Abstract
Drought stress is currently one of the major threats to the global food security as it primarily is the main cause behind yield loss and hence overall productivity. While conventional breeding, molecular breeding and genetic engineering approaches were widely used in developing drought tolerant varieties, but these techniques are laborious, time consuming and also transgenics developed have some ethical issues hence are not widely accepted. Plant breeders and biotechnologists are now keenly approaching towards genome editing and using various genome editing principles for improving various agronomically important traits in plants. Among all available genome editing principles, the clustered regularly interspaced short palindromic repeat-Cas (CRISPR/Cas) system is widely accepted due to its robust nature, simplicity, adaptability, flexibility and wide applicability. CRISPR Cas’s highly advanced technique of multiple sequence-specific nucleases has facilitated precise gene modification leading to development of novel climate resilient crops. In this review, we will try to understand the molecular mechanism of drought response in plants and the application of CRISPR/Cas genome-editing system for improving drought tolerance in plants for mitigating drought stress.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Improving characters like resistance to abiotic stresses has a deep root in agriculture globally. There are many ways which breeders and agriculturists used to overcome these stresses but adapting novel technologies is the need of the hour for sustaining crop yield and overcoming the ill effects of global warming and climate change. Among the various abiotic stresses, drought is the main agent beyond the yield related agricultural loses and hence to food security (Lobell and Gourdji 2012), so it the epic cause of undernourishment and arisen hunger index worldwide (Martignago et al. 2020). Drought varies differentially round territories of world and also in its effects too, so plants have developed a variable number of structural and physiological responses towards drought. These include various degrees of drought escape, drought avoidance and drought tolerance (Fahad et al. 2017). Conventional plant breeding and transgenic methods have successfully improved drought tolerance character in crops like rice, wheat, maize and soybean (Ashraf 2010), but problem with these drought tolerant lines is that many of the promising lines among them does not show high yield performances under drought conditions. It indicates that there is still a wide scope for novel tools and techniques which can enhance drought tolerance in such a manner that under stress conditions there is least effect on yield performance. Conventional plant breeding is delivering goods since its beginning but is highly time consuming, laborious and expensive. Also a tremendous role has been played by molecular markers in characterizing the specific sections of plant genome under drought conditions (Rao et al. 2016). With the aid of these molecular markers several drought tolerance responsible QTLs of different crops have been identified (Khan et al. 2016), but the certainty and appropriateness in recognizing the QTLs is tough and so much complicated. The another means of improving the weaklings of various agricultural crops is Genetic engineering, but due to some social and ethical issues related transgenics and the biosafety regulations, these genetically modified crops face a lot of hindrances (Prado et al. 2014). Bearing these facts in mind, there is a need of novel technologies to be developed and implied for improving plants against various biotic and abiotic stresses. The arrival of genome editing technologies and its allied principles has revolutionized the field of agriculture. Genome editing principles like ZFNs, TALENs, CRISPR Cas etc. use sequence-specific nucleases for the purpose of making precise genome modifications (Costa et al. 2017). Among all the available genome-editing platforms, CRISPR/ Cas system is widely accepted because of its robust nature and simplicity. The CRISPR/Cas system has a complex consisting of a Cas9 endonuclease and a single guide RNA (sgRNA), which moves along the DNA strand and creates double-stranded breaks (DSBs) on the DNA strand. These breaks are subsequently repaired by the cell’s endogenous repair mechanisms resulting in the development of novel mutants (Voytas and Gao 2014). In its recent progress, CRISPR/Cas technology has been efficiently used in modifying polygenic characters like salinity, submergence and drought in major crops (Shi et al. 2017; Zhang et al. 2019). In this review we will focus on the progress made by CRISPR Cas in mitigating drought and also the short comings and future aspects of CRISPR Cas system in mitigating abiotic stresses like drought effectively, for meeting the demands of globalization and increasing population (Fig. 1).
Depicting about why CRISPR Cas9 technology has an edge over other procedures of developing a variety with a specific character under study in 4–6 years compared to other procedures which require a large amount of time, labour and cost (Chen et al. 2019)
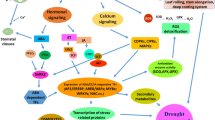
How plants respond to drought stress?
A climatic situation resulted due to low level of water available in soil brought up by lack of water and high rate of evaporation affecting the normal growth and development of the plant is drought stress. During plant growth, drought affects plant water–interactions, which in turn disrupt the entire metabolical pathways at the molecular as well as physiological levels, depending on the magnitude and duration of stress (Sharma et al. 2020). The remarkable signs of plants subject to drought stress at the early vegetative stages of their life cycle are reduced height, wilted leaves, and disrupted flowering (Zafar et al. 2020). For overcoming drought stress, plants have adopted a variable number of morphological, biochemical, physiological and molecular mechanisms (Fang and Xiong 2015). Plants resist drought possibly by four means like drought avoidance (DA), drought escape (DE), drought recovery (DR) and drought tolerance (DT) (Fang and Xiong 2015). Drought avoidance may be defined as a phenomenon implied by plants in which despite scarcity of water in soil plants by morphological adjustment are able to maintain relatively higher tissue water content and perform normal physiological processes (Luo 2010). Plants achieve drought avoidance via stomatal closure, wax accumulation on leaves, thick leaves, limited vegetative growth including number and size of leaves, modified root system, enhanced water uptake to avoid dehydration etc. Drought escape is achieved by plants via modifying their life cycle by either natural or artificial means during water stress before the onset of drought. Likewise, drought recovery (DR) mechanism involves plant’s ability to refurbish growth and vigour after exposure to severe drought stress. In contrast to these mechanisms, Drought tolerance (DT) marks the plant’s potential to carry on physiological activities despite severe drought conditions by regulating the stress responsive genes and signalling pathways (Fang and Xiong 2015). These physiological activities occur either individually or in cumilation and are witnessed in diverse plants under different developmental stages. Drought tolerance is a polygenic trait controlled by many genes, hence it is mandatory to understand the molecular as well as physiological mechanisms controlling this character. Various morpho-physiological traits used as indicators for evaluating drought tolerance in plants are water potential, proline content, abscisic acid content, root traits, osmatic adjustment etc. (Fang and Xiong 2015). In addition to these, other mechanisms like osmotic adjustment, antioxidation and osmo-protection also enable plants to tolerate water stress condition and act as tools for indicating drought (Luo 2010). As per Walter et al. (2009) leaf morphology, an important agronomic trait is useful in generating drought tolerant plants, genotypes having rolled leaves reduce water loss content and enhance drought tolerance character (Xiang et al. 2012). The Fig. 2 represents the generalized mechanism of drought tolerance by depicting out various signalling molecules like reactive oxygen species (ROS), calcium, ABA and allied Phytohormones plus different factors which play an important role during drought stress like condition (Hu and Xiong 2014). The biosynthesis of Phytohormones and the subsequent signalling triggers various dehydration responsive genes which encode ion transporters, calcium dependent protein kinases (CDPKs), calcineurin interacting protein kinases (CIPKs), mitogen-activated protein kinases (MAPKs), sucrose non fermenting protein (SNF1)-related kinase 2 (SnRK2), calcineurin B-like interacting protein kinase (CIPK) and transcription factors (Fang and Xiong 2015). Up-regulated expression of OsCDPK7 and OsCIPK23 positively regulate drought tolerance in rice (Yang et al. 2008). In Arabidopsis, drought tolerance is conferred by SnRK2C molecule as it regulates the stress responsive gene expression (Umezawa et al. 2004). According to Joshi et al. (2016), there are various transcription factors like AP2/EREBF, AREB/ABFs, MYB, NAC and zinc-finger transcription factors which are predicted to exhibit tolerance during drought conditions in plants (Joshi et al. 2016) (Fig. 1). In Arabidopsis, transcription factors like AREB1, AREB2 and AREB3 on validation in coordination with ABA mediated positive regulation provide drought tolerance (Yoshida et al. 2010), drought tolerance is enriched by AP2/EREBF TF SHN as it enhances wax biosynthesis (Aharoni et al. 2004). Among the phytohormones associated with signalling process, ABA signals stresses like drought, salinity etc. Drought tolerance governed by abscisic acid (ABA) is regulated by coordinative action of three classes of proteins namely (a) the Pyrabactin Resistance 1 (PYR1) and/or PYR1-like protein (PYL) and/or Regulatory component of the ABA receptor (RCAR) (here with referred as PYLs), (b) Protein phosphatase 2C (PP2C) and (c) SnRK2s (Joshi et al. 2016). When ABA is absent, PP2Cs associate with SnRK2s inactivates kinases by dephosphorylating the association loop. Under drought stress conditions, Abscisic acid binds PYLs and inhabits phosphate activity of PP2C due to which PP2C releases SnRK2s which are auto phosphorylated and phosphorylate downstream effectors too which add on drought tolerance. In ABA biosynthetic pathways, modification in key enzymes enhances yield as well as improves drought resistance character (Park et al. 2008). A combinative approach of Abscisic acid and Ca+ together with the osmolytes like sorbitol, proline, mannitol, glycine, betaine etc. and osmoprotective proteins also called as late embryogenesis abundant proteins enhance water efficiency by reducing water loss through stomatal closure. The effector proteins enhance drought tolerance by activating the antioxidant enzymes like superoxide dismutase (SOD), ascorbate peroxidase (APX) and glutathione peroxidase (GPX) which facilitates ROS detoxification and lead to overcoming drought. The understanding of these enzymes and there molecular mechanisms have enabled in improving drought resistance in plants using conventional breeding techniques and transgenic technologies. Since last few decades, various approaches like classical breeding techniques, molecular tools and genomic operations led by these tools have played a key role in deciphering the molecular mechanism of various genes controlling different traits and associated mechanisms like drought resistance mechanism, salinity tolerance mechanism etc. (Kulkarni et al. 2017; Cao et al. 2017; Sahebi et al. 2018). QTL mapping of QTLs linked to root, leaf structures and various physiological traits has been successfully performed, which liberated that there are as many as hundreds of drought responsive genes found by RNA sequencing which lead to drought resistance in plants (reviewed in Hu and Xiong 2014). As conventional breeding is intensive labours and time consuming and transgenics having ethical issues, the current scenario demands novel techniques like genome editing principles especially CRISPR-Cas9 to be implied by plant breeders which are labour as well as time efficient and have no grudges of GMO regulations and hence are a good prospectus for mitigating highly emerging abiotic stresses like drought brought up by global warming and climate change (Fig. 3).
A generalized scheme of drought tolerance in plants. (Rev by Joshi et al. 2020)
Structure of CRISPR Cas (Tomida et al. 2017)
CRISPR/Cas9
CRISPR/Cas9 was first used to edit plant genes in 2013 since then, it is the most prevalent genome editing tool (Li et al. 2013a, b; Nekrasov et al. 2013; Shan et al. 2013). It was discovered as a prokaryotic immune system which protects cells by selectively targeting and destroying foreign DNA like viruses or plasmids (Horvath and Barrangou 2010; Marraffini and Sontheimer 2010). The CRISPR–Cas system of genome editing has been highly acknowledged for its adaptability and ease of operation. CRISPR Cas uses a single guide RNA in addition to complex Cas endonuclease that alters along the DNA strand for enacting to arose double-stranded DNA breaks, which are subsequently repaired by endogenic cell mending mechanisms, leading to the expansion of novel mutations (Raza et al. 2020). The engineered CRISPR/Cas9 has three main domains/components i.e. the CRISPR associated protein 9 (Cas9), and two noncoding CRISPR RNAs (crRNAs): a trans-activating crRNA (tracrRNA), and a precursor crRNA (precrRNA) (Horvath and Barrangou 2010; Bhaya et al. 2011). Cas9 is an endonuclease (DNA endonuclease) containing a HNH nuclease domain and a RuvC-like nuclease domain and it is Cas9 that is involved in the crRNA maturation process plus crRNA-guided DNA cleavage (Horvath and Barrangou 2010; Bhaya et al. 2011). The tracrRNA, a small transencoded RNA which contains a sequence having almost perfect complementarity with the repeats within the pre-crRNA so as to allow the formation of an RNA duplex which is necessary for crRNA maturation and also the DNA cleavage guided by crRNA (Horvath and Barrangou 2010; Bhaya et al. 2011). The pre-crRNAs are transcribed from CRISPR loci consisting of a recurring repeat-spacer array with usually 23–47 bp typically identical in length as well as sequence within a loci but varying between different loci. Mostly the repeats are palindromes or short inverted repeats forming hairpin loop like structures (Horvath and Barrangou 2010). The spacers, derived from invading viral or plasmid DNA can guide Cas9 to cleave an invading protospacer (Horvath and Barrangou 2010; Bhaya et al. 2011). Within a CRISPR locus the spacer sequences are typically unique and there size is similar to that of the repeats in the same array (Grissa et al. 2007). The pre-crRNA is composed mainly of the CRSIPR repeat-spacer array and both RNAs I.e. pre-crRNA and tracrRNA which are transcribed together. Subsequently, the tracrRNA and pre-crRNA hybridize together resulting in an RNA duplex which then associates with Cas9. The RNase III enzyme acts on this RNA hetero duplex to produce mature crRNAs with a truncated spacer at one end. The 20 nucleotide spacer at 5’ end of crRNA direct Cas9 to target sequence consisting of protospacer adjacent motifs (PAM) and complementary protospacer sequence. The DNA strand that is complementary to RNA is cleaved by Cas9 HNH nuclease domain while as the DNA domain is cleaved by the RuvC-like nuclease domain and creates a DSB within the protospacer about 3–4 nucleotides upstream of PAM (Horvath and Barrangou 2010; Bhaya et al. 2011; Cong et al. 2013). Engineered CRISPR/Cas9 systems developed on the basis of the type II CRISPR (Jinek et al. 2012; Cong et al. 2013; Mali et al. 2013) which is having both the RNAs (crRNA and tracrRNA) fused giving single guide RNA (sgRNA) to make it technically simple with only two components: the Cas9 endonuclease, used for cleavage, and a sgRNA, for specificity and guidance of Cas9 to its target (Cong et al. 2013; Mali et al. 2013). The engineered CRISPR Cas9 system for targeting a specific sequence in genome with sgRNA requires an introduction of 20 bp spacer sequence. This makes CRISPR Cas9 much simpler and easier for manipulation compared to the other genome editing principles like ZFNs, TALENs etc., also most important character which CRISPR Cas9 possesses is that it has much higher efficiency in producing targeted mutations. Also multiple editing is possible by CRISPR Cas9 by using multiple sgRNAs with different target sequences simultaneously, giving CRISPR Cas9 an edge over rest genome editing tools. The various plant species were, CRISPR Cas9 has been successfully used since 2013 when it was first successfully used in plants are; rice, wheat, tobacco, Arabidopsis, sorghum, tomato, maize, potato, poplar, soybean, barley, moss, Brassica oleracea, sweet orange, apple, liverwort, grape, lettuce, cotton, Lotus japonicus, dandelion, flax, petunia, citrus, watermelon and mushroom (Jiang et al. 2013; Li et al. 2013a, b; Mao et al. 2013; Nekrasov et al. 2013; Shan et al. 2013; Brooks et al. 2014; Feng et al. 2017; Jia and Wang 2014; Sugano et al. 2014; Fan et al. 2015; Lawrenson et al. 2015; Li et al. 2015; Svitashev et al. 2015; Wang et al. 2015; Woo et al. 2015; Iaffaldano et al. 2016; Lopez-Obando et al. 2016; Nishitani et al. 2016; Ren et al. 2016; Sauer et al. 2016; Waltz 2016; Wang et al. 2016b; Zhang et al. 2016; Chen et al. 2017b, a; Jia et al. 2017; Tian et al. 2017).
CRISPR Cas based genome editing strategies for mitigating drought
The CRISPR–Cas system has been professionally used for inculcating resistance to multiple as well as diverse range of abiotic stresses likes of, heavy metals, salinity, drought, submergence etc. (Hyun 2020). Abiotic stresses like drought, soil salinity, heat stress, water lodging, frost etc. have significantly affected crop yields throughout the world by retarding growth and development of the plants (Pandey et al. 2017). The successful use of genome editing principles especially CRISPR Cas for inducing drought tolerance has been demonstrated since long. Overexpression of specific transcription factors and genes bound to drought stress signalling, promote the aggregation of signalling molecules as well as metabolites result ultimately in improving drought tolerance of crops (Tran et al. 2020). Even though, there are various means for tackling drought stress especially QTL mapping, but However, the accuracy and reliability of QTL identification remain problematic. In this light, genetic engineering has proven very successful in improving crops against abiotic and biotic stresses (Shinwari et al. 2020). As drought is a polygenic trait controlled by many genes so editing genes for drought stress was a catastrophe before the advanced genome editing tools were furnished. Hence the genome editing against drought has been demonstrated very recently (Table 1). Drought tolerance is enhanced in plants by the over expression of several genes and transcription factors which are associated with drought signalling, which results in the accumulation of signalling molecules and metabolites resulting in the enhancement of drought tolerance in plants (Fang and Xiong 2015) e, g drought treatment induced miR168 and miR396 expression in Arabidopsis (Li et al. 2021a, b) and tobacco (Frazier et al. 2011) but inhibited it in rice. A diverse family of endogenous, small RNA molecules I,e MicroRNAs (miRNAs), regulate the expression of genes involved in various developmental processes and signaling pathways (Zhang et al. 2021). In contrast, expression of some specific genes called sensitive (S) genes enhances drought conditions in plants through induced ROS production, reduced antioxidant activity and hormonal imbalance. For example, in case of cotton, the yield as well as quality is affected by drought. Promoters that respond to stress conditions and up-regulate transgenes are of great significance in crop improvement using genetic engineering approach. Dass et al. (2017) isolated and characterized dehydration responsive gene GhRDL1 promotor in cotton and found that under in-vitro conditions the GhRDL1 is upregulated in presence of polyethylene glycol which creates water stress condition; hence GhRDL1 promoter, might be useful in generating drought tolerant cotton.In sugarcane seedlings a Non-specific lipid transfer proteins I.e. ScNsLTP transcript levels varied differentially with signalling molecules. It decreased in response to SA, whereas it increased under MeJA treatment, suggesting variable regulatory mechanism between the signalling molecules of MeJA and SA. The varied transcription levels under stress conditions proved that the variation resulted by up regulation of ScNsLTP gene, increased expression of ScNsLTP suggests that the signalling pathway of MeJA-induced may mediate the ScNsLTP gene in response to drought stress (Chen et al. 2017b). In Populus tomentosa (Chinese white poplar) a transcription factor PtoMYB170 was found which controls lignin deposition and drought tolerance. Over expression of PtoMYB170 in transgenic popular plants resulted in high lignified and thick secondary walls of xylem compared to wild type plants, while as the CRISPR Cas9 generated mutants for PtoMYB170 showed feeble lignin deposition which results in flexible and collapsed xylem phenotypes. Also in Arabidopsis, heterologous expression of PtoMYB170 enhanced drought tolerance by closing the stomata in dark compared PtoMYB216 hence reflecting its diverse role. The study reveals that PtoMYB170-dependent positive transcriptional regulation on lignin deposition in poplar and its coordinated function in enhancing drought tolerance by prompting stomatal closure in dark (Xu et al. 2017). In addition, in case of rice (Oryza sativa) stress related ring finger protein 1 (OsSRFP1), drought and salt tolerant protein 1 (OsDST) and drought induced SINA protein 1 (OsDIS1) all perform as negative regulators for drought stress, on silencing of these proteins resulted drought tolerance capacity via enhancing antioxidant enzyme activity and lowering H2O2 levels (Huang et al. 2009; Ning et al. 2011; Fang et al. 2015; Kumar et al. 2020). Therefore, by targeting the drought sensitive (S) or negatively regulating genes in the plant genome, abiotic stresses can be countered and abiotic stress tolerance character can be incorporated. The proof to the above strategy was performed in Arabidopsis for stomatal response caused by prominent plasma membrane proton H + ATPase (AHAs), with the aid of CRISPR Cas technology novel alleles were incorporated into the gene encoding OPEN STOMATA 2 (OST2) (Osakabe et al. 2016). The Plasma membrane proton (H +) ATPases (AHAs) is involved in developing proton gradients which are required to initiate stomatal opening (Merlot et al. 2007). H + ATPase is inhabited as ABA binds to C-terminus under dehydration stress leading to the closure of Stomata. Interestingly in the OST2 locus two dominant mutations have been detected retarding response of stomata towards ABA resulting in disruption of proton pump and necrotic lesions (Merlot et al. 2007). In transgenic plants mutations were detected with high efficiency (> 32%) and no off target modifications using CRISPR Cas9 laced with truncated sgRNA (tru-sgRNA) and Cas9 combination. On evaluation for stomatal response under ABA induced conditions ost2-crispr mutants had high degree of stomatal closure coupled with low level of transcription compared wild type which reflected that drought tolerance through enhanced stomatal response was facilitated by the CRISPR Cas9 induced mutations. Most recently, in case of tomato for generating mutant lines in mitigating drought CRISPR Cas9 system was used to supress gene 1 (NPR1) I.e. a pathogenesis related gene so as to confirm its role in drought tolerance (Li et al. 2019). As NPR1 is a key regulator of biotic stresses and has a limited role in abiotic stresses. In drought responsive apple trees a reduced expression of MdNPR1 has been reported (Bassett et al. 2014), while as in rice against drought stress a hypersensitive response has resulted by overexpression of AtNPR1 (Quilis et al. 2008). In Tomato, suppressing the expression of S1NRP1 gene using CRISPR Cas principle of genome editing resulted in the production of mutants demonstrating drought susceptibility, stomatal aperture wider than wild type, electrolyte leakage prone, monoaldehyde (MDA) and hydrogen peroxide produce is high and antioxidant enzymes lower in concentration than wild type. The role of S1NPR1 was also confirmed by down regulating drought responsive genes like SIGST, SIDHN and SIDREB in the CRISPR Cas9 driven mutants. Hence it can be concluded that there is a key role played in mitigating drought stress by S1NPR1 gene, also using CRISPR Cas9 genome editing multiple S1NPR1 variants can be developed in tomato to enhance drought resistance character in it and other solinaceous crops. Comprehensive molecular analysis has revealed that the pramiry factor of drought response in plants is abscisic acid (ABA), a phytohormone which regulates the expression of stress related genes and also controls stomatal movements to prevent excess transpiration (Osakabe et al. 2014), in addition to ABA, ethylene plays a key role in drought and heat response displayed by diverse physiological pathways underlying abiotic stress response (Zia et al. 2020). The crucial elements of ABA signalling are ABA responsive element binding protein/ABRE binding factors (AREBs/ABTs) also called as bZIP group of TFs (Nakashima et al. 2014). High drought tolerance was demonstrated on regulating up the expression of AREB1 gene and on silencing the same gene resulted in high sensitivity to drought stress (Yoshida et al. 2010; Singh and Laxmi 2015). A huge set of genes downstream of the ABA signalling pathway are regulated by AREB1 gene. It also acts as a major determinant of ABA biosynthesis, antioxidant signalling and osmotic protection (Barbosa et al. 2013; Li et al. 2013a, b). Due to the above statistics, AREB1 could be used as an effective weapon in mitigating drought by improving drought tolerance trait in crop plants. A recent research suggested that a novel CRISPR Cas9 system comprising of a dead Cas9 (dCas9) combined with catalytic domain of enzyme histone acetyl transferase (HAT) derived from Arabidopsis. These two together in calibration with sgRNA were used in targeting the promoter region of AREB1 gene in Arabidopsis (Roca-Paixão et al. 2019). The upregulation of AREB1 elevated drought tolerance, while as AREB1 knockout amplified drought susceptibility. AREB1 regulates the expression of a wide range of genes throughout the ABA signalling pathway and serves as a key element for water deficit stress response, antioxidant signalling, and ABA biosynthesis (Bouzroud et al. 2020). The CRISPR dCasHAT system generated stable phenotypically dwarf lines for the proof of the above suggested novel character of CRISPER Cas system in Arabidopsis. The binding of catalytic domain of Arabidopsis HAT enzyme with dead Cas9 (dCas9) liberated in acetylation of core histone prompting it to higher exposure of promotor region of AREB1 to the transcriptional machinery for its transcription and expression of the transcribed material. Molecular and Physiological characterization of the said mutants revealed that they were laced with higher chlorophyll content, faster stomatal aperture variability along with higher expression of AREB1 gene and its regulated RD29A genes under drought conditions. The mutant lines generated by using CRISPR Cas9 showed better survival rate compared wild types under drought conditions. Compiling the whole in cracks suggests that CRISPR Cas system is an epic asset for induction of epigenetic modifications for enhancing drought tolerance by positive regulation of drought responsive genes. A key regulator of abscisic acid (ABA) dependent hyper osmotic stress signalling and development in plants is SNF1-related protein kinase 2 I.e. a family of plant specific protein kinases (Kobayashi et al. 2004). Different members of the SnRK2 are differently implicated in various physiological and biochemical processes like ABA signalling, ABA-mediated stomatal closure, hyper-osmotic response, drought tolerance, seed germination, seedling growth etc. (rev by Kulik et al. 2011). Particularly in Arabidopsis a distinct network of SnRK2 stress regulatory genes has been identified which in collaboration with AtSnRK2.8 demonstrate positive regulation for drought tolerance along with upregulated expression of the genes which show response on any sort of stress (Umezawa et al. 2004). Also, there is not a clear cut differentiation for survivability and stomatal damage between wild type and SnRK2.8 mutant, the SnRK2 gene commands regulation of AREB/ABF and there point of targets as shown by microarray analysis (Mizoguchi et al. 2010). In case of rice enhanced response towards the abiotic stresses have been demonstrated by members of subclass I and II of SnRK2 family (Kulik et al. 2011). Also OsPYL gene present upstream of OsSAPK2 on expression causes ABA-hypersensitive phenotype in rice especially during seedling growth (Kim et al. 2012). ABA mediated stress signalling is positively regulated by OsSAPK9 gene in rice (Dey et al. 2016). As per another study OsSAPK2 induces drought tolerance by phosphorylasing genes OsbZIP23 and OsbZIP46 in rice (Zong et al. 2016), hence suggesting OsSAPK2 being fundamental for ABA mediated stress tolerance in rice and was confirmed by developing mutants using CRISPR Cas9 with loss of function of OsSAPK2 (Lou et al. 2017). The mutants produced were more drought sensitive compared wild type having functional SAPK2 so the wild type were more tolerant to drought stress by compatible solute accumulation so as to prevent water loss, early stomatal closure, induced stress responsive gene expression and reduced antioxidant enzyme genes expression for promoting ROS scavenging. Bearing in mind all the above said findings reveal that SAPK2 could be highly significant molecule for developing drought tolerant character in future for welfare of plant breeding. Among various Phytohormones involved in mitigation of various abiotic stresses by network of physiological reactions, ethylene plays an important role in high temperature regulation and water deficit situations (Kawakami et al. 2010). In maize higher grain yield under drought conditions has been reported by silencing 1-aminocyclopropane-1-carboxylic acid synthase 6 (ACS6) an ethylene biosynthetic gene (Habben et al. 2014). A study recently reported negative regulation of ethylene signalling by AUXIN REGULATED GENE INVOLVED IN ORGAN SIZE (ARGOS) family genes, it also confers enhancement in drought tolerance and higher grain yield under drought conditions (Shi et al. 2017). CRISPR- Cas based genome editing enhanced shelf life of tomato without affecting rest of the agronomic parameters, such as plant growth and fruit firmness. Tomato plants with mutated fruit ripening gene (lnRNA1459) using a CRISPR–Cas9 mediated knockout of tomato ripening related gene lnRNA1459 (Li et al. 2021a, b). In maize, the endogenous ARGOS8 transcript expression is relatively low and fluctuating hence a group of scientists created a novel ARGOS8 variants at Dupont Pioneer for various beneficial traits likes of drought as well (Shi et al. 2017). The drought tolerant maize lines were generated by inserting promoter GOS into ARGOS8 gene’s 5′ untranslated region.
The ARGOS8 mRNA expression in a diverse population of above 400 maize inbreds for drought tolerance was analysed, which revealed that, the inbreds showed lower level of expression compared mutant ones, also an enhanced yield plus no yield loss under drought stress conditions. By the above studies reported, it is clear cut point to mark that CRISPR Cas is a great asset for creating allelic variation lines to mitigate drought and is hence a crucial weapon for the breeders to be used in future for breeding against drought stress. Researches have conveyed that MAPK signalling is a three tired process of MAPK, MAPK kinases and MAPK kinase kinases. The trio recognizes extracellular signals and accordingly regulate various physiological as well as biochemical responses, drought stress tolerance being one among them (Sinha et al. 2011). In various plants, specific genes have shown a significant response to abiotic stresses especially dehydration stress like in Arabidopsis AtMPK3 gene, in maize ZmMPK3 and in case of rice OsMSRMK2 and OsMPK5, this suggests that for analysing drought stress they could act as active target molecules. In a recent study on tomato for drought stress tolerance it was concluded that editing of SIMAPK3 gene enhances drought stress response (Wang et al. 2017a, b). The SIMAPK3 mutants created by CRISPR Cas9 displayed severe wilting symptoms, high accumulation of hydrogen peroxide, lower antioxidant enzyme activities and severe membrane damage under drought stress conditions compared to wild type plants. On knocking out of the same gene I.e. SIMAPK3 displayed a differential expression of other drought responsive important genes like SlLOX, SlGST, SlDREB etc. hence proved the role of SIMAPK3 in tomato for drought stress. By using CRISPR Cas genome editing tool, novel MAPK variants can be developed in other plants as well for modulating genes which are downstream to the said gene for enhancing drought tolerant characteristics. Drought adaptable plants have a significant feature of leaf rolling, as leaf rolling decreases stomatal conductance and simultaneously reduces transpiration rate (Fang and Xiong 2015). Therefore against drought, leaf rolling character is a good asset, hence developing genotypes with rolled leaves and heaving enhanced yield under drought like conditions will be and are already on priority. In rice the various genes controlling leaf phenotype, semi rolled leaf 1 (SRL1) and semi rolled leaf 2(SRL2) are more important as they encode glycosylphosphatidyl inositol-anchored proteins which are among the major determinants determining number, size and arrangement of bulliform cells of leaf tissue (Liu et al. 2016). On mutating SRL1 lesser lignin and cellulose contents were marked in bulliform cells while as SRL2 mutants demonstrated severe cuticular development (Liu et al. 2016; Li et al. 2017). For this purpose CRISPR Cas9 system is performing the job of modifying plants as has been done in rice by modifying the SRL1 and SRL2 genes (Liao et al. 2019). The modified lines having homozygous SRL1 and SRL2 were laced with features like retardation in various characters like stomatal number, stomatal conductance, transpiration rate, chlorophyll content, vascular bundles and other agronomic traits in comparison to wild type ones. On subjecting to drought stress, survival rate was higher in mutated ones supported by higher ABA content, super oxide dismutase, catalase activities and grain filling than wild type. On proteomic analysis it liberated that mutants derived from CRISPR-Cas9 possessed up regulated 107 proteins, major of them were abiotic stress responsive in nature. In addition to this, the hybrids developed from mutants, displayed semi rolled leaves and better agronomic traits like increased no. of panicles, increase in no. of grains and hence yield per plant. In rice CRISPR Cas was used for knocking out GCS1, result in fertilization failure and pollen tube-dependent ovule enlargement morphology (POEM). The POEMed-like rice ovule (‘endosperm-focused’) could develop to near-normal sized seed, contrary to that which was observed in Arabidopsis in which the gcs1 ovules (‘embryo- focused’) were aborted. The POEMed-like rice ovules contained a very high concentration of sucrose (98% of all sugars) (Honma et al. 2020). On conclusion, with this note I conclude that there is such a vast importance of genome editing and its scope in mitigating abiotic stresses like drought by creating leaf rolling genotypes.
Limitations suggested remedies and future prospectus
The CRISPR-Cas9 technology even though has revolutionized the genome engineering world due to its noble features but this technology still has some weaklings like, downstream of the target sites for CRISPR-Cas9 cleavage requires a 5′-NGG PAM sequence which may limit the range of available targets, but some Cas9 homologs from other strains of archaea or bacteria use quite different PAMs hence might overcome this limitation. The other limitation of CRISPR-Cas9 is off-target mutagenesis result by targeting homologous sequences in unintended loci 45–47, it can be overcomed by avoiding these sequences and also sequences having homology to many other sites. CRISPR Cas9 also shows wayward sgRNA/target as certain sgRNAs may have low efficiencies or may even fail to work properly, this limitation can be restricted by using highly appropriate sgRNAs. Still the future of genome engineering rests in the hands of the highly advanced genome editing technologies like CRISPR Cas9 which is drastically changing the fate of the genome engineering by providing scientists and breeders a tool to precisely modify the DNA of crop plants for welfare of mankind. Important aspect of the CRISPR-Cas9 led genome editing is that it enables genome modifications not only in field crops but also in potential crop plants like duckweed for which genetic manipulation has been a challenge, provided efficient high quality whole genome sequences and reliably good transformation procedures are available. The first CRISPR-Cas9 modified organism (Agaricus bisporus) for resistance to browning can be cultivated and sold without further oversight of US regulations. Interestingly, without signs of transgenics, CRISPR cas9 alters genome of crops by conveying preassembled Cas9-sgRNA ribonucleoproteins or by brief expression of the in vitro driven transcripts of Cas9-coding sequence plus sgRNA in such a manner that they can’t be summed up in GMOs (genetically modified organisms) category as per devised biosafety regulations by various world treaties. For crop improvement especially against abiotic stresses, CRISPR Cas technology has been widely used for genome modification by modifying specific genes and associated molecular mechanisms. However, among the mutants a large number of them especially transgenic ones are a sort of barrier to CRISPR Cas’s flare due to some ethical reasons and hence a catastrophe in the eye of the regulatory authorities, to overcome, transgene free integration is required to limit the off target mutations e.g. using preassembled CRISPR Cas9 based ribonucleoproteins (RNPs) can develop DNA free mutants possessing merely any off target activity in plants (Woo et al. 2015). It has been successfully used in crops like maize (Svitashev et al. 2016), wheat (Liang et al. 2017) and rice (Toda et al. 2019) etc. For monitoring transgene free plants researchers used a fluorescence dependent transgene module in crops like rice, tomato and wheat (Aliaga-Franco et al. 2019; Okada et al. 2019). The ribonucleoproteins (RNPs) can be efficiently delivered by nanoparticles into the meristematic cells of plants with least off targets (Glass et al. 2018). Since CRISPR Cas9 is having the potential of rapidly improving complex traits e.g. engineering drought tolerance comprises of manipulating multiple genes governing complex metabolic pathways concurrently. Practically for drought tolerance various molecular techniques are used like multiple sgRNAs driven by independent promoters multiplexed into single CRISPR Cas expression vector using Gibson assembly or Golden Gate method (Silva and Patron 2017). CRISPR Cas12a or multiplex CRISPR Cas9 developed by engineered endogenous tRNA processing system, can target multi sites simultaneously within a single polycistronic gene (Xie et al. 2015). As in rice, at least six target sites of three genes were mutated using CRSPR/Cas12a system having single crRNA array separated by mature direct repeats (Wang et al. 2017a, b). Hence in modifying the polygenic traits like drought, CRISPR Cas driven multi case system is a good asset. As per Liu et al. (2014), in the promotor region of genes several cis-regulatory sequences are found, which act as binding sites for multiple transcription factors as they serve as negative regulatory module, as in Arabidopsis ANA069 binds to a specific sequence CACGT in the promotor region of various genes responsible for osmotic stress tolerance and ROS scavenging and inhabit there expression (He et al. 2017a, b) it is genuine to conclude that these cis-elements in addition to sensitive and tolerant genes could act as sites to target for achieving abiotic stress tolerance. Studies suggest that very recently a CRISPR Cas9 genome editing approach has been devised as well as used for generating more than hundred regulatory mutations in tomato S1CLV3 promoters for creating novel phenotypic variations for polygenic traits like yield, vigour etc. (Rodríguez-Leal et al. 2017), hence CRISPR Cas system has a scope in novel promotor variant development via HDR driven gaining function mutations to modify polygenic traits. Due to low efficiency, HDR is technically a challenge in plants but with newly emerging CRISPR Cas toolbox, HDR can be used for achieving wide range drought tolerance. The cis-elements not only act as negative regulator but also as positive regulator by acting as a binding site for various transcription factors which in turn govern facilitating other responses to regulate positively e.g. as OsMYB2 increases drought and salinity tolerance in rice by binding to cis-regulatory element MYBR downstream stress responsive genes (Yang et al. 2012), other OsMYBs while binding the same sequence and regulating negatively stress tolerance and metabolic activities. Therefore, by carefully altering cis-regulatory sequences coupled with overexpression of complementary stress responsive genes to overcome odd effects of cis mutation could be having a great scope in mitigating drought. The vast genomic studies reflect the novel variations are the grants of polymorphism (Henikoff and Comai 2003). In maize, 77 SNPs associated with 10 drought responsive transcription factors have been found (Mittal et al. 2017), as inducing single base modifications by CRISPR Cas system is a tedious job as template DNA dependent HDR approach is less efficient compared template independent NHEJ approach hence a novel gene editing strategies is the need of hour for precise point mutations in plants and CRISPR Cas mediated precise nucleotide substitution without requiring donor template has emerged as an advanced tool in performing the dues (Komor et al. 2016). The catalytically-inactive CRISPR Cas9 domain (dCas9 or Cas9 nickase) recognises the sequence and substitution trick is performed by deaminase domain as, the C-G to T-A substitution results of the cytosine base editors while as A-T to G-C modification is governed by adenine base editors. Adding to the base editing tally, by using a catalytically inactive Cas13 (dCas13) in combination with adenosine deaminase acting on RNA (ADAR), an RNA base editor has been devised, such combination is for adenosine to inosine conversion in RNA sequences (Cox et al. 2017). The various crops were base editing approach has been successfully demonstrated include rice, wheat, maize, tomato etc. (Mishra et al. 2020) and has been successfully utilized in crops like wheat, rice, tomato and potato for herbicide tolerance (Zong et al. 2018; Veillet et al. 2019). In another study Ren et al. (2018) reported the modulated broad spectrum disease resistance in rice against blast disease via CRISPR Cas based base editing technique with the aid of rBE5 (hAID*∆-XTEN-Cas9n-UGI-NLS) blast resistance gene Pi-d2 was targeted and broad spectrum based disease resistance was achieved, but for complex traits like drought, salinity etc., none of the reports suggest yet its usage, but recently several ABE and CBE variants have been developed that could probably vasten the range of base editing and make its access valid in mitigating complex traits like drought effectively (Mishra et al. 2020). Summing up with this note that base editing has on offer new opportunities that can be comprehensively explored in upcoming times in mitigating complex traits.
Conclusion
The continuous climatic changes result in various abiotic stresses which are currently the major threats to the global food security as they primarily are the main cause behind yield loss and hence overall productivity. Over the past several decades, conventional breeding, molecular breeding and genetic engineering approaches have contributed significantly in mitigating these stresses. Since these stress conditions are highly spontaneous, diverse as well as dynamic in nature, these existing techniques are not efficient means to overcome these stresses within a short span of time. This limitation created a void which has been fulfilled by CRISPR Cas9 tool box, with its ability of precise genome editing and simple nature CRISPR Cas9 has revolutionized the field of plant breeding. Through targeted mutagenesis in sensitive as well as tolerant genes CRISPR/Cas technology has made it possible to develop drought tolerant plants. Besides this, in regulating expression of genes CRISPR/Cas based alteration of cis-regulatory sequences hold a great promise for gene regulation which is crucial for drought tolerance. Also with the arrival of novel tools like multiplex editing, RNP based DNA free editing and base editing CRISPR Cas technologies are effectively mitigating drought and other abiotic stresses in various crops (Fig. 4). Currently CRISPER Cas technology has some short comings like the inefficient regeneration ability of the edited plants and off-target mutations which hinder its path but by using edited pollens and immature embryo that can outdo the normal tissue culture and application of CRISPR Cas technique for stress inducing results in negligible off-targets (Nandy et al. 2019). Summing up, CRISPR/Cas based technologies undoubtedly are and will continue in transforming various breeding programs towards the development of climate resilient crops.
Genome editing strategies towards drought tolerance in plants. (Rev by Joshi et al. 2020)
References
Aharoni A, Dixit S, Jetter R, Thoenes E, van Arkel G, Pereira A (2004) The SHINE clade of AP2 domain transcription factors activates wax biosynthesis, alters cuticle properties, and confers drought tolerance when overexpressed in Arabidopsis. Plant Cell 16:2463–2480
Aliaga-Franco N, Zhang C, Presa S et al (2019) Identification of transgene-free CRISPR-edited plants of rice, tomato, and Arabidopsis by monitoring DsRED fluorescence in dry seeds. Front Plant Sci 10:1150
Ashraf M (2010) Inducing drought tolerance in plants: recent advances. Biotechnol Adv 28(1):169–183
Barbosa EGG, Leite JP, Marin SRR, Marinho JP, Carvalo JFX, Fuganti-Pagliarini R et al (2013) Overexpression of the ABA dependent AREB1 transcription factor from Arabidopsis thaliana improves soybean tolerance to water deficit. Plant Mol Rep 31:719–730
Bassett CL, Baldo AM, Moore JT, Jenkins RM, Soffe DS, Wisniewski ME, Norelli JL, Farrell RE (2014) Genes responding to water deficit in apple (Malus domestica Borkh.) roots. BMC Plant Biol 14:182
Bhaya D, Davison M, Barrangou R (2011) CRISPR-Cas systems in bacteria and archaea: versatile small RNAs for adaptive defense and regulation. Annu Rev Genet 45:273
Bouzroud S, Gasparini K, Hu G et al (2020) Down regulation and loss of auxin response factor 4 function using CRISPR/Cas9 alters plant growth, stomatal function and improves tomato tolerance to salinity and osmotic stress. Genes 11(3):272
Brooks C, Nekrasov V, Lippman ZB, Van Eck J (2014) Efficient gene editing in tomato in the first generation using the clustered regularly interspaced short palindromic repeats/CRISPR-Cas9-associated system. Plant Physiol 166:1292–1297
Cao Y, Luo Q, Tian Y, Meng F (2017) Physiological and proteomic analyses of the drought stress response in Amygdalus mira (Koehne) Yüet Lu roots. BMC Plant Biol 17:53
Chen X, Lu X, Shu N, Wang S, Wang J, Wang D, Guo L, Ye W (2017a) Targeted mutagenesis in cotton (Gossypium hirsutum L.) using the CRISPR/Cas9 system. Sci Rep 7:44304
Chen Y, Ma J, Zhang X, Yang Y, Zhou D, Yu Q, Que Y, Xu L, Guo J (2017b) A novel non-specific lipid transfer protein gene from sugarcane (NsLTPs), obviously responded to abiotic stresses and signaling molecules of SA and MeJA. Sugar Technol 19:17–25
Chen K, Wang Y, Zhang R, Zhang H, Gao C (2019) CRISPR/Cas genome editing and precision plant breeding in agriculture. Ann rev plant biol 70:667–697
Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Hsu PD, Wu X, Jiang W, Marraffini LA, Zhang F (2013) Multiplex genome engineering using CRISPR/Cas systems. Science 339:819–823
Costa JR, Bejcek BE, McGee JE, Fogel AI, Brimacombe KR, Ketteler R (2017) Genome editing using engineered nucleases and their use in genomic screening. In: Sittampalam GS, Grossman A, Brimacombe K et al (eds) Assay guidance manual. Eli Lilly & Company and the National Center for Advancing Translational Sciences, Bethesda
Cox DBT, Gootenberg JS, Abudayyeh OO, Franklin B, Kellner MJ (2017) RNA editing with CRISPR-Cas13. Science 358:1019–1027
Dass A, Abdin MZ, Reddy VS, Leelavathi S (2017) Isolation and characterization of the dehydration stress-inducible GhRDL1 promoter from the cultivated upland cotton (Gossypium hirsutum). J Plant Biochem Biotechnol 26:113–119
Dey A, Samanta MK, Gayen S, Maiti MK (2016) The sucrose non-fermenting 1-related kinase 2 gene SAPK9 improves drought tolerance and grain yield in rice by modulating cellular osmotic potential, stomatal closure and stress-responsive gene expression. BMC Plant Biol 16:158
Fahad S, Bajwa AA, Nazir U et al (2017) Crop production under drought and heat stress: plant responses and management options. Front Plant Sci 8:1147
Fan D, Liu T, Li C, Jiao B, Li S, Hou Y, Luo K (2015) Efficient CRISPR/Cas9-mediated targeted mutagenesis in Populus in the first generation. Sci Rep 5:12217
Fang Y, Xiong L (2015) General mechanisms of drought response and their application in drought resistance improvement in plants. Cell Mol Life Sci 72:673–689
Fang H, Meng Q, Xu J, Tang H, Tang S, Zhang H, Huang J (2015) Knock-down of stress inducible OsSRFP1 encoding an E3 ubiquitin ligase with transcriptional activation activity confers abiotic stress tolerance through enhancing antioxidant protection in rice. Plant Mol Biol 87:441–458
Feng Y, Xu P, Li B, Li P, Wen X, An F, Gong Y, Xin Y, Zhu Z, Wang Y, Guo H (2017) Ethylene promotes root hair growth through coordinated EIN3/EIL1 and RHD6/RSL1 activity in Arabidopsis. Proc National Acad Sci 114(52):13834–13839
Frazier TP, Sun G, Burklew CE, Zhang B (2011) Salt and drought stresses induce the aberrant expression of microRNA genes in tobacco. Mol Biotechnol 49(2):159–165
Glass Z, Lee M, Li Y, Xu Q (2018) Engineering the delivery system for CRISPR-based genome editing. Trends Biotechnol 36:173–185
Grissa I, Vergnaud G, Pourcel C (2007) The CRISPRdb database and tools to display CRISPRs and to generate dictionaries of spacers and repeats. BMC Bioinform 8:172
Habben JE, Bao X, Bate NJ, DeBruin JL, Dolan D, Hasegawa D, Helentjaris TG (2014) Transgenic alteration of ethylene biosynthesis increases grain yield in maize under field drought stress conditions. Plant Biotechnol J 12:685–693
He L, Shi X, Wang Y, Guo Y, Yang K, Wang Y (2017a) Arabidopsis ANAC069 binds to C[A/G]CG[T/G] sequences to negatively regulate salt and osmotic stress tolerance. Plant Mol Biol 93:369–387
He P, Zhao P, Wang L, Zhang Y, Wang X, Xiao H, Jianing Yu, Xiao G (2017b) The PIN gene family in cotton (Gossypium hirsutum): genome-wide identification and gene expression analyses during root development and abiotic stress responses. BMC Genom 18:507
Henikoff S, Comai L (2003) Single-nucleotide mutations for plant functional genomics. Annu Rev Plant Biol 54:375–401
Honma Y, Adhikari PB, Kuwata K et al (2020) High-quality sugar production by osgcs1 rice. Commun Biol 3(1):1–8
Horvath P, Barrangou R (2010) CRISPR/Cas, the immune system of bacteria and archaea. Science 327:167–170
Hu H, Xiong L (2014) Genetic engineering and breeding of drought resistant crops. Annu Rev Plant Biol 65:715–741
Huang XY, Chao DY, Gao JP, Zhu MZ, Shi M, Lin HX (2009) A previously unknown zinc finger protein, DST, regulates drought and salt tolerance in rice via stomatal aperture control. Genes Dev 23:1805–1817
Hyun TK (2020) CRISPR/Cas-based genome editing to improve abiotic stress tolerance in plants. Botanica Serbica 44(2):121–127
Iaffaldano B, Zhang Y, Cornish K (2016) CRISPR/Cas9 genome editing of rubber producing dandelion Taraxacumkok-saghyz using Agrobacterium rhizogenes without selection. Ind Crops Prod 89:356–362
Jia H, Wang N (2014) Targeted genome editing of sweet orange using Cas9/sgRNA. PLoS ONE 9:e93806
Jia H, Zhang Y, Orbovic V, Xu J, White F, Jones J, Wang N (2017) Genome editing of the disease susceptibility gene CsLOB1 in citrus confers resistance to citrus canker. Plant Biotechnol J 15:817–823
Jiang W, Zhou H, Bi H, Fromm M, Yang B, Weeks DP (2013) Demonstration of CRISPR/Cas9/sgRNA-mediated targeted gene modification in Arabidopsis, tobacco, sorghum and rice. Nucl Acids Res 41:e188
Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E (2012) A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 337:816–821
Joshi R, Wani SH, Singh B, Bohra A, Dar ZA, Lone AA, Pareek A, Singla-Pareek SL (2016) Transcription factors and plants response to drought stress: current understanding and future directions. Front Plant Sci 7:1029
Joshi RK, Bharat SS, Mishra R (2020) Engineering drought tolerance in plants through CRISPR/Cas genome editing. 3 Biotech 10(9):1–14
Kawakami EM, Oosterhuis DM, Snider JL (2010) Physiological effects of 1-methylcyclopropene on well-watered and water-stressed cotton plants. J Plant Growth Regul 29:280–288
Khan A, Sovero V, Gemenet D (2016) Genome-assisted breeding for drought resistance. Curr Genom 17:330–342
Kim H, Hwang H, Hong JW, Lee YN, Ahn IP, Yoon IS (2012) A rice orthologue of the ABA receptor, OsPYL/RCAR5, is a positive regulator of the ABA signal transduction pathway in seed germination and early seedling growth. J Exp Bot 63:1013–1024
Kobayashi Y, Yamamoto S, Minami H, Kagaya Y, Hattori T (2004) Differential activation of the rice sucrose nonfermenting1-related protein kinase2 family by hyperosmotic stress and abscisic acid. Plant Cell 16:1163–1177
Komor AC, Kim YB, Packer MS, Zuris JA, Liu DR (2016) Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature 533:420–424
Kulik A, Wawer I, Krzywińska E, Bucholc M, Dobrowolska G (2011) SnRK2 protein kinases: key regulators of plant response to abiotic stresses. Omics J Integr Biol 15:859–872
Kulkarni M, Soolanayakanahally R, Ogawa S, Uga Y, Selvaraj MG, Kagale S (2017) Drought response in wheat: key genes and regulatory mechanisms controlling root system architecture and transpiration efficiency. Front Chem 5:106
Kumar VS, Verma RK, Yadav SK et al (2020) CRISPR-Cas9 mediated genome editing of drought and salt tolerance (OsDST) gene in indica mega rice cultivar MTU1010. Physiol Mol Biol Plants. https://doi.org/10.1007/s12298-020-00819-w
Lawrenson T, Shorinola O, Stacey N, Li C, Ostergaard L, Patron N, Uauy C, Harwood W (2015) Induction of targeted, heritable mutations in barley and Brassica oleracea using RNA-guided Cas9 nuclease. Genome Biol 16:258
Li JF, Norville JE, Aach J, Mccormack M, Zhang D, Bush J, Church GM, Sheen J (2013a) Multiplex and homologous recombination-mediated genome editing in Arabidopsis and Nicotiana benthamiana using guide RNA and Cas9. Nat Biotechnol 31:688
Li XY, Liu X, Yao Y, Li YH, Liu S, He CY, Li JM, Lin YY, Li L (2013b) Overexpression of Arachis hypogaea AREB1 gene enhances drought tolerance by modulating ROS scavenging and maintaining endogenous ABA content. Int J MolSci 14:12827–12842
Li Z, Liu ZB, Xing A, Moon BP, Koellhoffer JP, Huang L, Ward RT, Clifton E, Falco SC, Cigan AM (2015) Cas9-guide RNA directed genome editing in soybean. Plant Physiol 169:960–970
Li WQ, Zhang MJ, Gan PF, Qiao L, Yang SQ, Miao H, Wang GF, Zhang MM, Liu WT, Li HF (2017) CLD 1/SRL 1 modulates leaf rolling by affecting cell wall formation, epidermis integrity and water homeostasis in rice. Plant J 92:904–923
Li R, Liu C, Zhao R, Wang L, Chen L, Yu W, Zhang S, Shen J, Shen L (2019) CRISPR/Cas9-mediated SlNPR1 mutagenesis reduces tomato plant drought tolerance. BMC Plant Biol 19:38
Li C, Brant E, Budak H et al (2021a) CRISPR/Cas: a Nobel Prize award-winning precise genome editing technology for gene therapy and crop improvement. J Zhejiang Univ Sci B 22(4):253–284
Li J, Jiao G, Sun Y et al (2021b) Modification of starch composition, structure and properties through editing of TaSBEIIa in both winter and spring wheat varieties by CRISPR/Cas9. Plant Biotechnol J 19(5):937–951
Liang Z, Chen K, Li T, Zhang Y, Wang Y, Zhao Q, Liu J, Zhang H, Liu C, Ran Y, Gao C (2017) Efficient DNA-free genome editing of bread wheat using CRISPR/Cas9 ribonucleoprotein complexes. Nat Commun 8:14261
Liao S, Qin X, Luo L, Han Y, Wang X, Usman B et al (2019) CRISPR/ Cas9-induced mutagenesis of semi-rolled leaf 1,2 confers curled leaf phenotype and drought tolerance by influencing protein expression patterns and ROS scavenging in rice (Oryza sativa L.). Agronomy 9:728
Liu J, Peng T, Dai W (2014) Critical cis-acting elements and interacting transcription factors: key players associated with abiotic stress responses in plants. Plant Mol Biol Rep 32:303–317
Liu X, Li M, Liu K, Tang D, Sun M, Li Y, Shen Y, Du G, Cheng Z (2016) Semi-Rolled Leaf2 modulates rice leaf rolling by regulating abaxial side cell differentiation. J Exp Bot 67:2139–2150
Lobell DB, Gourdji SM (2012) The influence of climate change on global crop productivity. Plant Physiol 160(4):1686–1697
Lopez-Obando M, Hoffmann B, Gery C, Guyon-Debast A, Teoule E, Rameau C, Bonhomme S, Nogue F (2016) Simple and efficient targeting of multiple genes through CRISPR-Cas9 in Physcomitrella patens. G3 6:3647–3653
Lou D, Wang H, Liang G, Yu D (2017) OsSAPK2 confers abscisic acid sensitivity and tolerance to drought stress in rice. Front Plant Sci 8:993
Luo LJ (2010) Breeding for water-saving and drought-resistance rice (WDR) in China. J Exp Bot 61(13):3509–3517
Mali P, Yang L, Esvelt KM, Aach J, Guell M, Dicarlo JE, Norville JE, Church GM (2013) RNA-guided human genome engineering via Cas9. Science 339:823
Mao Y, Zhang H, Xu N, Zhang B, Gou F, Zhu JK (2013) Application of the CRISPR-Cas system for efficient genome engineering in plants. Mol Plant 6:2008–2011
Marraffini LA, Sontheimer EJ (2010) CRISPR interference: RNA-directed adaptive immunity in bacteria and archaea. Nat Rev Genet 11:181–190
Martignago D, Rico-Medina A, Blasco-Escámez D et al (2020) Drought resistance by engineering plant tissue-specific responses. Front Plant Sci 10:1676
Merlot S, Leonhardt N, Fenzi F, Valon C, Costa M, Piette L, Vavasseur A, Genty B, Boivin K, Müller A, Giraudat J, Leung J (2007) Constitutive activation of a plasma membrane H(+)-ATPase prevents abscisic acid-mediated stomatal closure. EMBO J 26:3216–3226
Mishra R, Joshi RK, Zhao K (2020) Base editing in crops: current advances, limitations and future implications. Plant Biotechnol J 18:20–31
Mittal S, Kanika A, Rao AR, Mallikarjuna MG, Gupta HS, Nepolean T (2017) Genomic selection for drought tolerance using genome-wide SNPs in maize. Front Plant Sci 8:550
Mizoguchi M, Umezawa T, Nakashima K, Kidokoro S, Takasaki H, Fujita Y (2010) Two closely related subclass II SnRK2 protein kinases cooperatively regulate drought-inducible gene expression. Plant Cell Physiol 51:842–847
Nakashima K, Yamaguchi-Shinozaki K, Shinozaki K (2014) The transcriptional regulatory network in the drought response and its crosstalk in abiotic stress responses including drought, cold, and heat. Front Plant Sci 5:170
Nandy S, Pathak B, Zhao S, Srivastava V (2019) Heat-shock-inducible CRISPR/Cas9 system generates heritable mutations in rice. Plant Direct 3:e00145
Nekrasov V, Staskawicz B, Weigel D, Jones JD, Kamoun S (2013) Targeted mutagenesis in the model plant Nicotiana benthamiana using Cas9 RNA-guided endonuclease. Nat Biotechnol 31:691–693
Ning Y, Jantasuriyarat C, Zhao Q, Zhang H, Chen S, Liu J, Liu L, Tang S, Park CH, Wang X, Liu X, Dai L, Xie Q, Wang GL (2011) The SINA E3 ligase OsDIS1 negatively regulates drought response in rice. Plant Physiol 157:242–255
Nishitani C, Hirai N, Komori S, Wada M, Okada K, Osakabe K, Yamamoto T, Osakabe Y (2016) Efficient genome editing in apple using a CRISPR/Cas9 system. Sci Rep 6:31481
Okada A, Arndell T, Borisjuk N et al (2019) CRISPR/Cas9-mediated knockout of Ms1 enables the rapid generation of male-sterile hexaploid wheat lines for use in hybrid seed production. Plant Biotechnol J 17:1905–1913
Osakabe Y, Osakabe K, Shinozaki K, Tran LS (2014) Response of plants to water stress. Front Plant Sci 5:86
Osakabe Y, Watanabe T, Sugano SS, Ueta R, Ishihara R, Shinozaki K, Osakabe K (2016) Optimization of CRISPR/Cas9 genome editing to modify abiotic stress responses in plants. Sci Rep 6:26685
Pandey P, Irulappan V, Bagavathiannan MV, Senthil-Kumar M (2017) Impact of combined abiotic and biotic stresses on plant growth and avenues for crop improvement by exploiting physio-morphological traits. Front Plant Sci 8:537
Park HY, Seok HY, Park BK, Kim SH, Goh CH, Lee BH, Lee CH, Moon YH (2008) Overexpression of Arabidopsis ZEP enhances tolerance to osmotic stress. Biochem Biophys Res Commun 375:80–85
Prado JR, Segers G, Voelker T, Carson D, Dobert R, Phillips J, Cook K, Cornejo C, Monken J, Grapes L, Reynolds T, Martino-Catt R (2014) Genetically engineered crops: from idea to product. Annu Rev Plant Biol 65:769–790
Quilis J, Peñas G, Messeguer J, Brugidou C, Segundo BS (2008) The Arabidopsis AtNPR1 inversely modulates defense responses against fungal, bacterial, or viral pathogens while conferring hypersensitivity to abiotic stresses in transgenic rice. Mol Plant Microbe Interact 21:1215–1231
Rao GJN, Reddy JN, Variar M, Mahender A (2016) Molecular breeding to improve plant resistance to abiotic stresses. In: Al-Khayri J, Jain S, Johnson D (eds) Advances in plant breeding strategies: agronomic, abiotic and biotic stress traits. Springer, Cham
Raza A, Charagh S, Razzaq A et al (2020) Brassicaceae plants response and tolerance to drought stress: physiological and molecular interventions. In: The plant family brassicaceae. Springer, Singapore, pp 229–261. https://doi.org/10.1007/978-981-15-6345-4_7
Ren C, Liu X, Zhang Z, Wang Y, Duan W, Li S, Liang Z (2016) CRISPR/Cas9-mediated efficient targeted mutagenesis in Chardonnay Vitis vinifera L. Sci Rep 6:32289
Ren B, Yan F, Kuang Y, Li N, Zhang D, Zhou X, Lin H, Zhou H (2018) Improved base editor for efficiently inducing genetic variations in Rice with CRISPR/Cas9-guided hyperactive hAID Mutant. Mol Plant 11:623–626
Roca-Paixão JF, Gillet FX, Ribeiro TP, Bournaud C, Lourenco-Tessutti T, Noriega DD, Melo B, Almeida- Engler J, Grossi-de-Sa MF (2019) Improved drought stress tolerance in Arabidopsis by CRISPR/dCas9 fusion with a histone acetyl transferase. Sci Rep 9:8080
Rodríguez-Leal D, Lemmon ZH, Man J, Bartlett ME, Lippman ZB (2017) Engineering quantitative trait variation for crop improvement by genome editing. Cell 171:470-480.e8
Sahebi M, Hanafi MM, Rafii MY, Mahmud TMM, Azizi P, Osman M, Abiri R, Taheri S, Kalhori N, Shabanimofrad M, Miah G, Atabaki N (2018) Improvement of drought tolerance in rice (Oryza sativa L.): genetics, genomic tools, and the WRKY gene family. Biomed Res Int 2018:3158474
Sauer NJ, Narvaez-Vasquez J, Mozoruk J, Miller RB, Warburg ZJ, Woodward MJ, Mihiret YA, Lincoln TA, Segami RE, Sanders SL (2016) Oligonucleotide mediated genome editing provides precision and function to engineered nucleases and antibiotics in plants. Plant Physiol 170:1917–1928
Shan Q, Wang Y, Li J, Zhang Y, Chen K, Liang Z, Zhang K, Liu J, Xi JJ, Qiu JL (2013) Targeted genome modification of crop plants using a CRISPR-Cas system. Nat Biotechnol 31:686–688
Sharma A, Wang J, Xu D et al (2020) Melatonin regulates the functional components of photosynthesis, antioxidant system, gene expression, and metabolic pathways to induce drought resistance in grafted Carya cathayensis plants. Sci Total Environ 713:136675
Shi J, Gao H, Wang H, Lafitte HR, Archibald RL, Yang M et al (2017) ARGOS8 variants generated by CRISPR-Cas9 improve maize grain yield under field drought stress conditions. Plant Biotechnol J 15:207–216
Shinwari ZK, Jan SA, Nakashima K et al (2020) Genetic engineering approaches to understanding drought tolerance in plants. Plant Biotechnol Rep 14:151–162
Silva NV, Patron NJ (2017) CRISPR-based tools for plant genome engineering. Emerg Top Life Sci 1:135–149
Singh D, Laxmi A (2015) Transcriptional regulation of drought response: a tortuous network of transcriptional factors. Front Plant Sci 6:895
Sinha AK, Jaggi M, Raghuram B, Tuteja N (2011) Mitogen-activated protein kinase signaling in plants under abiotic stress. Plant Signal Behav 6:196–203
Sugano SS, Shirakawa M, Takagi J, Matsuda Y, Shimada T, Hara-Nishimura I, Kohchi T (2014) CRISPR/Cas9-mediated targeted mutagenesis in the liverwort Marchantia polymorpha L. Plant Cell Physiol 55:475–481
Svitashev S, Young JK, Schwartz C, Gao H, Falco SC, Cigan AM (2015) Targeted mutagenesis, precise gene editing and site-specific gene insertion in maize using Cas9 and guide RNA. Plant Physiol 169:931–945
Svitashev S, Schwartz C, Lenderts B, Young JK, Mark Cigan A (2016) Genome editing in maize directed by CRISPR-Cas9 ribonucleoprotein complexes. Nat Commun 7:13274
Tian S, Jiang L, Gao Q, Zhang J, Zong M, Zhang H, Ren Y, Guo S, Gong G, Liu F (2017) Efficient CRISPR/ Cas9-based gene knockout in water melon. Plant Cell Rep 36:399–406
Toda E, Koiso N, Takebayashi A, Ichikawa M, Kiba T, Osakabe K et al (2019) An efficient DNA- and selectable-marker-free genome editing system using zygotes in rice. Nat Plants 5:363–368
Tomida J, Morita Y, Shibayama K, Kikuchi K, Sawa T, Akaike T, Kawamura Y (2017) Diversity and microevolution of CRISPR loci in Helicobacter cinaedi. PLoS ONE 12(10):e0186241
Tran MT, Doan DTH, Kim J et al (2020) CRISPR/Cas9- based precise excision of SlHyPRP1 domain (s) to obtain salt stress-tolerant tomato. Plant Cell Rep 40(6):999–1011
Umezawa T, Yoshida R, Maruyama K, Yamaguchi-Shinozaki K, Shinozaki K (2004) SRK2C, a SNF1-related protein kinase 2, improves drought tolerance by controlling stress-responsive gene expression in Arabidopsis thaliana. Proc Natl Acad Sci USA 101:17306–17311
Veillet F, Perrot L, Chauvin L, Kermarrec MP, Guyon-Debast A, Chauvin JE, Nogue F, Mazier M (2019) Transgene-free genome editing in tomato and potato plants using Agrobacterium-mediated delivery of a CRISPR/Cas9 cytidine base editor. Int J MolSci 20:402
Voytas DF, Gao C (2014) Precision genome engineering and agriculture: opportunities and regulatory challenges. PLoS Biol 12:e1001877
Walter A, Silk WK, Schurr U (2009) Environmental effects on spatial and temporal patterns of leaf and root growth. Annu Rev Plant Biol 60:279–304
Waltz E (2016) CRISPR-edited crops free to enter market, skip regulation. Nat Biotechnol 34:582
Wang S, Zhang S, Wang W, Xiong X, Meng F, Cui X (2015) Efficient targeted mutagenesis in potato by the CRISPR/Cas9 system. Plant Cell Rep 34:1473–1476
Wang L, Chen L, Li R, Zhao R, Yang M, Sheng J, Shen L (2017a) Reduced drought tolerance by CRISPR/Cas9-mediated SlMAPK3 mutagenesis in tomato plants. J Agric Food Chem 65:8674–8682
Wang M, Mao Y, Lu Y, Tao X, Zhu JK (2017b) Multiplex gene editing in rice using the CRISPR- Cpf1 system. Mol Plant 10:1011–1013
Woo JW, Kim J, Kwon SI, Corvalan C, Cho SW, Kim H, Kim SG, Kim ST, Choe S, Kim JS (2015) DNA-free genome editing in plants with preassembled CRISPR-Cas9 ribonucleoproteins. Nat Biotechnol 33:1162–1164
Xiang JJ, Zhang GH, Qian Q, Xue HW (2012) Semi-rolled leaf1 encodes a putative glycosylphosphatidyl inositol-anchored protein and modulates rice leaf rolling by regulating the formation of bulliform cells. Plant Physiol 159:1488–1500
Xie K, Minkenberg B, Yang Y (2015) Boosting CRISPR/Cas9 multiplex editing capability with the endogenous tRNA-processing system. Proc Natl Acad Sci USA 112:3570–3575
Xu C, Fu X, Liu R, Guo L, Ran L, Li C, Tian Q, Jiao B, Wang B, Luo K (2017) PtoMYB170 positively regulates lignin deposition during wood formation in poplar and confers drought tolerance in transgenic Arabidopsis. Tree Physiol 37(12):1713–1726
Yang W, Kong Z, Omo-Ikerodah E, Xu W, Li Q, Xue Y (2008) Calcineurin B-like interacting protein kinase OsCIPK23 functions in pollination and drought stress responses in rice (Oryza sativa L.). J Genet Genomics 35(9):531-S2
Yang A, Dai X, Zhang WH (2012) A R2R3-type MYB gene, OsMYB2, is involved in salt, cold, and dehydration tolerance in rice. J Exp Bot 63:2541–2556
Yoshida T, Fujita Y, Sayama H, Kidokoro S, Maruyama K, Mizoi J, Shinozaki K, Yamaguchi-Shinozaki K (2010) AREB1, AREB2, and ABF3 are master transcription factors that cooperatively regulate ABRE-dependent ABA signaling involved in drought stress tolerance and require ABA for full activation. Plant J 61:672–685
Zafar SA, Zaidi SSEA, Gaba Y et al (2020) Engineering abiotic stress tolerance via CRISPR/Cas-mediated genome editing. J Exp Bot 71(2):470–479
Zhang B, Yang X, Yang C, Li M, Guo Y (2016) Exploiting the CRISPR/Cas9 system for targeted genome mutagenesis in petunia. Sci Rep 6:20315
Zhang A, Liu Y, Wang F, Li T, Chen Z, Kong D, Bi J, Zhnag F, Luo X, Wang J, Tang J, Yu X, Liu G, Luo L (2019) Enhanced rice salinity tolerance via CRISPR/Cas9-targeted mutagenesis of the OsRR22 gene. Mol Breed 39:47
Zhang CC, Li Y, Feng XZ et al (2021) Circular RNA circ_0001287 inhibits the proliferation, metastasis, and radiosensitivity of non-small cell lung cancer cells by sponging microRNA miR-21 and up-regulating phosphatase and tensin homolog expression. Bioengineered 12(1):414–425
Zhao Y, Zhang C, Liu W, Gao W, Liu C, Song G, Li WX, Mao L, Chen B, Xu Y, Li X (2016) An alternative strategy for targeted gene replacement in plants using a dualsgRNA/Cas9 design. Sci Rep 6(1):1–11
Zia R, Nawaz MS, Siddique MJ et al (2020) Plant survival under drought stress: implications, adaptive responses, and integrated rhizosphere management strategy for stress mitigation. Microbiol Res 242:126626
Zong W, Tang N, Yang J, Peng L, Ma S, Xu Y, Li G, Xiong L (2016) Feedback regulation of ABA signaling and biosynthesis by a bZIP transcription factor targets drought resistance related genes. Plant Physiol 171:2810–2825
Zong Y, Song Q, Li C, Jin S, Zhang D, Wang Y, Qiu JL, Gao C (2018) Efficient C-to-T base editing in plants using a fusion of nCas9 and human APOBEC3A. Nat Biotechnol 36:950–953
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Ali, Z., Rai, S.K., Jan, S. et al. An assessment on CRISPR Cas as a novel asset in mitigating drought stress. Genet Resour Crop Evol 69, 2011–2027 (2022). https://doi.org/10.1007/s10722-022-01364-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10722-022-01364-z