Abstract
The high biological diversity of tropical forests has been attributed to various biogeographic mechanisms promoting diversification. Among these, the riverine barrier hypothesis postulates that populations of a species that become isolated on opposite sites of a major river may gradually diverge to form separate lineages. Brown spider monkeys (Ateles hybridus) are Critically Endangered primates are currently distributed along both banks of the Magdalena River in Colombia. Based on their pelage coloration, populations of A. hybridus on opposite sides of the river have been proposed to belong to two different subspecies: A. h. brunneus on the west bank and A. h. hybridus on the east bank. We sequenced portions of the noncoding HVI region of the mitochondrial D-loop (N = 41) and the COII gene (N = 35) from a total of 51 individuals from populations along both banks of the Magdalena River with the goal of evaluating the role of the river as a barrier to gene flow in this endangered primate. Mitochondrial DNA haplotypes were shared between populations on both banks and we found no evidence of highly structured populations occupying opposite banks of the river, suggesting that the Magdalena River has not acted as an insurmountable barrier for brown spider monkeys. Population genetic analyses also reveal likely gene flow between banks, and only a minor portion of the genetic variation currently found in brown spider monkeys can be explained by the river acting as a barrier or by isolation by distance. Our study provides evidence suggesting that the Magdalena River has not acted as a major barrier influencing the evolution of brown spider monkeys and suggests that the subspecific taxonomy of one of the most endangered primates in the world may need reexamination.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Explaining the striking diversity of species found in tropical forests requires an understanding of the various processes that might account for the presence of a high number of species in a single place and for the extensive turnover of species between habitats and regions. Accordingly, evolutionary biologists often attribute the high diversity seen in the tropics to a variety of ecological processes that facilitate species coexistence and to various historical, evolutionary, and biogeographical processes that promote diversification through time and space (Ricklefs 2004). In the latter category, many studies have focused on the geographic context of speciation and —in the case of allopatric speciation models— on the physical causes of population isolation (Haffer 1969; Moritz et al. 2000; Wallace 1852).
Large rivers are some of the most obvious physical barriers to the dispersal of terrestrial and arboreal animals. As first described by Wallace (1852), the distributional boundaries of closely related animal taxa in Amazonia often coincide with major rivers. The riverine barrier hypothesis posits a major role for large rivers acting as barriers to gene flow, such that populations that become isolated on either side of these rivers gradually diverge from one another, eventually forming separate lineages. The riverine barrier hypothesis predicts 1) that sister taxa or phylogroups should occupy opposite banks of large rivers rather than meeting in contact zones within interfluvia or across habitat gradients; 2) that levels of genetic differentiation between populations on opposite river banks should increase with river width and flow rate (because wider and faster rivers are presumably more difficult to cross) and thus should be greater at greater distances from headwaters; and 3) that taxa occurring in upland forest should show higher levels of differentiation across rivers than taxa occurring in seasonally flooded forests located adjacent to a river (Ayres and Clutton-Brock 1992; Capparella 1988, 1991; Gascon et al. 1998; Gehring et al. 2012; Peres et al. 1996). Alternatively, rivers may serve as barriers limiting secondary contact between previously isolated, expanding populations (Moritz et al. 2000; Vences et al. 2009).
Studies of tropical birds (Capparella 1991; Cheviron et al. 2005; Maldonado-Coelho et al. 2013; Ribas et al. 2011; Smith et al. 2014; Voelker et al. 2013), anurans (Pellegrino et al. 2005; Torres-Perez et al. 2007), and mammals (Ayres and Clutton-Brock 1992; Hershkovitz 1977; Nicolas et al. 2011; Patton and da Silva 1998) provide empirical support for the importance of large rivers as barriers to gene flow and as boundaries between the distributions of closely related species and subspecies. However, not all such studies support the notion that large rivers are drivers of diversification. For example, Gascon et al. (2000) suggested a significant influence of the Andean orogenic axis and the associated thrust-and-fold morphology it generated in the lowlands for explaining patterns of genetic diversity in small mammals and frogs along the Juruá River in Brazil, rather than the river itself acting as a barrier.
Although the role of rivers as barriers to dispersal might vary from taxon to taxon, several researchers have argued that the distributions of living primates seem to be largely constrained by large rivers (Amazonia: Ayres and Clutton-Brock 1992; Boubli et al. 2015; Merces et al. 2015; Wallace 1852; Madagascar: Goodman and Ganzhorn 2004; Africa: Grubb 1990; Harcourt and Wood 2012). Among primates, rivers have been shown to correlate with genetic differentiation in a number of cases. For example, orangutans (Pongo pygmaeus) show significant mitochondrial differentiation across the Kinabatangan River in Sabah (Arora et al. 2010; Goossens et al. 2005; Jalil et al. 2008). The situation is similar for chimpanzees (Pan troglodytes) across the Sanaga River in Cameroon, which appears to separate the ranges of two currently recognized subspecies (Gonder et al. 2006). Likewise, a significant proportion of the mitochondrial DNA variation found in bonobos (Pan paniscus) inhabiting the Democratic Republic of Congo exists between geographical regions divided by rivers; however, strong differentiation was observed only between populations divided by the Lomami River (Eriksonn et al. 2004). Rivers seem also to have played a major role in shaping genetic diversity and speciation in Malagasy lemurs (Markolf and Kappeler 2013; Olivieri et al. 2007; Quemere et al. 2010).
In the Neotropics, support for the importance of rivers in shaping population structure in primates comes from a seminal study showing that similarity between Amazonian primate communities on opposite banks of rivers correlated negatively with river width, rate of discharge, and distance from the headwaters (Ayres and Clutton-Brock 1992). Similarly, Peres et al. (1996) found evidence of gene exchange between populations of saddleback tamarins (Saguinus fuscicollis) located on opposite banks of the headwaters of the Juruá River and increasing divergence between banks when progressing from the headwaters to the mouth. However, the Juruá River is not a primary barrier causing the diversification of these tamarins because populations occurring on the two banks are not reciprocally monophyletic, likely as a result of occasional passive transfer of individuals and their genes across the river owing to shifting river courses (Peres et al. 1996). A more recent study on Geoffroyi’s tamarins (Saguinus geoffroyi) provided further evidence for the role of riverine barriers and large bodies of water in shaping the genetic structure of these small-bodied primates (Diaz-Muñoz 2012), and similar results were found in a recent study of squirrel monkeys on both banks of the Amazon River (Merces et al. 2015). Finally, in a comprehensive assessment of the influence of large rivers in the evolution of diurnal Amazonian primates, Boubli et al. (2015) recently documented evidence consistent with the hypothesis that the formation of the Negro River promoted diversification across taxa, whereas the role of the Branco River as a vicariance agent was less clear.
The diversification, phylogenetic relationships, and range limits of spider monkeys (Ateles spp.) have traditionally been explained on the basis of mechanisms such as the formation of forest refugia in the Pleistocene, riverine barriers, geological events, and ecological changes (Collins and Dubach 2000). However, because the majority of speciation events among spider monkeys occurred during the middle to late Pliocene, there is little evidence to support the idea that either Pleistocene refugia or the recent hydrogeographic development of the Amazon river system acted as main drivers of speciation in the group (Collins and Dubach 2000; Morales-Jimenez et al. 2015).
In brown spider monkeys (Ateles hybridus), the Magdalena River —the largest inter-Andean river in Colombia— has been proposed to separate the ranges of two subspecies differing subtly in pelage coloration (Defler 2003), raising the possibility that the river might have acted as a barrier to gene flow and promoted their genetic divergence. Populations with dark brown pelage on the west bank have been classified as Ateles hybridus brunneus whereas those in the east bank have been said to exhibit lighter brown pelage and are classified as A. h. hybridus (Defler 2003). A. h. brunneus occurs between the lower Cauca and Magdalena Rivers in the Colombian departments of Bolívar, Antioquia, and Caldas, whereas A. h. hybridus is distributed in northern Colombia, the inter-Andean lowland forests of the middle Magdalena valley, the Catatumbo region, and several disjunct areas in Venezuela (Defler 2003). Brown spider monkeys are currently considered Critically Endangered by the IUCN and have been listed among the 25 most endangered primate species in the world (Mittermeier et al. 2012) due to loss and fragmentation of their natural habitats (Link et al. 2013) and subsistence hunting pressures (Defler et al. 2003). These threats, coupled with their slow reproductive rates, large body size, and highly frugivorous diet, pose an imminent risk to their persistence (Johns and Skorupa 1987; Milton 1981).
Here, we evaluate the influence of the Magdalena River as a barrier to gene flow between populations of Ateles hybridus in Colombia, particularly between those assigned to different putative subspecies. Specficially, we used phylogenetic and population genetic analyses to evaluate two predictions following from the hypothesis that the Magdalena River has acted as a barrier between populations of brown spider monkeys: 1) haplotypes on opposite river banks should form reciprocally monophyletic groups (assuming time has been sufficient for complete lineage sorting); and 2) genetic differentiation should be greater between populations from opposite river banks rather than among populations from the same bank. In addition, we examined trends in effective population size through time; evidence of demographic expansion in differentiated populations from each bank would suggest the river has acted as a barrier limiting secondary sympatry, whereas no demographic changes would be expected if the river has been a primary barrier to gene flow (Cheviron et al. 2005). In addition to providing a test of the riverine barrier hypothesis, by evaluating the phylogenetic relationships and the degree of genetic differentiation among populations of A. hybridus we provide valuable information relevant to the conservation and management of one of the most world’s most endangered primates.
Materials and Methods
Sample Collection and DNA Extraction
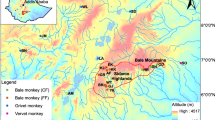
We collected 51 samples from 12 distinct wild populations of brown spider monkeys sampled from across the species’ current distribution in Colombia and Venezuela (Fig. 1; Table I). We followed free-ranging groups and collected fresh fecal samples noninvasively from as many individuals as possible during each field session. Approximately 10 g of feces were stored in ca. 5 ml of RNALater™ (Ambion) nucleic acid preservation buffer in the field at room temperature for up to several weeks before being transferred to the lab and stored at –20°C until extraction.
We extracted DNA from ca. 200 μl of homogenized feces–RNA Later slurry using QIAamp® DNA Stool Mini Kits (Qiagen) following the manufacturer’s instructions, with slight modifications as follows. First, we extended the lysis step of the extraction protocol by placing each sample with ASL buffer on an orbital rocker at room temperature for 1+ h before addition of the InhibitEX tablet. Second, after addition of the InhibitEX tablet, we centrifued the sample at full speed for 6 min to fully pellet stool particles and polymerase chain reaction (PCR) inhibitors bound to the InhibitEX matrix. Third, we extended the proteinase K digestion at 70°C from 10 min to 30 min (with the sample being vortex mixed briefly every 10 min during incubation). Finally, we eluted purified DNA into a final volume of 100 μl of Buffer AE, which was heated to 70°C and allowed to sit on the spin column for 15–20 min.
Mitochondrial DNA Amplification
We amplified roughly 330 bp of the hypervariable region 1 (HVI) of the mitochondrial D-loop using primers H16340 and L15926 (Di Fiore 2009). We performed PCR amplifications in 25-μl reactions with final concentrations of 1X reaction buffer (Promega), 0.16 mM dNTPs, 1.25 mM MgCl2, 0.48 μM for each primer, and 1 U of Taq polymerase (Promega), with 3 μl of a 1:10 dilution of unquantified DNA extract used as a template. In all cases, we supplemented the reaction with bovine serum albumin (BSA) to a final concentration of 1 μg/μl. The PCR cycling conditions consisted of an initial denaturation for 5 min at 94°C, followed by 43 cycles of denaturation at 94°C for 30 s, annealing at 54°C for 1 min, and extension for 1 min at 72°C, with a final extension at 72°C for 5 min. We also amplified ca. 560 bp of the cytochrome c oxidase subunit II (COII) gene using primers L6955 and H7766 (Ashley and Vaughn 1995). In this case, PCR amplifications were also perfomed in 25-μl reactions with final concentrations of 1X reaction buffer (Promega), 0.16 mM dNTPs, 3.5 mM MgCl2, 0.48 μM for each primer, and 1 U of Taq polymerase (Promega). Again, we used 3 μl of a 1:10 dilution of unquantified DNA extract as a template and supplemented the reaction with BSA to a final concentration of 1 μg/μl. The PCR cycling profile consisted of an initial denaturation for 3 min at 94°C, followed by 43 cycles of denaturation at 94°C for 45 s, annealing at 50°C for 45 s, and extension for 90 min at 72°C, with a final extension at 72°C for 3 min.
We used an Exo-SAP (USB Corporation) procedure to clean PCR products and remove unincorporated single-strander primers and then sequenced each amplicon in both directions using the same primers used for amplification either in the Molecular Primatology Laboratory at New York University or by Macrogen, Inc. We aligned DNA sequences from both strands using either Sequencher 4.1 (GeneCodes Corporation) or Geneious 4.8.5 (Biomatters Ltd.) and reconciled any differences by eye.
Population Genetic Analyses
To visualize the relationships between sequences and examine possible geographic associations among haplotypes, we constructed haplotype networks using the package pegas (Paradis 2010) for R v3.0.2 (R Core Team 2012). We used analysis of molecular variance (AMOVA, Excoffier et al. 1992), as implemented in Arlequin 3.1 (Excoffier et al. 2005), to assess the effect of the Magdalena River on the partitioning of genetic variation among populations of brown spider monkeys. We defined two hierarchical levels at which we characterized population structure: 1) among populations from opposite river banks and 2) among populations along each bank. We assessed the statistical significance of indices of population differentiation based on 20,000 random permutations. We calculated corrected pairwise genetic distances in Mega 5.0 (Tamura et al. 2007) and characterized haplotype and nucleotide diversity using DNAsp 5.0 (Rozas and Rozas 1995).
We estimated the extent of gene flow between riverbanks under a Bayesian coalescent framework by using Migrate-n version 3.6.4 (Beerli 2006; Beerli and Felsenstein 2001) to obtain the posterior distribution of the number of immigrants per generation (Nm) for both the COII and HVI loci. Migrate-n allows one to estimate gene flow under different migration models and to evaluate the suitability of such models to the data (Beerli and Palczewski 2010). We ran preliminary analyses for two models of migration: one with symmetric rates of migration between banks and other one with asymmetric rates. We assessed the suitability of the models by using the marginal likelihood (values under Bezzier approximation) to calculate the Bayes factor (Kass and Raftery 1995). Because we found greater support for the symmetric model of migration for both loci, we report results obtained under the symmetric model. For all the runs, we set a static heating strategy with four short chains (1, 1.5, 3, and 1,000,000 temperature values) and a single long chain; 200,000 steps were recorded every 100 generations, discarding the first 200,000 steps as burn-in. Initial values for the analysis were calculated based on FS T values. We assesed the convergence of the Markov chain Monte Carlo (MCMC) run and the suitability of the burn-in length by a visual examination of trace plots in Tracer 1.5 (Rambaut and Drummond 2007); also, this program confirmed that effective sample size of the parameters was in all cases >2000.
To examine whether the Magdalena River may have acted as a barrier to populations of spider monkeys expanding from previously separate areas rather than dividing a formerly contiguous population, we assessed the historical demography of populations on both banks. We examined mismatch distributions, i.e., the distribution of observed pairwise nucleotide site differences, and the significance of the raggedness index using R v3.0.0 (Librado and Rozas 2009) assuming a constant population size and default values for theta initial, theta final, and Tau. Using DNAsp 5.0 (Librado and Rozas 2009), we also calculated Fu’s FS (Fu 1997), Fu and Li’s D* (Fu and Li 1993), Fu and Li’s F* (Fu and Li 1993), R 2 (Ramos-Onsins and Rozas 2002), and Tajima’s D as different measures of possible deviations from neutrality and to determine whether the populations better fit a stationary demographic scenario or one of expanding population size. We assesed the statistical significance of D* and F* statistics using the critical values obtained by Fu and Li (1993), as implemented in DnaSP, and for the statistical significance of Tajima’s D, the confidence limits of D were obtained assuming that D fits a beta distribution (Tajima 1989). For the other parameters, statistical significance was assessed using a coalescent algorithm as implemented in DnaSP.
To further infer possible demographic contractions or expansions, we used the extended Bayesian skyline plot (EBSP) method implemented in BEAST v1.8.0, which estimates posterior probabilities of effective population size over time (Heled and Drummond 2008). This method detects likely demographic changes by estimating population size as a function of time using sequence data from multiple loci (Heled and Drummond 2008). Using jModelTest 2.1.3 (Darriba et al. 2012), we determined that the HKY model of nucleotide substitution (Hasegawa et al. 1985) best fit the data for both loci according to the Akaike information criterion (AIC; Akaike 1973). Each EBSP run extended for 1 × 107 generations, sampling every 1000 steps, with 1,000,000 steps discarded as burn-in. Operators were modified to enhance mixing following Heled (2010). We performed a preliminary run under the “coalescent: constant size” setting and then used the resulting values as a prior in the final run (as in Valderrama et al. 2014). We assesed the convergence of the MCMC run by a visual examination of chains in Tracer 1.5 (Rambaut and Drummond 2007). EBSP plots were then constructed using R v3.0.0. We did not attempt to assess effective population sizes at specific moments in time; thus, we did not calibrate EBSP plots using estimates of substitution rates. Accordingly, we report demographic patterns of change in population size over relative, i.e., not absolute, time since the most recent common ancestor of samples in our data set.
To decompose the relative contribution of the historical effect of the Magdalena River versus isolation by distance in explaining genetic structure in Ateles hybridus, we used a partial Mantel test (Legendre and Legendre 1998; Manly 2007) following de Campos Telles and Diniz-Filho (2005) and Maldonado-Coelho et al. (2013). In simple Mantel tests, geographic distance and isolation by the river were considered as predictors of genetic distance separately, whereas in multiple Mantel regression, they were combined using three matrices: 1) a matrix of pairwise corrected genetic distances between populations, 2) a matrix of pairwise geographic distances between populations, and 3) a pairwise binary matrix coding the position of each population pair relative to the river (populations on the same river bank were scored as 0 and those on different river banks as 1). Mantel tests were performed using FSTAT (Goudet 1995) with 10,000 permutations. Geographic distances were measured as straight lines between populations. Sites with only one sample were excluded from this analysis.
Ethical Note
This research adhered to the legal requirements of the Colombian Government and the Ministerio de Ambiente, Vivienda y Desarrollo Territorial (research permit # IBD0049).
Results
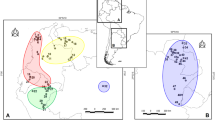
Overall, none of the genes reflected a clear clustering of haplotypes with respect to river bank (Fig. 2). However, we found some differences between the HVI and the COII haplotype networks. In the network based on the less-variable COII sequences, there was no evident division of haplotypes corresponding to east and west banks of the Magdalena River, and individual haplotypes were recovered in multiple populations from both sides of the river. In networks based on the more variable HVI sequences, however, haplotypes from the west bank tended to be more clustered together. For the HVI network, only one of the western haplotypes was shared with populations from the east bank, and sharing occurred only with two populations from the east (Quinchas and San Juan; Fig. 2).
Networks showing relationships among haplotypes of brown spider monkeys (Ateles hybridus) for COII (top) and HVI (bottom). On the left side of each panel, the circles are colored according to riverbank (west bank of the Magdalena River = blue, east bank of the Magdalena River = red), and on the right side of each panel, the various colors correspond to different sampling localities visited during 2009 to 2011. Each circle represents a unique haplotype, and the diameter of each circle reflects the frequency of that haplotype in the overall sample. The number of tickmarks along the lines connecting haplotypes represents the number of mutational steps.
For the HVI data, the mean pairwise genetic distance between banks (0.048) was not different from that within the east bank (0.048), but was higher than the mean pairwise distance among populations within the west bank (0.004). For the COII sequences, the mean pairwise genetic distance between banks (0.006) was slightly higher than that within both the west bank (0.003) and the east bank (0.005; Table II). Nucleotide diversity for all datasets was relatively high for populations on both banks, but was generally higher on the east bank. Likewise, we found higher haplotype diversity among the east bank samples (Table II).
The AMOVA revealed that a greater proportion of the overall genetic variation exists within populations (46.8% and 67.8%) than either between populations from opposite banks (30.1% and 23.9%) or among populations from the same river bank (23.1% and 8.3%) for HVI and COII, respectively (Table III). Most of the genetic variation was attributable to differences observed within populations rather that among populations between river banks (ΦCT = 0.3, P = 0.018 and ΦCT = 0.23, P =0.057 for HVI and COII, respectively). However, in the HVI dataset, the proportion of the total overall variation seen within populations (46.8%) was not as different from the variation between banks (30.1%). Nonetheless, we caution that our estimates of variation within populations may be relatively inaccurate because of the small number of samples in some populations.
Migrate-n analysis estimated a high rate of gene flow across the Magdalena River. The posterior distribution of the Nm parameter peaked at moderate to high rates of migration (mode equal to 1.3 and 6.1 migrants per generation for HVI and COII, respectively). In the case of the COII data, high posterior densities were observed even for very high migration rates (Fig. 3).
We found no support for expanding population sizes as predicted by the hypothesis that the river acts as a barrier where expanding populations meet in secondary contact. Mismatch distribution analyses for both mitochondrial regions suggest that populations of Ateles hybridus from both banks of the Magdalena River have had patterns consistent with a history of constant rather than expanding population size (Fig. 4). Tajima’s D values were positive for the east bank of the River indicating stable (or declining) population sizes, but were significant only for the COII dataset (Table II). For the west bank, all Tajima’s D values were negative, which suggests population expansion. However, R 2 and Fu values implied constant population sizes for all datasets and sets of populations (Table II). These latter results are consistent with those of our ESBP analysis, which showed no signal of demographic expansion, although credibility intervals of estimates of effective population size were wide (Fig. 5).
Mismatch distributions showing the frequency spectrum of the number of pairwise differences between haplotypes for COII (top) and HVI (bottom) in brown spider monkeys (Ateles hybridus). The dashed lines represent expected frequencies assuming a model of constant population size, while the solid lines represent a density estimate derived from the observed distribution.
Effective population size over time for populations of brown spider monkeys for each bank of the Magdalena River in Colombia and Venezuela. The solid line is the median and the dashed lines show the 95% highest posterior densities. Gray lines correspond to the trend of effective population size over time for each of the genealogies that were used to estimate the 95% highest posterior density. Time is given in a relative scale (scaled by mutation rate) where 0 represents the present.
Based on the results of the partial Mantel test, only a limited portion of the variation in corrected pairwise genetic distances can be explained either by the historical isolation caused by the Magdalena River acting as a barrier to gene flow (24.5% and 28.8%) or by isolation by distance (0.9% and 6.9%), using the COII (P = 0.55) and HVI (P = 0.70) data, respectively. Most of the variation remains unexplained (72.7% and 67.2% for COII and HVI, respectively), and only a small fraction of the variation can be explained by the combined effect of a river barrier plus isolation by distance (1.9% and –2.9% for COII and HVI, respectively). There was no correlation between corrected genetic distances and geographical distances for COII (r = 0.045, P = 0.84) or HV1 (r = 0.063, P = 0.75). If the Magdalena River had acted as a barrier to gene flow, one would have expected genetic differences between populations within the same bank of the river to be smaller than those between populations on different banks, independent of the geographical distance among them, but we did not find this pattern (Fig. 6).
Relationship between corrected genetic distances and geographical distances (km) in brown spider monkeys (Ateles hybridus) in Colombia and Venezuela. Black and empty circles represent pairs of locations from the same bank of the river, east and west respectively. X represent pairs of locations from opposite banks of the river.
Discussion
The lack of any significant phylogeographic structure in the mtDNA haplotype networks of brown spider monkeys associated with the Magdalena River suggests that this river has not played a major role in shaping patterns of mitochondrial genetic variation within the species. Moreover, genetic distances between populations on opposite banks were not greater than those within either the east or west bank, and most of the genetic variation observed in brown spider monkeys exists within populations rather than among populations from within the same bank or from opposite banks. Migrate-n analyses suggest a high rate of gene flow between populations of brown spider monkeys found on opposite banks. Distinguishing migration from incomplete lineage sorting due to recent divergence is challenging, but we believe our results likely reflect recurrent gene flow between populations across the river and not recent divergence resulting from the river acting as a vicariant barrier because geological and paleobotanical evidence suggest that the Magdalena River has existed in its current location since the Pliocene, i.e., the Andes of Colombia reached their current elevation between 5 and 2 MYa (Gregory-Wodzicky 2000). Finally, populations on both banks appear to have been demographically stable, which suggests that the river has not acted as a barrier at which expanding populations have met in secondary contact following allopatric divergence.
Previous studies have shown that Neotropical primate faunas from opposite banks of fast-running black water rivers tend to be less similar than those on opposite sides of slower moving, white-water rivers, suggesting that dispersal across the latter is not as restricted (Ayres and Clutton-Brock 1992). Accordingly, the Magdalena River might not represent a major dispersal barrier for brown spider monkeys because this is a relatively slow-flowing and meandering river that carries large amounts of suspended matter and often forms oxbows, which when cut off can allow organisms to spread passively from one side of a river to the other (Jackson and Austin 2013). Peres et al. (1996) proposed a similar scenario to account for patterns of genetic and phenotypic variation in saddleback tamarins (Saguinus fuscicollis) in Amazonia. In addition, most of the middle Magdalena River basin is surrounded by extensive interconnected wetlands that might increase the rate of fluctuations in the main river flow, also allowing land masses to frequently change banks.
In sum, our results do not support the predictions of the riverine barrier hypothesis in brown spider monkeys. It remains to be seen, however, whether the Magdalena River represents a barrier to gene flow for other lowland organisms. For example, the range of silvery-brown tamarins (Saguinus leucopus) is limited to the western bank of the Magdalena River and the river may indeed have served as a barrier preventing the eastward expansion of that species. Moreover, studies on montane lineages of birds and frogs suggests that the Magdalena River valley is an important barrier to gene flow (Cadena et al. 2007; Gutierrez-Pinto et al. 2012; Muñoz-Ortiz et al. 2015; Valderrama et al. 2014), although the effect documented in these studies may be more due to the valley´s elevation and climate than to the river itself.
In contrast with studies on other primate species showing that differences in pelage color are concordant with mitochondrial gene divergence (Ashley and Vaughn 1995; Peres et al. 1996; van der Kuyl et al. 2000), our results reveal a discordance between mtDNA variation and the phenotypic variation in brown spider monkeys because populations occurring on different banks of the Magdalena River have been assigned to different subspecies based on pelage coloration (Defler 2003) yet we found no genetic divergence. A lack of correlation between pelage color and the genetic structure of populations has also recently been found for woolly monkeys (Botero et al. 2010, 2015) and Mesoamerican spider monkeys (Morales-Jimenez et al. 2015), suggesting that discordance between pelage color and genetic differentiation might be relatively frequent, at least within ateline primates.
Given that females are the dispersing sex in spider monkeys (Symington 1987, 1988), the weak to nonexistent genetic structure we observed in maternally inherited mtDNA might not be representative of patterns of population structure across the river for other genomic regions. Specifically, pelage coloration in Ateles hybridus might be a sex-linked trait (Lyon 1962), and therefore, analyses of the genetic structure of markers reflecting male-mediated gene flow, i.e., genes linked to the Y chromosome, may show correspondence with phenotypic variation, a possibility that remains to be explored. The above notwithstanding, pelage color is highly variable within populations of several species of spider monkeys including A. hybridus (Link and Di Fiore, unpubl. data). Therefore, we suggest that quantitative analyses are necessary to determine whether the recognition of populations of A. hybridus from different river banks as separate subspecies is truly warranted based on phenotypic differences.
In addition to its relevance for testing biogeographic hypotheses, understanding how genetic variation is apportioned in brown spider monkeys is important for conservation initiatives considering the status of this highly endangered taxon. Evolutionary significant units (ESU) for conservation are often designated on the basis of reciprocal monophyly at mitochondrial markers and significant divergence at nuclear loci (Moritz 1994). Therefore, owing to the lack of clear phylogeographic structure in Ateles hybridus, it appears that defining distinct ESUs within this species is not possible based on the mitichondrial markers used in this study. However, as with many other species lacking phylogroups (Zink 2004; Zink et al. 2000), we believe that given that brown spider monkeys are critically endangered (Link et al. 2013), preservation efforts are warranted for all viable populations regardless of their apparent genetic homogeneity based on mtDNA (Moritz 2002), pending analyses of variation at other loci.
In conclusion, although the Magdalena River might have influenced the population genetic structure of brown spider monkeys based on mtDNA data to some extent, i.e., all populations in the west bank are closely related to each other, it appears that the Magdalena River has not been an insurmountable barrier to female-mediated gene flow. Analyses of loci with other modes of inheritance and of loci involved in adaptive variation will be important to provide further insights into the role of the river as a barrier to genetic exchange and into the evolutionary history of these critically endangered atelines.
References
Akaike, H. (1973). Information theory and extension of the maximum likelihood principle. In Second international symposium on information theory (pp. 267–281). Budapest: Akademiai Kiado.
Arora, N., Nater, A., van Schaik, C. P., Willems, E. P., van Noordwijk, M. A., Goossens, B., Morf, N., Bastian, M., Knott, C., & Morrogh-Bernard, H. (2010). Effects of Pleistocene glaciations and rivers on the population structure of Bornean orangutans (Pongo pygmaeus). Proceedings of the National Academy of Sciences of the USA, 107, 21376–21381.
Ashley, M. V., & Vaughn, J. L. (1995). Owl monkeys (Aotus) are highly divergent in mitochondrial cytochromec oxidase (COII) sequences. International Journal of Primatology, 16, 793–806.
Ayres, J., & Clutton-Brock, T. (1992). River boundaries and species range size in Amazonian primates. American Naturalist, 140, 531–537.
Beerli, P. (2006). Comparison of Bayesian and maximum-likelihood inference of population genetic parameters. Bioinformatics, 22, 341–345.
Beerli, P., & Felsenstein, J. (2001). Maximum likelihood estimation of a migration matrix and effective population sizes in n subpopulations by using a coalescent approach. Proceedings of the National Academy of Sciences of the USA, 98, 4563–4568.
Beerli, P., & Palczewski, M. (2010). Unified framework to evaluate panmixia and migration direction among multiple sampling locations. Genetics, 185(1), 313–326.
Botero, S., Rengifo, L. Y., Bueno, M. L., & Stevenson, P. R. (2010). How many species of woolly monkeys inhabit Colombian Forests? American Journal of Primatology, 72, 1131–1140.
Botero, S., Stevenson, P. R., & Di Fiore, A. D. (2015). A primer on the phylogeography of Lagothrix lagotricha (sensu Fooden) in northern South America. Molecular Phylogenetics and Evolution, 82, 511–517.
Boubli, J. P., Ribas, C., Lynch-Alfaro, J. W., Alfaro, M. E., Nazareth, M., da Silva, F., Oihno, G. M., & Farias, I. P. (2015). Spatial and temporal patterns of diversification on the Amazon: A test of the riverine hypothesis for all diurnal primates of Rio Negro and Rio Branco in Brazil. Molecular Phylogenetics and Evolution, 82, 400–412.
Cadena, C. D., Klicka, J., & Ricklefs, R. E. (2007). Evolutionary differentiation in the Neotropical montane region: Molecular phylogenetics and phylogeography of Buarremon brush-finches (Aves, Emberizidae). Molecular Phylogenetics and Evolution, 44, 993–1016.
Capparella, A. P. (1988). Genetic variation in neotropical birds: Implications for the speciation process. Acta Congressus Internationalis Ornithologici, 19, 1658–1664.
Capparella, A. P. (1991). Neotropical avian diversity and riverine barrier. Acta Congressus Internationalis Ornithologici, 20, 307–316.
Cheviron, Z. A., Hackett, S. J., & Capparella, A. P. (2005). Complex evolutionary history of a Neotropical lowland forest bird (Lepidothrix coronata) and its implications for historical hypotheses of the origin of Neotropical avian diversity. Molecular Phylogenetics and Evolution, 36, 338–357.
Collins, A. C., & Dubach, J. M. (2000). Biogeographic and ecological forces responsible for speciation in Ateles. International Journal of Primatology, 21, 421–444.
Darriba, D., Taboada, G. L., Doallo, R., & Posada, D. (2012). jModelTest 2: More models, new heuristics and parallel computing. Nature Methods, 9, 772–772.
de Campos Telles, M. P., & Diniz-Filho, J. A. F. (2005). Multiple Mantel tests and isolation-by-distance, taking into account long-term historical divergence. Genetic Molecular Research, 4, 742–748.
Defler, T. R. (2003). Primates de Colombia. Bogota: Conservacion Internacional Colombia.
Defler, T. R., Rodriguez-Mahecha, J. V., & Hernandez-Camacho, J. I. (2003). Conservation priorities for Colombian primates. Primate Conservation, 19, 10–18.
Diaz-Muñoz, S. L. (2012). Role of recent and old riverine barriers in fine‚Äêscale population genetic structure of Geoffroy's tamarin (Saguinus geoffroyi) in the Panama Canal watershed. Ecology and Evolution, 2, 298–309.
Di Fiore, A. (2009). Genetic approaches to the study of dispersal and kinship in New World primates. In P. Garber, A. Estrada, J. C. Bicca-Marques, E. K. Heymann, & K. B. Strier (Eds.), South American primates (pp. 211–250). New York: Springer Science + Business Media.
Eriksonn, J., Hohmann, G., Boesch, C., & Vigilant, L. (2004). Rivers influence the population genetic structure of bonobos (Pan paniscus). Molecular Ecology, 13, 3425–3435.
Excoffier, L., Laval, G., & Schneider, S. (2005). Arlequin ver. 3.0: An integrated software package for population genetics data analysis. Evolutionary Bioinformatics Online, 1, 47–50.
Excoffier, L., Smouse, P. E., & Quattro, J. M. (1992). Analysis of molecular variance inferred from metric distances among DNA haplotypes: Application to human mitochondrial DNA restriction data. Genetics, 131, 479–491.
Fu, Y. (1997). Statistical tests of neutrality against population growth, hitchhiking, and background selection. Genetics, 147, 915–925.
Fu, Y., & Li, W. (1993). Statistical test of neutrality of mutations. Genetics, 133, 693–709.
Gascon, C., Lougheed, S. C., & Bogart, J. P. (1998). Patterns of genetic population differentiation in four species of Amazonian frogs: A test of the riverine barrier hypothesis. Biotropica, 30, 104–119.
Gascon, C., Malcolm, J. R., Patton, J. L., da Silva, M. N. F., James, P. B., Lougheed, S. C., Peres, C. A., Neckel, S., & Boag, P. T. (2000). Riverine barriers and the geographic distribution of Amazonian species. Proceedings of the National Academy of Sciences of the USA, 97, 13672–13677.
Gehring, P. S., Pabijan, M., Randrianirina, J. E., Glaw, F., & Vences, M. (2012). The influence of riverine barriers on phylogeographic patterns of Malagasy reed frogs (Heterixalus). Molecular Phylogenetics and Evolution, 64, 618–632.
Gonder, M. K., Disotell, T. R., & Oates, J. F. (2006). New genetic evidence on the evolution of chimpanzee populations and implications for taxonomy. International Journal of Primatology, 27, 1103–1127.
Goodman, S. M., & Ganzhorn, J. U. (2004). Biogeography of lemurs in the humid forests of Madagascar: The role of elevational distribution and rivers. Journal of Biogeography, 31, 47–55.
Goossens, B., Chikhi, L., Jalil, M. F., Ancrenaz, M., Lackmann‐Ancrenaz, I., Mohamed, M., Andau, P., & Bruford, M. W. (2005). Patterns of genetic diversity and migration in increasingly fragmented and declining orang‐utan (Pongo pygmaeus) populations from Sabah, Malaysia. Molecular Ecology, 14, 441–456.
Goudet, J. (1995). FSTAT (version 1.2): A computer program to calculate F-statistics. Journal of Heredity, 86, 458–486.
Gregory-Wodzicky, K. M. (2000). Uplift history of the Central and Northern Andes: A review. Geological Society of America Bulletin, 112, 1091–1105.
Grubb, P. (1990). Primate geography in the Afro-tropical rain forest biome. In G. Peters & R. Hutterer (Eds.), Vertebrates in the tropics Bonn: Alexander Koenig Zoological Research Institute and Zoological Museum
Gutierrez-Pinto, N., Cuervo, A. M., Miranda, J., Perez-Eman, J. L., Brumfield, R. T., & Cadena, C. D. (2012). Non-monophyly and deep genetic differentiation across low-elevation barriers in a Neotropical montane bird (Basileuterus tristriatus; Aves: Parulidae). Molecular Phylogenetics and Evolution, 64, 156–165.
Haffer, J. (1969). Speciation in Amazonian forest birds. Science, 165, 131–137.
Harcourt, A., & Wood, M. (2012). Rivers as barriers to primate distributions in Africa. International Journal of Primatology, 33, 168–183.
Hasegawa, M., Kishino, H., & Yano, T. A. (1985). Dating of the human-ape splitting by a molecular clock of mitochondrial DNA. Journal of Molecular Evolution, 22, 160–174.
Heled, J. (2010). Extended Bayesian skyline plot tutorial. Available from: tutorial east bio ed ac uk/Tutorials
Heled, J., & Drummond, A. J. (2008). Bayesian inference of population size history from multiple loci. BMC Evolutionary Biology, 8, 289.
Hershkovitz, P. (1977). Living new world monkeys (Platyrrhini). Chicago: University of Chicago Press.
Jackson, N. D., & Austin, C. C. (2013). Testing the role of meander cutoff in promoting gene flow across a riverine barrier in ground skinks (Scincella lateralis). PloS one, 8, e62812.
Jalil, M. F., Cable, J., Sinyor, J., Lackman-Ancrenaz, I., Ancrenaz, M., Bruford, M. W., & Goossens, B. (2008). Riverine effects on mitochondrial structure of Bornean orang-utans (Pongo pygmaeus) at two spatial scales. Molecular Ecology, 17, 2898–2909.
Johns, A. D., & Skorupa, J. P. (1987). Responses of rain-forest primates to habitat disturbance: A review. International Journal of Primatology, 8, 157–191.
Kass, R. E., & Raftery, A. E. (1995). Bayes factors. Journal of the American Statistical Association, 90(430), 773–795.
Legendre, P., & Legendre, L. (1998). Numerical ecology. Amsterdam: Elsevier.
Librado, P., & Rozas, J. (2009). DnaSP v5: A software for comprehensive analysis of DNA polymorphism data. Bioinformatics, 25, 1451–1452.
Link, A., De Luna, A. G., & Burbano-Giron, J. I. (2013). Conservation status in Colombia of one of the most threatened primates: The brown spider monkey (Ateles hybridus). In T. R. Defler, P. R. Stevenson, M. L. Bueno, & D. Guzman (Eds.), Primates Colombianos en peligro de extinción (pp. 87–117). Bogota, Colombia: Asociación Primatológica Colombiana.
Lyon, M. F. (1962). Sex chromatin and gene action in the mammalian X-chromosome. American Journal of Human Genetics, 14, 135.
Maldonado-Coelho, M., Blake, J., Silveira, L., Batalha-Filho, H., & Ricklefs, R. (2013). Rivers, refuges and population divergence of fire-eye antbirds (Pyriglena) in the Amazon Basin. Journal of Evolutionary Biology, 26, 1090–1107.
Manly, B. F. (2007). Randomization, bootstrap and Monte Carlo methods in biology. Boca Raton, FL: CRC/Chapman and Hall.
Markolf, M., & Kappeler, P. M. (2013). Phylogeographic analysis of the true lemurs (genus Eulemur) underlines the role of river catchments for the evolution of micro-endemism in Madagascar. Frontiers in Zoology, 10, 70.
Merces, M. P., Lynch-Alfaro, J. W., Ferreira, W. A. S., Harada, M. L., & Silva-Junior, J. S. (2015). Morphology and mitochondrial phylogenetics reveal that the Amazon River separates two eastern squirrel monkey species: Saimiri sciureus and S. collinsi. Moleculas Phylogenetics and Evolution, 82, 426–435.
Milton, K. (1981). Estimates of reproductive parameters for free-ranging Ateles geoffroyi. Primates, 22, 574–579.
Mittermeier, R. A., Schwitzer, C., Rylands, A. B., Taylor, L. A., Chiozza, F., Williamson, E. A., & Wallis, J. (2012). Primates in peril: The world’s 25 most endangered primates 2012–2014 (pp. 1–40). IUCN/SSC Primate Specialist Group (PSG), International Primatological Society (IPS), Conservation International (CI), and Bristol Conservation and Science Foundation (BCSF).
Morales-Jimenez, A. L., Disotell, T., & Di Fiore, A. (2015). Revisiting the phylogenetic relationships, biogeography and taxonomy of spider monkeys (genus Ateles) in light of new molecular data. Molecular Phylogenetics and Evolution, 82, 467–483.
Moritz, C. (1994). Defining "evolutionary significant units" for conservation. Trends in Ecology & Evolution, 9, 373–375.
Moritz, C. (2002). Strategies to protect biological diversity and the evolutionary processes that sustain it. Systematic Biology, 51, 238–254.
Moritz, C., Patton, J., Schneider, C., & Smith, T. (2000). Diversification of rainforests faunas: An integrated molecular approach. Annual Review of Ecology and Systematics, 31, 533–563.
Muñoz-Ortiz, A., Velásquez-Álvarez, A. A., Guarnizo, C. E., & Crawford, A. J. (2015). Of peaks and valleys: Testing the roles of orogeny and habitat heterogeneity in driving allopatry in mid-elevation frogs (Aromobatidae: Rheobates) of the northern Andes. Journal of Biogeography, 42, 193–205.
Nicolas, V., Missoup, A. D., Denys, C., Kerbis Peterhans, J., Katuala, P., Couloux, A., & Colyn, M. (2011). The roles of rivers and Pleistocene refugia in shaping genetic diversity in Praomys misonnei in tropical Africa. Journal of Biogeography, 38, 191–207.
Olivieri, G., Zimmermann, E., Randrianambinina, B., Rasoloharijaona, S., Rakotondravony, D., Guschanski, K., & Radespiel, U. (2007). The ever-increasing diversity in mouse lemurs: Three new species in north and northwestern Madagascar. Molecular Phylogenetics and Evolution, 43, 309–327.
Paradis, E. (2010). pegas: An R package for population genetics with an integrated-ìmodular approach. Bioinformatics, 26, 419–420.
Patton, J. L., & da Silva, M. N. F. (1998). Rivers, refuges, and ridges: The geography of speciation of Amazonian mammals. In D. J. Howard & S. H. Berlocher (Eds.), Endless forms: Species and speciation (pp. 202–213). New York: Oxford University Press.
Pellegrino, K. C. M., Rodriges, M. T., Waite, A. N., Morando, M., Yassuda, Y. Y., & Sites, J. W. J. (2005). Phylogeography and species limits in the Gymnodactylus darwinii complex (Gekkonidae, Squamata): Genetic structure coincides with river systems in the Brazilian Atlantic Forest. Biological Journal of the Linnean Society, 85, 13–26.
Peres, C. A., Patton, J. L., & da Silva, M. N. F. (1996). Riverine barriers and gene flow in Amazonian saddle back tamarins. Folia Primatologica, 67, 113–124.
Quemere, E., Crouau-Roy, B., Rabarivola, C., Louis, E. E., & Chikhi, L. (2010). Landscape genetics of an endangered lemur (Propithecus tattersalli) within its entire fragmented range. Molecular Ecology, 19, 1606–1621.
R Core Team. (2012). R: A language and environment for statistical computing.
Rambaut A., & Drummond, A. J. (2007). Tracer v1.5. Available from: http://beast.bio.edu.ac.uk/software/tracer/
Ramos-Onsins, S., & Rozas, J. (2002). Statistical properties of new neutrality tests against population growth. Molecular Biology and Evolution, 19, 2092–2100.
Ribas, C. C., Aleixo, A., Nogueira, A. C., Miyaki, C. Y., & Cracraft, J. (2011). A palaeobiogeographic model for biotic diversification within Amazonia over the past three million years. Proceedings of the Royal Society B: Biological Sciences. doi:10.1098/rspb.2011.1120.
Ricklefs, R. E. (2004). A comprehensive framework for global patterns in biodiversity. Ecology Letters, 7, 1–15.
Rozas, J., Rozas, R. (1995). DnaSP, DNA sequence polymorphism: an interactive program for estimating population genetics parameters from DNA sequence data. Computer Applications in the Biosciences, 11, 621–625.
Smith, B. T., McCormack, J. E., Cuervo, A. M., Hickerson, M. J., Aleixo, A., Cadena, C. D., Pérez-Emán, J. L., Burney, C. W., Xie, X., Harvey, M. G., Faircloth, B. C., Glenn, T. C., Derryberry, E. P., Prejean, J., Fields, S., & Brumfield, R. T. (2014). The drivers of tropical speciation. Nature, 515, 406–409.
Symington, M. M. (1987). Sex ratio andmaternal rank in wild spidermonkeys:When daughters disperse. Behavioral Ecology and Sociobiology, 20, 421–425.
Symington, M. M. (1988). Demography, ranging patterns, and activity budgets of Black spider monkeys (Ateles paniscus chamek) in the Manu National Park. American Journal of Primatology, 15, 45–67.
Tajima, F. (1989). The effect of change in population size on DNA polymorphism. Genetics, 123(3), 597–601.
Tamura, K., Dudley, J., Nei, M., & Kumar, S. (2007). MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Molecular Biology and Evolution, 24, 1596–1599.
Torres-Perez, F., Lamborot, M., Boric-Bargetto, D., Hernandez, C. E., Ortiz, J. C., & Palma, R. E. (2007). Phylogeography of a mountain lizard species: An ancient fragmentation process mediated by riverine barriers in the Liolaemus monticola complex (Sauria: Liolaemidae). Journal of Zoological Systematics and Evolutionary Research, 45, 72–81.
Valderrama, E., Pérez-Emán, J. L., Brumfield, R. T., Cuervo, A. M., & Cadena, C. D. (2014). The influence of the complex topography and dynamic history of the montane Neotropics on the evolutionary differentiation of a cloud forest bird (Premnoplex brunnescens, Furnariidae). Journal of Biogeography, 41(8), 1533–1546.
van der Kuyl, A. C., Dekker, J. T., & Goudsmit, J. (2000). Primate genus Miopithecus: Evidence for the existence of species and subspecies of dwarf guenons based on cellular and endogenous viral sequences. Molecular Phylogenetics and Evolution, 14, 403–413.
Vences, M., Wollenberg, K. C., Vieites, D. R., & Lees, D. C. (2009). Madagascar as a model region of species diversification. Trends in Ecology & Evolution, 24, 456–465.
Voelker, G., Marks, B. D., Kahindo, C., A'genonga, U., Bapeamoni, F., Duffie, L. E., Huntley, J. W., Mulotwa, E., Rosenbaum, S. A., & Light, J. E. (2013). River barriers and cryptic biodiversity in an evolutionary museum. Ecology and Evolution, 3, 536–545.
Wallace, A. R. (1852). On the monkeys of the Amazon. Proceedings of the Zoological Society of London, 20, 107–110.
Zink, R. M. (2004). The role of subspecies in obscuring avian biological diversity and misleading conservation policy. Proceeding of the Royal Society of London, 271, 561–564.
Zink, R. M., Barrowclough, G. F., & Atwood, J. L. (2000). Genetics, taxonomy, and conservation of the threatened California gnatcatcher. Conservation Biology, 14, 1394–1405.
Acknowledgments
We are grateful to the Colombian government and the Ministerio de Ambiente, Vivienda y Desarrollo Territorial for permission to conduct this research (permit # IBD0049). This project received financial support from the Mohamed Bin Zayed Species Conservation Fund, Conservation International Primate Action Fund, Fundación Proyecto Primates, New York University, New York Consortium in Evolutionary Primatology, and National Science Foundation Grant No. 1025675. We are also grateful to the Laboratorio de Ecología de Bosques Tropicales y Primatología, led by Dr. Pablo Stevenson, and the Molecular Primatology Laboratory at New York University, with co-PIs Dr. Todd Disotell and Clifford Jolly, for allowing us to work in their facilities. WCS Colombia and Nestor Roncancio provided logistic support in the field. Marcos Maldonado-Coelho provided helpful guidance on the analyses and interpretation of partial Mantel tests. We thank members of Fundación Proyecto Primates who helped us in the field and lab, especially Sergio Botero, Mateo Marquez, and Nelson Galvis. Natalia Gutierrez-Pinto, Angela Navas, and Sebastian Gonzalez helped with data management and population genetic analyses. We also thank Eugenio Valderrama, who provided us with the script to construct the EBSP in R. Finally, we thank two anonymous reviewers and the editor-in-chief for the comments and insights raised in earlier versions of the manucript.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Link, A., Valencia, L.M., Céspedes, L.N. et al. Phylogeography of the Critically Endangered Brown Spider Monkey (Ateles hybridus): Testing the Riverine Barrier Hypothesis. Int J Primatol 36, 530–547 (2015). https://doi.org/10.1007/s10764-015-9840-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10764-015-9840-6