for 48 h, the relative transcription levels of Sjβ-CA in S. japonica female and male gametophytes were estimated by qRT-PCR at every 8 h. When S. japonica gametophytes were bubbled with filtered air for the culture as a control, the mRNA levels of Sjβ-CA tended towards a decrease but there were no any significant differences among them (Fig. 7). While using 3% CO2 instead of filtered air for the culture, the gene transcripts in the cultured gametophytes fluctuated between 0.4 and 1.5 without any significant differences neither (Fig. 7). It was concluded that CO2 had few effects on the transcription of Sjβ-CA in S. japonica gametophytes no matter what they were female or male ones.
Changes in transcription patterns over a 48 h incubation of Sjβ-CA in Saccharina japonica gametophytes cultured in PES medium supplemented with different carbon sources. (A) is for female gametophyte of S. japonica, and (B) is for male one. The different lowercase letters on the columns denote the significant difference among them (P < 0.05). Standard deviation bars (n = 3) are shown
When NaHCO3 was supplemented to cultivate S. japonica gametophytes in a shaker, it was found that there was no significant variation in the transcription of Sjβ-CA before 16 h (Fig. 7). Subsequently, the mRNA levels of Sjβ-CA began to increase gradually. By the end of this experiment, the transcription levels of Sjβ-CA reached the maximum and were 7.80 ± 0.94 for the female and 8.05 ± 0.81 for the male, which were approximately eight times as that at the initial stage (0 h, Fig. 7) of this experiment. Obviously, the kelp gametophytes could gradually increase the transcripts of Sjβ-CA in response to the addition of NaHCO3 into the PES medium. In combination with the aforementioned subcellular location of periplasmic Sjβ-CA, this gene Sjβ-CA was proposed to make a positive contribution to adapt the kelp gametophytes to the elevated NaHCO3.
To understand this, the carbonate system parameters including HCO3- concentration in the medium were determined. When 18 mM NaHCO3 was supplemented to the PES medium, the HCO3- concentration was elevated from the lowest 13.988 mg L−1 without addition of any inorganic carbon for the male gametophyte culture to the highest 276.268 mg L−1 also for the male gametophyte culture (Supplementary Table 3). In the process of cultivation the HCO3- concentrations in the medium showed a gradually decreasing trend over time (Supplementary Table 3). By the end of this experiment, the HCO3- concentrations in the medium lowered to 173.122 mg L−1 for the male gametophyte culture (Supplementary Table 3). Regarding the capacity of periplasmic Sjβ-CA as illustrated by Fig. 4, it was expected to be responsible for the consumption of HCO3-. This was reflected by the established negative correlation (r = − 0.943, P = 0.001 in female; r = − 0.925, P = 0.003 in male) between the transcription levels of Sjβ-CA and the contents of remaining HCO3- in the medium (Table 1). In other words, the expression levels of Sjβ-CA were significantly positive correlated to the utilized HCO3- in the gametophytes of S. japonica. Surely, the non-enzymatic conversion of CO2 from HCO3- cannot be excluded. In such a high value of pH (> 7.7 as shown in Supplementary Table 3) supplied with NaHCO3, however, this conversion was restricted to some extent, so that HCO3- was the predominant form of inorganic carbon (Millero 2013). Accordingly, the non-enzymatic conversion of CO2 from HCO3- possibly made few contributions to the reduction of HCO3- in the medium.
Without supplying with any inorganic carbon for culture, the CO2 concentrations ranged from 0.119 to 0.162 mg L−1 (incubation for 0 h excluding NaHCO3 treatment in Supplementary Table 3). These were equivalent to 2.70 μM and 3.68 μM CO2, respectively. They were higher than the CO2 compensation point 1.635 μM as reported by Yue et al. (2000). Nevertheless, these CO2 concentrations were much lower than the Michaelis–Menten affinity constant (Km = 19.4 ± 1.2 μM) of RuBisCO for CO2 which was obtained from S. latissima by Iñiguez et al. (2019), since the data was not available from S. japonica. These comparisons pointed out that supplying with inorganic carbon could help S. japonica for photosynthesis and growth, which has already been evidenced in the gametophytes by Yue et al. (2000) and in the sporophytes by Zhang et al. (2020).
Whenthe kelp gametophytes were aerated continuously with filtered air (low CO2), the CO2 concentrations in the PES medium were improved well, ranging from 0.224 to 0.762 mg L−1 (Supplementary Table 3) equivalent to 5.09 μM and 17.31 μM CO2, respectively. However, they were still lower than the Km of RuBisCO for CO2. It was reasonable to cultivate the kelp gametophytes with elevated CO2. Once 3% CO2 (high CO2) was bubbled continuously into the medium, the CO2 concentrations ranged as expected from 32.799 to 87.159 mg CO2 L−1 (Supplementary Table 3) corresponding to 745.27 μM and 1980.46 μM CO2. At the same time, pH decreased from about alkaline 7.9 to acidic 5.4 (Supplementary Table 3), also reflecting the increase in CO2 concentration in the medium. (Millero 2013). In this case, CO2 might become a non-limiting factor for S. japonica gametophyte photosynthesis. It thus seemed that the existence of periplasmic CA proteins was unnecessary. As a result, no significant correlation between the changed levels of any carbonate system parameters and the gene transcription levels of either Sjβ-CA (Table 1) or Sjα-CA2 (Supplementary Table 4) was obtained while bubbling the the of kelp gametophyte cultures with 3% CO2.
Discussion
From the high-throughput RNA-sequencing data of the juvenile sporophytes of S. japonica (Deng et al. 2012; Wang et al. 2023) and the unigene sequences of the kelp gametophytes (Ye et al. 2015), 12 genes coding for S. japonica CA were retrieved and compiled by Bi and Zhou (2016). Eleven of these CA genes from the kelp gametophytes were sequenced and annotated by Bi et al. (2019a) using single-molecule real-time sequencing technique. Of these 11 CA genes, two belonging to α-CA (Ye et al. 2014; Bi et al. 2021c) and one to γ-CA (Bi et al. 2021a) classes have been characterized in detail. The present study reports another gene of CA which has two Cys and one His residues that coordinate Zn2+ in the highly conserved Pro_CA domain (Fig. 2), a feature of the β-CA family reported for the first time in this kelp in brown seaweeds.
According to Rowlett (2014), the β-CA genes were first recognized as an evolutionarily distinct class of carbonic anhydrase in 1990 when the DNA sequence of the gene coding for this enzyme from Spinacea oleracea was obtained (Burnell et al. 1990). Afterwards, β-CA has been found at least from both the green alga Chlamydomonas reinhardtii (Eriksson et al. 1996; Ynalvez et al. 2008; Yu et al. 2020; Rai et al. 2021) and red algae such as Porphyridium purpureum (Mitsuhashi et al. 2000), Gracilariopsis chorda (Razzak et al. 2019), and Neopyropia yezoensis (Wang et al. 2020; Zhang et al. 2022). Although several papers have discussed the origin and evolution of general β-CA genes (Hewett-Emmett and Tashian 1996; Hewett-Emmett 2000; Banerjee and Deshpande 2016), higher plant β-CA genes (Ludwig 2011, 2016), and bacterial β-CA genes (Capasso and Supuran 2015), there is little information on algal β-CA genes possibly due to fewer functionally identified genes available. The present study thereby attempts to unveil the phylogenetic relationship of algal β-CA genes.
Origin and evolution of algal β-CA genes
The ancestors of modern cyanobacteria or cyanophytes evolved O2-generating photosynthesis some 3 500 million years ago (MYA) (Dyall et al. 2004; Falcón et al. 2010). It is now generally accepted that an ancestral cyanobacterium was engulfed by a non-photosynthetic protist via primary endosymbiosis to give rise to three photosynthetic lineages, i.e. red algae, glaucophyte algae, and green algae and their land plant descendants (Moreira et al. 2000; Rodríguez-Ezpeleta et al. 2005; Keeling 2010; Sibbald and Archibald 2020). This endosymbiosis event has occurred between about 1 500 to 900 MYA as estimated by Yoon et al. (2004) and Shih and Matzke (2013). Approximately 1 300 MYA (Yoon et al. 2004), a new eukaryotic host took up up a red algal ancestor to form a diverse range of red-algal-derived photosynthetic eukaryotes, such as Ochrophyta, Cryptophyta, Haptophyta, and Alveolata, by secondary endosymbiosis (Janouškovec et al. 2010; Green 2011; Archibald 2012; Gentil et al. 2017), though this hypothesis is under debate. Regardless of horizontal or lateral gene transfer, the endosymbiont genome is thus thought to have two donors: one from a cyanobacterium-like or rhodophyte-like ancestor and the other from a eukaryote cell. As illustrated by the constructed ML phylogenetic tree (Fig. 3), three separate clades are supported by higher bootstrap proportions, thus suggesting that algal β-CA genes could have been donated at least by two different contributors.
As shown by Fig. 3 and Supplementary Fig. 2, most extant cyanobacteria are grouped in the two closer clades, Clade II and Clade III, and these two clades are positioned at the root of this constructed phylogenetic tree. Therefore, green algal β-CA genes such as the model organism C. reinhardtii Cah4, Cah5, and Cah6 in Clade III are suggested to originate from a cyanobacterial ancestor. Compared to the primitively diverged Cah6, the co-existence of Cah4 and Cah5 in C. reinhardtii possibly occurred recently by duplication because of 96% identity in their cDNA sequences (Eriksson et al. 1996). These green algal β-CA genes pass from lower plants such as mosses to seed plants such as Arabidopsis thaliana. Nevertheless, the β-CA genes of three cyanobacteria (Coleofasciculus chthonoplastes, Synechocystis sp. PCC 6803, and Synechococcus elongatus PCC 6301) are surprisingly closer to those of two species of seed plants (Ensete ventricosum and Tanacetum cinerariifolium) by a 100% bootstrap support (Fig. 3) than to charophytes, mosses, and liverworts. This anomalous dendrogram suggests a horizontal gene transfer event could take place recently between these cyanobacteria and seed plants. Such a gene transfer possibly speeds up the divergence of streptophyte β-CA genes (Clade II) from the green algal ones (Clade III) (Fig. 3). Accordingly, the β-CA genes of these two clades, especially Clade II, are considered to acclimate themselves to terrestrial environmental conditions since horizontal gene transfer is thought to drive the evolution and adaption of land plants (Schönknecht et al. 2014; Wang et al. 2023).
The algal β-CA genes in Clade I have diverged so greatly that they can be divided into three distinct sub-groups (Fig. 3) with 94%, 66%, and 99% bootstrap proportion supports. Interestedly, these three sub-groups correspond well to the primary endosymbiosis-derived green algae and red algae including the descendants of the latter one, showing that they are monophyletic. Fewer extant cyanobacteria are grouped in this clade, indicating that these algal β-CA genes might not be of cyanobacterial origin. Regarding to the two contributors to the endosymbiont genome as earlier described, the algal β-CA genes in Clade I are supposed to be donated by the non-photosynthesis eukaryotic host. It is of interest that the red algae and their descendants possess the eukaryote-donated β-CA genes alone, whereas the green lineage have both eukaryote-donated and cyanobacteria-donated β-CA genes. Accordingly, the cyanobacteria-donated β-CA genes present in Clades II and III were possibly lost for some unknown reason while forming the red algae lineage through primary endosymbiosis. Such an evolutionary process explains to some extent why the green and red algae lineages differ from each other. But how the non-photosynthesis eukaryotic host in the secondary endosymbiosis affects the evolution of algal β-CA genes remains to be investigated further.
In the sub-group where Sjβ-CA resides two red algae, Porphyra umbilicalis and Porphyridium purpureum, stand at the root of the constructed NJ phylogenetic tree by a 96% bootstrap support (Supplementary Fig. 2). Based upon this, the β-CA genes of S. japonica and the other red-algal-derived organisms as illustrated in Fig. 3 in this sub-group are proposed to descend from a unicellular ancestral rhodophyte through secondary endosymbiosis. It is worth noting that only a cyanobacterium Nostoc linckia z16 CA (GenBank accession No. PHK25687.1) is positioned at the root of red-algal-derived algae (Fig. 3), reflecting that a gene transfer from this cyanobacterium to those eukaryotic algae might occur.
Unlike the evolution from green algae in Clade III to land plants in Clade II, the green algal β-CA genes in Clade I stop succeeding to streptophytes due to the absence of land plants (Fig. 3). Instead, their homologues in red algae can pass to the red algal derivatives (Supplementary Fig. 2) via the aforementioned secondary endosymbiosis. Since these red-algal-derived algae are now the predominant species of marine primary producers (Falkowski et al. 2004; Archibald 2012), it is presumed that this evolution adapted these β-CA genes to ocean environments. Moreover, the gene transfer from a cyanobacterium N. linckia z16 in this sub-group is supposed to facilitate these red algal derivatives to the modern marine environmental conditions such as alkaline but CO2-poor seawater.
Elucidated role played by periplasmic Sjβ-CA in HCO3- utilization.
Using either labeled 14CO2 gas or NaH14CO3-containing seawater for the culture of S. japonica sporophytes, Ji et al. (1980) found that the radioactivities in alcohol soluble fractions increases generally with the time of illumination. This is firm evidence that this kelp can use both CO2 and HCO3- as a substrate for photosynthesis, like the majority of brown seaweeds (see reviews by Johnston 1991, Badger 2003, and Bi et al. 2019b and references therein for details) in inorganic carbon utilization. Treating the juvenile kelp sporophytes with DIDS and SITS, Yue et al. (2001) have not detected any changes of photosynthesis in comparison with the control without any treatment. This result seems to rule out the possibility that the kelp could take up HCO3- directly through plasmalemma-located AE proteins. Instead, S. japonica sporophytes can use HCO3- from seawater principally via external CA enzymes, as reflected by the finding that AZ significantly ihibits kelp photosynthesis (Yue et al. 2001). Such a strategy for HCO3- utilization has also been examined in the other Laminariales (Surif and Raven 1989; Haglund et al. 1992; Axelsson et al. 2000; García-Sánchez et al. 2016), suggesting that all could use exogenous HCO3- for photosynthesis in the same manner.
In the gametophyte, a haploid generation of S. japonica, Bi et al. (2021b) detected comparable external CA activity, though AZ has been reported by Yue et al. (2000) to have little effect on the photosynthesis of the female gametophyte. Recently, a periplasmic space-located Sjα-CA2 has been determined by immuno-electron microscopy with the prepared anti-Sjα-CA2 polyclonal antibody in the kelp gametophytes (Bi et al. 2021c), thus settling the dispute whether periplasmic CA occurs in S. japonica gametophytes. Unfortunately, no positive correlation between Sjα-CA2 gene transcription and the consumed HCO3- has been established (Supplementary Table 4), though it has a transcription peak between 16 and 24 h after a supply of HCO3- (Supplementary Fig. 5). Alternatively, Sjβ-CA might be one of candidates to reside on the periplasmic space of the kelp gametophytes due to the presence of a predicted signal peptide (Fig. 2 and Supplementary Table 2).
To understand this, Sjβ-CA cDNA and DNA sequences were cloned (Fig. 1) in the present study. Then, it was expressed heterologously in E. coli (Supplementary Fig. 1) to obtain fusion protein (Supplementary Fig. 2) for functional identification and polyclonal antibody preparation. Using immuno-electron microscopy with the prepared anti-Sjβ-CA polyclonal antibody, we provide direct evidence that Sjβ-CA is localized in periplasmic space (Fig. 6). This subcellular localization resembles C. reinhardtii CAH8 (Ynalvez et al. 2008) and Microcoleus (Coleofasciculus) chthonoplastes (an alkaliphilic cyanobacterium) β-CA (Kupriyanova et al. 2011) but differs from most reported plant ones. In higher plants, β-CA proteins are documented to reside in mitochondria, chloroplasts, cytoplasm, and plasma membrane (see reviews by DiMario et al. 2017; Rudenko et al. 2021 and references therein for details). Although the CO2 hydration activity of recombinant mSjβ-CA (Fig. 4) is not higher than those of M. chthonoplastes β-CA (53.47 U mg−1 protein, Kupriyanova et al. 2011) and C. reinhardtii CAH8 (4.2 U mg−1 protein, Ynalvez et al. 2008), the detected activity of recombinant mSjβ-CA indicates that this gene is active and can act as a CA in this kelp. Consequently, the periplasmic Sjβ-CA is expected to be responsible for the interconversion of environmental CO2 and HCO3-.
The data obtained (Fig. 7 and Supplementary Table 3) demonstrate that there is no significant correlation between Sjβ-CA transcripts and CO2 levels in the PES medium no matter what is supplied with CO2 or NaHCO3 (Table 1). In contrast, the gene transcription levels of Sjβ-CA are negatively correlated with the remaining contents of HCO3- in the medium (Table 1). That is to say, the expression of Sjβ-CA is positively correlated to the utilized HCO3- (in the female r = 0.964, P = <0.001; in the male r = 0.936, P = 0.002). Regarding to the ability of HCO3- dehydration as illustrated by Fig. 4, Sjβ-CA is proposed to be responsible for the consumption of HCO3- by the kelp gametophytes from the medium. However, the role in HCO3- utilization played by the periplasmic Sjα-CA2 as documented by Bi et al. (2021c) cannot be underestimated, since this gene transcription has also been stimulated by treating with bicarbonate especially for 16 to 24 h (Supplementary Fig. 5). This change might be a shock response to the abruptly changed environment since no significant correlation has been established as earlier mentioned. Accordingly, both of these two periplasmic CA proteins make a contribution to the comparable activity of external CA detected by Bi et al. (2021b), but Sjβ-CA is higher than Sjα-CA2 in gene transcription suggesting it might play a principal role in HCO3- utilization. Similarly, two periplasmic α-CA proteins, i.e. CAH1 and CAH2, have been shown to be required for high bicarbonate tolerance in the halo-tolerant green alga D. salina HTBS (Hou et al. 2016). These CA proteins may detoxify HCO3- by facilitating its use in photosynthesis as reviewed by Polishchuk (2021).
During the consumption of HCO3- catalyzed by periplasmic CA enzymes, the generated CO2 could diffuse via the plasmalemma into the cell for photosynthesis but the formed OH− could be left so as to increase the medium pH (Prins and Elzenga 1989; Raven et al. 2014). On the contrary, the detected carbonate system parameters of the PES medium (Supplementary Table 3) points out that the pH tends to decline gradually in the entire experiment while supplying with HCO3-. Such a tendency has been observed in previous studies (Bi et al. 2021b, c). In addition to the bulk buffer, one possible explanation is that another component such as energy-consuming H+-ATPase could participate in the process of HCO3- utilization as documented by Klenell et al. (2004) in the congener S. latissima. Unfortunately, several genes coding for vacuole-type and F-type rather than plasmalemma-type H+-ATPase are found in the kelp genome (Ye et al. 2015) and transcriptome (Bi et al. 2019a) database. Instead, it is of interest that a few of genes encoding glycerol:H+ symporter are found from the kelp transcriptome database (Bi et al. 2019a). Whether and how these symporters function in the utilization of HCO3- remains to be investigated.
Recently, a gene-editing platform has been established for S. japonica (Shen et al. 2023). It is believed that functions of the components of CO2-concentrating mechanism (CCM) of this important kelp will be interpreted with the help of gene-editing techniques in the near future.
Data availability
The datasets generated during and/or analyzed during the current study are available in the GenBank repository [https://www.ncbi.nlm.nih.gov/protein/ARM53418.1/] and [https://www.ncbi.nlm.nih.gov/nuccore/KY041784.1/].
References
Archibald JM (2012) The evolution of algae by secondary and tertiary endosymbiosis. Adv Bot Res 64:87–118
Axelsson L, Mercado JM, Figueroa FL (2000) Utilization of HCO3 at high pH by the brown macroalga Laminaria saccharina. Eur J Phycol 35:53–59
Badger MR (2003) The roles of carbonic anhydrases in photosynthetic CO2 concentrating mechanisms. Photosynth Res 77:83–94
Banerjee S, Deshpande PA (2016) On origin and evolution of carbonic anhydrase isozymes: A phylogenetic analysis from whole-enzyme to active site. Comput Biol Chem 61:121–129
Bendayan M, Zollinger M (1983) Ultrastructural localization of antigenic sites on osmium-fixed tissues applying the protein A-gold technique. J Histochem Cytochem 31:101–109
Bernal M, Testillano PS, Alfonso M, Risueño MD, Picorel CR, Yruela I (2007) Identification and subcellular localization of the soybean copper P1B-ATPase GmHMA8 transporter. J Struct Biol 158:46–58
Bi Y-H, Zhou Z-G (2016) Absorption and transport of inorganic carbon in kelps with emphasis on Saccharina japonica. In: Najafpour MM (ed) Applied Photosynthesis-New Progress. InTech, Rijeka pp 111−131
Bi Y-H, Li J-L, Zhou Z-G (2019a) Full-length mRNA sequencing in Saccharina japonica and identification of carbonic anhydrase genes. Aquacult Fish 4:53–60
Bi Y-H, Wei N-N, Li J-L, Wang Z, Xu L, Zhou Z-G (2019b) The role of carbonic anhydrase in inorganic carbon acquisition and utilization by brown seaweeds. Chin Bull Life Sci 31:296–309 (in Chinese with English abstract)
Bi Y-H, Du A-Y, Li J-L, Zhou Z-G (2021a) Isolation and characterization of a γ-carbonic anhydrase localized in the mitochondria of Saccharina japonica. Chemosphere 266:129162
Bi Y-H, Liang C-L, Li J-L, Yin H, Tian R-T, Zhou Z-G (2021b) Effects of inorganic carbon concentration and pH on carbonic anhydrase activity of gametophytes of Saccharina japonica. Aquacult Fish 6:51–55
Bi Y-H, Qiao Y-M, Wang Z, Zhou Z-G (2021c) Identification and characterization of a periplasmic α-carbonic anhydrase (CA) in the gametophytes of Saccharina japonica (Phaeophyceae). J Phycol 57:295–310
Bradford MM (1976) A rapid and sensitive method for the quantification of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72:248–254
Burnell JN, Gibbs MJ, Mason JG (1990) Spinach chloroplastic carbonic anhydrase: nucleotide sequence analysis of cDNA. Plant Physiol 92:37–40
Capasso C, Supuran CT (2015) An overview of the alpha-, beta- and gamma-carbonic anhydrases from Bacteria: can bacterial carbonic anhydrases shed new light on evolution of bacteria? J Enzyme Inhib Med Chem 30:325–332
Choo KH, Tan TW, Ranganathan S (2005) SPdb−a signal peptide database. BMC Bioinformatics 6:249
Deng Y, Yao J, Wang X, Guo H, Duan D (2012) Transcriptome sequencing and comparative analysis of Saccharina japonica (Laminariales, Phaeophyceae) under blue light induction. PLoS One 7:e39704
DiMario RJ, Clayton H, Mukherjee A, Ludwig M, Moroney JV (2017) Plant carbonic anhydrases: structures, locations, evolution, and physiological roles. Mol Plant 10:30–46
Dyall SD, Brown MT, Johnson PJ (2004) Ancient invasions: From endosymbionts to organelles. Science 304:253–256
Emanuelsson O, Brunak S, von Heijne G, Nielsen H (2007) Locating proteins in the cell using TargetP, SignalP and related tools. Nat Protoc 2:953–971
Eriksson M, Karlsson J, Ramazanov Z, Gardeström P, Samuelsson G (1996) Discovery of an algal mitochondrial carbonic anhydrase: Molecular cloning and characterization of a low-CO2-induced polypeptide in Chlamydomonas reinhardtii. Proc Natl Acad Sci USA 93:12031–12034
Falcón LI, Magallón S, Castillo A (2010) Dating the cyanobacterial ancestor of the chloroplast. ISME J 4:777–783
Falkowski PG, Katz ME, Knoll AH, Quigg A, Raven JA, Schofield O, Taylor FJ (2004) The evolution of modern eukaryotic phytoplankton. Science 305:354–360
Gao K, McKinley KR (1994) Use of macroalgae for marine biomass production and CO2 remediation: a review. J Appl Phycol 6:45–60
García-Sánchez MJ, Delgado-Huertas A, Fernández JA, Flores-Moya A (2016) Photosynthetic use of inorganic carbon in deep-water kelps from the Strait of Gibraltar. Photosynth Res 127:295–305
Gentil J, Hempel F, Moog D, Zauner S, Maier UG (2017) Review: origin of complex algae by secondary endosymbiosis: a journey through time. Protoplasma 254:1835–1843
Green BR (2011) After the primary endosymbiosis: an update on the chromalveolate hypothesis and the origins of algae with Chl c. Photosynth Res 107:103–115
Haglund K, Ramazanov Z, Mtolera M, Pedersén M (1992) Role of external carbonic anhydrase in light-dependent alkalization by Fucus serratus L. and Laminaria saccharina (L.) Lamour. (Phaeophyta). Planta 188:1−6
Hanahan D (1983) Studies on transformation of Escherichia coli with plasmids. J Mol Biol 166:557–580
Hewett-Emmett D (2000) Evolution and distribution of the carbonic anhydrase gene families. In: Chegwidden WR, Carter ND, Edwards YH (eds) The Carbonic Anhydrases: New Horizons. Springer, Baselpp, pp 29–76
Hewett-Emmett D, Tashian RE (1996) Functional diversity, conservation, and convergence in the evolution of the α-, β-, and γ-carbonic anhydrase gene families. Mol Phylogenet Evol 5:50–77
Hou Y, Liu Z, Zhao Y, Chen S, Zheng Y, Chen F (2016) CAH1 and CAH2 as key enzymes required for high bicarbonate tolerance of a novel microalga Dunaliella salina HTBS. Enzyme Microb Technol 87–88:17–23
Hu Y-J, Zhou Z-G (2001) Extraction of RAPD-friendly DNA from Laminaria japonica (Phaeophyta) after enzymatic dissociation of the frozen sporophyte tissue. J Appl Phycol 13:415–422
Iñiguez C, Galmés J, Gordillo FJ (2019) Rubisco carboxylation kinetics and inorganic carbon utilization in polar versus cold-temperate seaweeds. J Exp Bot 70:1283–1297
Janouškovec J, Horák A, Oborník M, Lukeš J, Keeling PJ (2010) A common red algal origin of the apicomplexan, dinoflagellate, and heterokont plastids. Proc Natl Acad Sci USA 107:10949–10954
Ji M, Pu S, Ji X (1980) Studies on the initial products of 14C metabolism in Laminaria japonica. Oceanol Limnol Sinica 11:229–240 (in Chinese with English abstract)
Johnston AM (1991) The acquisition of inorganic carbon by marine macroalgae. Can J Bot 69:1123–1132
Keeling PJ (2010) The endosymbiotic origin, diversification and fate of plastids. Phil Trans R Soc B 365:729–748
Kikutani S, Nakajima K, Nagasato C, Tsuji Y, Miyatake A, Matsuda Y (2016) Thylakoid luminal θ-carbonic anhydrase critical for growth and photosynthesis in the marine diatom Phaeodactylum tricornutum. Proc Natl Acad Sci USA 113:9828–9833
Klenell M, Snoeijs P, Pedersén M (2004) Active carbon uptake in Laminaria digitata and L. saccharina (Phaeophyta) is driven by a proton pump in the plasma membrane. Hydrobiologia 514:41–53
Kupriyanova EV, Sinetova MA, Markelova AG, Allakhverdiev SI, Los DA, Pronina NA (2011) Extracellular β-class carbonic anhydrase of the alkaliphilic cyanobacterium Microcoleus chthonoplastes. J Photochem Photobiol B 103:78–86
Kussmann M, Roepstorff P (2000) Sample preparation techniques for peptides and proteins analyzed by MALDI-MS. Meth Mol Biol 146:405–424
Laemmli UR (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680–685
Lane CE, Mayes C, Druehl LD, Saunders GW (2006) A Multi-gene molecular investigation of the kelp (Laminariales, Phaeophyceae) supports substantial taxonomic re-organization. J Phycol 42:493–512
Langella E, Di Fiore A, Alterio V, Monti SM, De Simone G, D’Ambrosio K (2022) α-CAs from photosynthetic organisms. Int J Mol Sci 23:12045
Larsson C, Axelsson L (1999) Bicarbonate uptake and utilization in marine macroalgae. Eur J Phycol 34:79–86
Liu Y, Bao H, Zhu M-L, Hu C-X, Zhou Z-G (2022) Subcellular localization and identification of acyl-CoA: lysophosphatidylethanolamine acyltransferase (LPEAT) in the arachidonic acid-rich green microalga, Myrmecia incisa Reisigl. J Appl Phycol 34:837–855
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25:402–408
Ludwig M (2011) The molecular evolution of β-carbonic anhydrase in Flaveria. J Exp Bot 62:3071–3081
Ludwig M (2016) (2016) Evolution of carbonic anhydrase in C4 plants. Curr Opin Plant Biol 31:16–22
Maberly SC, Gontero B (2017) Ecological imperatives for aquatic CO2-concentrating mechanisms. J Exp Bot 68:3797–3814
Majeau N, Coleman JR (1991) Isolation and characterization of a cDNA coding for pea chloroplastic carbonic anhydrase. Plant Physiol 95:264–268
Miller SE, Howell DN (2006) Immunoelectron microscopy. In: Howard GC, Kaser MR (eds) Making and Using Antibodies: A Practical Handbook. CRC Press, Boca Raton, pp 315–337
Millero FJ (2013) Chemical Oceanography (4th Ed.). CRC Press, Boca Raton, pp. 259−333
Missner A, Kügler P, Saparov SM, Sommer K, Mathai JC, Zeidel ML, Pohl P (2008) Carbon dioxide transport through membranes. J Biol Chem 283:25340–25347
Mitsuhashi S, Mizushima T, Yamashita E, Yamamoto M, Kumasaka T, Moriyama H, Ueki T, Miyachi S, Tsukihara T (2000) X-ray structure of β-carbonic anhydrase from the red alga, Porphyridium purpureum, reveals a novel catalytic site for CO2 hydration. J Biol Chem 275:5521–5526
Moreira D, Guyader HL, Philippe H (2000) The origin of red algae and the evolution of chloroplasts. Nature 405:69–72
Moroney JV, Ma Y, Frey WD, Fusilier KA, Pham TT, Simms TA, DiMario RJ, Yang J, Mukherjee B (2011) The carbonic anhydrase isoforms of Chlamydomonas reinhardtii: intracellular location, expression, and physiological roles. Photosynth Res 109:133–149
Nesvizhskii AI (2007) Protein identification by tandem mass spectrometry and sequence database searching. Meth Mol Biol 367:87–119
Ouyang L-L, Du D-H, Yu S-Y, Li C-Y, Zhang C-W, Gao H-J, Zhou Z-G (2012) Expressed sequence tags analysis revealing the taxonomic position and fatty acid biosynthesis in an oleaginous green microalga, Myrmecia incisa Reisigl (Trebouxiophyceae, Chlorophyta). Chin Sci Bull 57:3342–3352
Polishchuk OV (2021) Stress-related changes in the expression and activity of plant carbonic anhydrases. Planta 253:58
Prins H, Elzenga JT (1989) Bicarbonate utilization: function and mechanism. Aquat Bot 34:59–83
Rai AK, Chen T, Moroney JV (2021) Mitochondrial carbonic anhydrases are needed for optimal photosynthesis at low CO2 levels in Chlamydomonas. Plant Physiol 187:1387–1398
Raven JA, Beardall J, Giordano M (2014) Energy costs of carbon dioxide concentrating mechanisms in aquatic organisms. Photosynth Res 121:111–124
Razzak MA, Lee JM, Lee DW, Kim JH, Yoon HS, Hwang I (2019) Expression of seven carbonic anhydrases in red alga Gracilariopsis chorda and their subcellular localization in a heterologous system, Arabidopsis thaliana. Plant Cell Rep 38:147–159
Rodríguez-Ezpeleta N, Brinkmann H, Burey SC, Roure B, Burger G, Löffelhardt W, Bohnert HJ, Philippe H, Lang BF (2005) Monophyly of primary photosynthetic eukaryotes: green plants, red algae, and glaucophytes. Curr Biol 15:1325–1330
Rowlett RS (2014) Structure and catalytic mechanism of β-carbonic anhydrases. Subcell Biochem 75:53–76
Rudenko NN, Ignatova LK, Nadeeva-Zhurikova EM, Fedorchuk TP, Ivanov BN, Borisova-Mubarakshina MM (2021) Advances in understanding the physiological role and locations of carbonic anhydrases in C3 plant cells. Protoplasma 258:249–262
Schönknecht G, Weber AP, Lercher MJ (2014) Horizontal gene acquisitions by eukaryotes as drivers of adaptive evolution. BioEssays 36:9–20
Shen Y, Motomura T, Ichihara K, Matsuda Y, Yoshimura K, Kosugi C, Nagasato C (2023) Application of CRISPR-Cas9 genome editing by microinjection of gametophytes of Saccharina japonica (Laminariales, Phaeophyceae). J Appl Phycol 35:1431–1441
Shih PM, Matzke NJ (2013) Primary endosymbiosis events date to the later Proterozoic with cross-calibrated phylogenetic dating of duplicated ATPase proteins. Proc Natl Acad 110:12355–12360
Sibbald SJ, Archibald JM (2020) Genomic insights into plastid evolution. Genome Biol Evol 12:978–990
Smith KS, Ferry JG (2000) Prokaryotic carbonic anhydrases. FEMS Microbiol Rev 24:335–366
Starr RC, Zeikus JA (1993) UTEX-The culture collection of algae at the University of Texas at Austin 1993 list of cultures. J Phycol 29(S2):1–106
Stumm W, Morgan JJ (1996) Aquatic Chemistry: Chemical Equilibria and Rates in Natural Waters, 3rd edn. John Wiley & Sons, New York, p 1022
Surif MB, Raven JA (1989) Exogenous inorganic carbon sources for photosynthesis in seawater by members of the Fucales and the Laminariales (Phaeophyta): ecological and taxonomic implications. Oecologia 78:97–105
Suzuki S, Furuya K, Takeuchi I (2006) Growth and annual production of the brown alga Laminaria japonica (Phaeophyta, Laminariales) introduced into the Uwa Sea in southern Japan. J Exp Mar Biol Ecol 339:15–29
Tamura K, Stecher G, Kumar S (2021) MEGA11: molecular evolutionary genetics analysis version 11. Mol Biol Evol 38:3022–3027
Thiry A, Supuran CT, Masereel B, Dogné J-M (2008) Recent developments of carbonic anhydrase inhibitors as potential anticancer drugs. J Med Chem 51:3051–3056
Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 25:4876–4882
Tseng CK (1981) Commercial cultivation. In: Lobban CS, Wynne MJ (eds) The Biology of seaweeds. Blackwell, Oxford, pp 680–725
Uehlein N, Kai L, Kaldenhoff R (2017) Plant aquaporins and CO2. In: Chaumont F, Tyerman SD (eds) Plant Aquaporins: From Transport to Signaling. Springer, Cham, pp 255–265
Wang W-J, Wang F-J, Sun X-T, Liu F-L, Liang Z-R (2013) Comparison of transcriptome under red and blue light culture of Saccharina japonica (Phaeophyceae). Planta 237:1123–1133
Wang D, Yu X, Xu K, Bi G, Cao M, Zelzion E, Fu C, Sun P, Liu Y, Kong F, Du G, Tang X, Yang R, Wang J, Tang L, Wang L, Zhao Y, Ge Y, Zhuang Y, Mo Z, Chen Y, Gao T, Guan X, Chen R, Qu W, Sun B, Bhattacharya D, Mao Y (2020) Pyropia yezoensis genome reveals diverse mechanisms of carbon acquisition in the intertidal environment. Nat Commun 11:4028
Wang H, Li Y, Zhang Z, Zhong B (2023) Horizontal gene transfer: Driving the evolution and adaptation of plants. J Integr Plant Biol 65:613–616
Wilbur KM, Anderson NG (1948) Electrometric and colorimetric determination of carbonic anhydrase. J Biol Chem 176:147–154
Ye N, Zhang X, Miao M, Fan X, Zheng Y, Xu D, Wang J, Zhou L, Wang D, Gao Y, Wang Y, Shi W, Ji P, Li D, Guan Z, Shao C, Zhuang Z, Gao Z, Qi J, Zhao F (2015) Saccharina genomes provide novel insight into kelp biology. Nat Commun 6:6986
Ye R-X, Yu Z, Shi W-W, Gao H-J, Bi Y-H, Zhou Z-G (2014) Characterization of α-type carbonic anhydrase (CA) gene and subcellular localization of α-CA in the gametophytes of Saccharina japonica. J Appl Phycol 26:881–890
Ynalvez RA, Xiao Y, Ward AS, Cunnusamy K, Moroney JV (2008) Identification and characterization of two closely related β-carbonic anhydrases from Chlamydomonas reinhardtii. Physiol Plant 133:15–26
Yoon HS, Hackett JD, Ciniglia C, Pinto G, Bhattacharya D (2004) A molecular timeline for the origin of photosynthetic eukaryotes. Mol Biol Evol 21:809–818
Yu K, Liu P, Venkatachalam D, Hopkinson BM, Lechtreck KF (2020) The BBSome restricts entry of tagged carbonic anhydrase 6 into the cis-flagellum of Chlamydomonas reinhardtii. PLoS One 15:e0240887
Yue G-F, Ji Y-C, Wang J-F, Zhou B-C (2000) Inorganic carbon acquisition by the female gametophytes of Laminaria japonica. Mar Sci 24:33–36 (in Chinese with English abstract)
Yue G-F, Wang J-X, Wang J-F, Zhou B-C, Zeng C-K (2001) Inorganic carbon acquisition by juvenile sporophyte of Laminarials (L. japonica×L. longissima). Oceanol Limnol Sin 32:647−652 (in Chinese with English abstract)
Zhang B, Liu X, Huan L, Shao Z, Zheng Z, Wang G (2022) Carbonic anhydrase isoforms of Neopyropia yezoensis: intracellular localization and expression profiles in response to inorganic carbon concentration and life stage. J Phycol 58:657–668
Zhang J, Yao J-T, Sun Z-M, Fu G, Galanin DA, Nagasato C, Motomura T, Hu Z-M, Duan D-L (2015) Phylogeographic data revealed shallow genetic structure in the kelp Saccharina japonica (Laminariales, Phaeophyta). BMC Evol Biol 15:237
Zhang X, Xu D, Guan Z, Wang S, Zhang Y, Wang W, Zhang X, Fan X, Li F, Ye N (2020) Elevated CO2 concentrations promote growth and photosynthesis of the brown alga Saccharina japonica. J Appl Phycol 32:1949–1959
Zhou Z-G, Wu C-Y (1998) Clone culture of Laminaria japonica and induction of its sporophytes. Chin J Biotech 14:109–111
Acknowledgements
This research was supported by the National Key Research and Development Program of China (Grant No. 2018YFD0901500), the National Natural Science Foundation of China (Grant No. 32172963) and the Double First-Class Discipline of Fisheries Science of China.
Funding
The National Key Research and Development Program of China (Grant No. 2018YFD0901500)
The National Natural Science Foundation of China (Grant No. 32172963)
Author information
Authors and Affiliations
Contributions
Z.-G. Zhou has made a significant contribution to the work's conceptual design and this manuscript's writing. H.-M. Hao has carried out most experiments, for example, heterologous expression, enzyme activity, Western blotting experiment, real-time quantitative PCR, and she has written most of this draft. Y.-H. Bi has revised the draft. N.-N. Wei and S.-H. Mei has conducted gene cloning experiment and bioinformatics analysis. P.-C. Lin has performed partial data processing. All authors have read and agreed to the published version of the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Hao, HM., Bi, YH., Wei, NN. et al. Expression of a periplasmic β-carbonic anhydrase (CA) gene is positively correlated with HCO3- utilization by the gametophytes of Saccharina japonica (Phaeophyceae, Ochrophyta). J Appl Phycol 35, 3021–3040 (2023). https://doi.org/10.1007/s10811-023-03088-8
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10811-023-03088-8