Abstract
Key message
ANAC069 binds to the DNA sequence of C[A/G]CG[T/G] to regulate the expression of genes, resulting in decreased ROS scavenging capability and proline biosynthesis, which contribute to increased sensitivity to salt and osmotic stress.
Abstract
NAM-ATAF1/2 and CUC2 (NAC) proteins are plant-specific transcription factors that play important roles in abiotic stress responses. In the present study, we characterized the physiological and regulatory roles of Arabidopsis thaliana ANAC069 in response to abiotic stresses. Arabidopsis plants overexpressing ANAC069 displayed increased sensitivity to abscisic acid, salt, and osmotic stress. Conversely, ANAC069 knockdown plants showed enhanced tolerance to salt and osmotic stress, but no change in ABA sensitivity. Further studies showed that ANAC069 inhibits the expression of SOD, POD, GST, and P5CS genes. Consequently, the transcript level of ANAC069 correlated negatively with the reactive oxygen species (ROS) scavenging ability and the proline level. The genes regulated by ANAC069 were further studied using a gene chip on a genome-wide scale, and 339 and 226 genes up- and downregulated by ANAC069 were identified. Analysis of the promoters of the genes affected by ANAC069 suggested that ANAC069 regulates the expression of genes mainly through interacting with the DNA sequence C[A/G]CG[T/G] in response to abiotic stresses. Collectively, our data suggest that ANAC069 could recognize C[A/G]CG[T/G] sequences to regulate the expression of genes that negatively regulates salt and osmotic stress tolerance by decreasing ROS scavenging capability and proline biosynthesis.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
NAM-ATAF1/2 and CUC2 (NAC) proteins belong to a large family of plant-specific transcription factors (TFs) that are expressed at various developmental stages in different tissues. To date, 105 NAC genes have been identified in Arabidopsis thaliana (Ooka et al. 2003), 157 in maize (Lu et al. 2015), 138 in rice (Fang et al. 2008), 152 in soybean (Le et al. 2011) and 163 in poplar (Hu et al. 2010). All proteins in the NAC family possess an NAC domain at the N-terminus, comprising a highly conserved stretch of ~160 amino acids, which serves as a platform for DNA binding and is involved in the formation of homo- or hetero-dimers with other NAC domain proteins (Ooka et al. 2003; Lindemose et al. 2014; Welner et al. 2012; Fang et al. 2014). By contrast, the C-terminal region is a diversified domain of variable sequence and length, which serves as a transcriptional activator (Xie et al. 2000) or transcriptional repressor (Hao et al. 2011). Furthermore, some NAC TFs contain a transmembrane domain (TMD) in the C-terminal region, which may determine their subcellular localization and conditional TF activity (Seo et al. 2010).
Plant NAC proteins play essential roles in a diverse range of processes, including leaf senescence (Balazadeh et al. 2010, 2011; Liang et al. 2014; Takasaki et al. 2015), lateral root formation (Xie et al. 2000; Hao et al. 2011), shoot apical meristem development (Aida et al. 1997), axillary meristem (Hibara et al. 2006), cotyledon formation (Souer et al. 1996), flowering (Kim et al. 2007), vascular development (Duval et al. 2014), xylem development (Ohashi-Ito et al. 2010), programmed cell death (Lee et al. 2014) and secondary cell wall synthesis (Yao et al. 2012).
Increasing evidence indicates that some NAC TFs play crucial roles in protecting plants against abiotic stresses. Certain NACs are involved in cold or thermal stress tolerance. For instance, a banana NAC protein (MaNAC1) is the target of the ICE transcription factor (MaICE1), which is involved in cold tolerance by interacting with the ICE1-CBF cold signaling pathway (Shan et al. 2014). NAC019 binds to the promoters of HSFA1b, HSFA6b, HSFA7a, and HSFC1 to regulate their expression, and overexpression of NAC019 in Arabidopsis increases thermotolerance (Guan et al. 2014). NAC proteins also regulate abiotic stress tolerance positively or negatively. For instance, JUNGBRUNNEN1 (JUB1) from Arabidopsis regulates salt tolerance positively. Plants overexpressing JUB1 exhibit a salt-tolerant phenotype, and jub1 knockout mutants are hypersensitive to salt and heat stresses (Wu et al. 2012; Shahnejat-Bushehri et al. 2012). In addition, JUB1 regulates the expression of DREB2A (DEHYDRATION RESPONSIVE ELEMENT BINDING 2A) directly, which is required for the expression of many other water stress-inducible genes (Sakuma et al. 2006a, b). In contrast to JUB1, ORESARA1 (ORE1) acts as a negative regulator of plant salt tolerance: ore1 mutants are tolerant to high salinity stress (Balazadeh et al. 2010). Soybean NAC proteins GmNAC2 and GmNAC20 are induced by drought, and further studies showed that GmNAC2 functions as a negative regulator in abiotic stress, whereas GmNAC20 overexpression in plants induced enhanced salt and freezing tolerance (Hao et al. 2011; Jin et al. 2013). A stress-responsive NAC1 (SNAC1) from rice was induced by osmotic stress, specifically in guard cells; a large number of stress-related genes are upregulated by SNAC1, and plants overexpressing SNAC1 showed significantly improved osmotic and salt tolerance (Hu et al. 2006). Additionally, SNAC1 also interacted with either OsNAC5 or OsNAC6 to induce the expression of stress-responsive genes (Nakashima et al. 2007; Takasaki et al. 2010).
In addition, certain membrane-anchored NAC transcription factors, such as NTL4 and NTL6, play important roles in abiotic stress tolerance. As NAC TFs that contain a C-terminal TMD, NTL6 and NTL4 are also involved in drought stress signaling. Plants overexpressing NTL6-OX exhibit drought tolerance, whereas ntl4 null mutant plants show drought tolerance (Kim et al. 2012; Lee and Park 2012), indicating that the functions of NTL4 and NTL6 are all involved in drought stress-responsive signaling. Notably, their C-terminal TMDs regulate the TF activities of these two NTLs conditionally: under water deficit conditions, cleavage of their TMDs allows NTL6 and NTL4 to translocate from the cytoplasm to the nucleus (Kim et al. 2012; Lee and Park 2012).
Previously, Park et al. (2011) showed that the plasma membrane-bound NAC transcription factor ANAC069 protein (also known as NTM2), integrates salt and auxin signals to regulate Arabidopsis seed germination. In that pathway, auxin delays seed germination under salinity stress conditions via cross talk with NTM2-mediated salt signaling. However, there are still many grey areas that need to be further studied in the ANAC069-mediated stress response, such as the gene regulatory networks through which ANAC069 exerts its functions during the stress response and its physiological integration. In this work, we demonstrated that ANAC069 regulates salt, osmotic and abscisic acid (ABA) stress tolerance negatively, and proposed a working model for ANAC069 in response to abiotic stress conditions.
Materials and methods
Plant materials and growth conditions
All A. thaliana plants used in this study were Columbia ecotype background. The ANAC069 mutant Arabidopsis plants (ANAC069 T-DNA inserted mutant line), SALK_082353 and SALK_095231C were obtained from the Arabidopsis Biological Resource Center (ABRC). Genotyping was determined by PCR detection using gene specific primers 095231C-LP/095231C-RP and 082353-LP/082353-RP together with LBa1 (Table S3). Plants were grown in a controlled environment at 22 °C and 16/8 h light/dark cycles with a light intensity of 100 μmol photons m−2 s−1.
Generation of lines overexpressing ANAC069
The CDS of ANAC069 (AT4G01550) was cloned into pROK II vector under the control of the CaMV35S promoter, and was transformed into Arabidopsis using the floral-dip method. Homozygous transgenic lines (OE lines) were obtained by selection on Kanamycin for three consecutive generations (Table S3 for primers).
Expression patterns of ANAC069
The 598 bp promoter region of ANAC069 together with its 5′UTR was amplified and cloned into pCAMBIA1301 (Table S3 for primers) to drive the GUS reporter gene (ANAC069pro:GUS), and used to transform Arabidopsis by the floral-dip method. Different tissues of 5-week-old ANAC069pro:GUS transgenic plants were harvest for GUS staining. At the same time, 4-day-old ANAC069pro:GUS transgenic seedlings grown on 1/2MS agar plates were incubated for 24 h in 1/2MS liquid media supplemented with 10 μM ABA or 200 mM mannitol. GUS staining was performed as previously described (Jefferson 1989), and plants were imaged with a stereomicroscope (Olympus, Japan).
For assessment of ANAC069 expression in response to ABA and abiotic stress, 4-week-old wild type plants were subjected to 50 µM ABA or 200 mM mannitol for 24 h, respectively. A fresh water-only control was conducted in parallel. After these treatments, the roots and leaves were harvested at the indicated times and stored at −80 °C for real-time RT-PCR assays.
Real-time RT-PCR
Real-time PCR was performed on a MJ Research OpticonTM2 instrument (Bio-Rad, Hercules, USA). The reaction mixture contained 12.5 μl of 2 × SYBR Green Realtime PCR Master Mix (Toyobo), 0.5 μM of each primer, and 2 μl of cDNA template (equivalent to 100 ng of total RNA) in a total volume of 25 μl. The reaction conditions were: 94 °C for 30 s followed by 45 cycles at 94 °C for 12 s, 58 °C for 30 s, 72 °C for 30 s and 80 °C for 1 s. ACT7 (AT5G09810) and TUB2 (AT5G62690) were used as internal controls. Melting curves were generated for each reaction to evaluate amplification specificity. The relative expression levels were calculated by the delta–delta Ct method (Pfaffl et al. 2002). Three biological replicates, each with three technical replicates, were analyzed for each sample. All primers used for real-time RT-PCR are shown in Table S3.
The recognition of ANAC069 to NACRS and ORS1S11 motif
One copy of NACRS (AGCTCTTCTTCTGTAACACGCATGT), its mutants (Ma: AAAAAA TC AAAAAAAACACGCATGT, Mg: AGGGGG TC GGGGGGAACACGCATGT, Mcacg: AGCTCTTCTTCTGTAATTTT CATGT), ORS1S11(CGGGGTTACGTACGGCACACGCAACCGTGC), its mutants ORS1S11M1(CGGGGTTAAAAACGGCACACGCAACCGTGC) or ORS1S11M2(CGGGGTTACGTACGGCAAAAACAACCGTGC) was cloned into a pHIS2 vector (Table S3 for primers), respectively. Y1H screening analysis was performed to study their interactions with ANAC069 following the protocol provided in the kit (BD Matchmaker One-Hybrid Library Construction & Screening Kit (#K1617-1)). For plant transient expression assays, one copy of the NACRS and mutant sequences Ma, Mg and Mcacg were fused to the minimal 35S promoter (−46 to +1) to drive GUS (constructs containing NACRS and mutant sequences were named pCAM-NACRS, pCAM-Ma, pCAM-Mg and pCAM-Mcacg, respectively) in a reformed pCAMBIA1301 (in which the region of 35S:Hygromycin was deleted and a 46 bp minimal promoter was inserted in the region between HindIII and GUS). The effector vector was constructed by cloning the truncated ANAC069ΔC form, which was similar in size to the transcriptionally active nuclear forms of NTM1 (Park et al. 2011), consisting of residues 1–287 of ANAC069into pROK II driven by the 35S promoter (pROK II-ANAC069ΔC) (Table S3 for primers). The reporter constructs pCAM-NACRS, pCAM-Ma, pCAM-Mg and pCAM-Mcacg were respectively cotrans formed with the effector construct pROK II-ANAC069ΔC into tobacco leaves using the particle bombardment method (Ueki et al. 2009). GUS activity was determined according to the method of Jefferson et al. (1987). The cotransformation of the reporter plasmid pCAM-NACRS and the empty pROK II were used as negative controls and the empty pCAMBIA1301 as a positive control. The 35S:LUC was also cotransformed to normalize the transgenic efficiencies. Three independent biological replications were performed.
Identification of the genes affected by ANAC069
Four-week-old OE (OE-3 line) and KO (KO-2 line) plants were treated with 200 mM NaCl for 3 h, which are independent from treated seedlings used in qRT-PCR. Four-week-old OE (OE-3 line) and KO (KO-2 line) plants were treated with 200 mM NaCl for 24 h, then the gene expression between OE and KO lines was compared using Affymetrix Arabidopsis gene chips (ATH1 Genome Array) with three independent biological replicates. P and Q values used for the comparison of genotypes were calculated using the EDGE software (http://www.genomine.org/edge/), and P = 0.05 was used as cutoff to select genes significantly differentially regulated in OE vs. KO lines. To validate the gene chip data, 10 differentially expressed genes identified in gene chip were randomly selected for real-time RT-PCR analyses (Table S3 for primers).
Identification of the motif recognized by ANAC069
To study whether ANAC069 regulated the expression of genes via recognizing other motifs besides NACRS, we randomly selected 59 genes that are highly regulated by ANAC069 (including up- and down-regulated genes), and the sequences of their promoter regions (from −1 to −1000) were obtained from TAIR (http://www.Arabidopsis.org). Conserved sequences among these promoters were searched using MEME (Multiple EM for Motif Elicitation) program (http://meme.nbcr.net/meme/cgi-bin/meme.cgi). The conserved sequences among the promoters of genes whose expression affected by ANAC069 were established and displayed using WebLogo.
The conserved sequences identified by MEME were “C[A/G]CG[T/G]”. Three tandem copies of different types of “C[A/G]CG[T/G]” sequences, including CACGT (C1), CACGG (C2), CGCGT (C3), CGCGG (C4), and their mutants AACGT (M1), CAAGT (M2), CACTT (M3) and AAATT (M4), were respectively cloned into a pHIS2 vector (Table S3 for primers), and used for Y1H analysis. For study the interaction between DNA and protein in plants, three tandem copies of C1-4 and M1-4 were respectively fused to the minimal 35S promoter to drive GUS in a reformed pCAMBIA1301 (in which the region of 35S:Hygromycin was deleted and a 46 bp minimal promoter was inserted in the region between HindIII and GUS) to generate reporters (Table S3 for primers). Each of these reporters was cotransformed with effector (pROK II-ANAC069ΔC) into tobacco leaves using the particle bombardment method (Ueki et al. 2009). The 35S:LUC was also cotransformed to normalize the transgenic efficiencies. Three independent biological replications were performed.
ABA and abiotic stress tolerance assays
Seeds of WT, ANAC069 overexpression (OE-2 and OE-3 lines), and ANAC069 knockdown (KO-1 and KO-2 lines) plants were plated on 1/2MS medium supplemented with different concentrations of ABA (0, 0.5, 1, and 2 μM), NaCl (0, 50, 100, and 150 mM), or mannitol (0, 100, 200, and 300 mM). Germination rates were averaged over three experiments at sowing for 4 days using emergence of visible radicles as a morphological marker for germination, and approximately 100–110 seeds were used for each measurement. For root length and fresh weight assessments, 5-days-old seedlings were transferred to vertical agar plates supplied with ABA (0, 10, 20, and 50 μM), NaCl (0, 50, 100, and 150 mM), or mannitol (0, 200, 300, and 400 mM). After 10 day of growth, root lengths and fresh weights of seedlings were measured. For survival rates assays of soil plants, 4-week-old plantlets of WT, OE-2, OE-3, KO-1, and KO-2 grown on soil were treated with water (control), 100 μM ABA, 200 mM NaCl and 300 mM mannitol, respectively. Photographs were taken on the 10th day of stress treatment. After treatment, plants were placed under normal growth conditions and grown for 4 additional days and survival rates were determined. All experiments were repeated at least three times, and 40–50 plants were used in each comparison.
Physiological roles of ANAC069
For physiological analysis, 4-week-old plantlets of WT, OE-2, OE-3, KO-1, and KO-2 were irrigated with water (control), ABA (100 μM), NaCl (200 mM), or mannitol (300 mM) for 3 days, respectively. SOD, POD activities, and electrolyte leakage were measured according to Wang et al. (2012). MDA amount, proline content, and GST activity were determined as described by Song et al. (2011), Bates et al. (1973) and Rezaeia et al. (2013), respectively. At least 30 seedlings were included in each sample and all experiments were run on three biological replicates.
To measure the SOD, POD and GST gene expression regulated by ANAC069, 4-week-old WT, OE and KO lines subjected with ABA (100 μM), NaCl (200 mM), or mannitol (300 mM) for 24 h were harvested. To compare the expression of proline-related genes among different lines under abiotic stress, WT, OE and KO lines treated as above for 3 day were harvested. The transcript levels of the target genes in WT plants under the same condition were used as a calibrator 1 to calculate their expression in other kinds of plants. Seven GST, ten POD, nine SOD and two P5CS genes from Arabidopsis were analyzed by real-time RT-PCR (the loci of these genes in Tair and primers are shown in Table S3). Three independent biological replicates were performed to ensure the accuracy of analyses.
Detection of ROS levels and cell death
Leaves from 4-week-old WT, OE and KO plantlets were subjected to treatments by ABA (50 μM), NaCl (200 mM) or mannitol (300 mM) for 2 h, and respectively infiltrated with 3, 30-diaminobenzidine (DAB) or nitroblue tetrazolium (NBT) following the procedures described by Zhanget al. (2011). Cell death was evaluated by Evans blue staining as described by Kim et al. (2003) with minor modifications. ROS production in intact guard cells was assessed using 2,7-dichlorofluorescin diacetate (H2DCF-DA, Fluka) as described by Zhang et al. (2011).
Water loss assays
Rosette leaves were detached from 4-week-old plants, weighed immediately (fresh weight, FW), and placed on a clean bench (relative humidity of about 50%, 25 °C). Desiccated weights were measured at designated time intervals, as indicated in Fig. 5c. Leaves were finally oven-dried for 24 h at 80 °C to a constant dry weight (DW) and water loss rates (%) measured according to the formula: water loss rate (%) = 1–(desiccated weight–DW)/(FW−DW) × 100.
Chromatin immunoprecipitation (ChIP) assays
The coding region of ANAC069ΔC, was ligated in frame to the N-terminus of GFP driven by the CaMV 35S promoter to generate the ANAC069ΔC-GFP fusion construct (35S::ANAC069ΔC-GFP), which was transformed into Arabidopsis plants. A ChIP assay was performed following the procedures of Haring et al. (2007). Briefly, protein and DNA were cross-linked using formaldehyde and purified. After purification, the cross-linked chromatins were fragmented into 0.3–1.0 kb by sonication, and 1/10 volume of the fragmented chromatin was used as the input. The remaining material was separated into two equal aliquots, and incubated with anti-GFP antibody (ChIP+) or a rabbit anti-hemagglutinin (HA) antibody, as a negative control (ChIP−). The antibody-bound complexes were precipitated by protein A + G Agarose beads, and the enriched DNA fragments were eluted by incubating the cross-linking complex at 65 °C for 5 h. PCR was performed to assay the bindings of ANAC069 to promoters of its target genes, which were visualized by gel electrophoresis. The primer binding regions in these promoters are shown in Fig. 8, and the primers used are shown in Table S3.
Statistical analyses
Statistical analyses were carried out using SPSS 16.0 software (SPSS Inc, Chicago, IL, USA). Data were compared using one-way ANOVA test and differences were considered statistically significant at P < 0.05.
Results
Tissue localization and stress-specific induction of ANAC069
To examine the expression patterns of ANAC069, the promoter sequence of ANAC069 and its full 5′ UTR sequence was fused to GUS (ANAC069pro:GUS), and the construct was transformed into Arabidopsis (Col-0) plants. In 5-day-old seedlings, GUS activity was high in shoots and petioles, and relatively lower GUS activity was detected in young leaves; the lowest GUS activity was observed in roots and stems. In 5-week old plants, GUS activity was detected in all tissues, although it was higher in rosette leaves, cauline leaves, stems and mature siliques, and relatively low in flowers (Fig. 1A). To further confirm these findings, real-time RT-PCR was performed (Fig. 1B), and the ANAC069 expression level in roots of 5-week old plants was used as a calibrator. Consistent with the GUS staining, ANAC069 was highly expressed in rosette leaves, followed by cauline leaves, stems, siliques and young leaves. The expression of ANAC069 in roots of 5-day-old seedlings was lower than that of 5-week-old plants. These results suggested that the expression of ANAC069 is tissue and developmental stage-specific.
Tissue localization and stress specific induction of ANAC069. A GUS staining analysis of ANAC069 expression. a 5-week-old T3 Arabidopsis plant; b root; c stem; d rosette-leaf; e cauline-leaf; f in florescences; g flower; h an inflorescence at anthesis; i sepal; j silique; k stamen; l and m mature silique; n 5-day-old plant. B Real time RT-PCR analysis of ANAC069 expression in different tissues. ANAC069 transcript levels in root were used as a calibrator (set to 0 after log2 transformation) to calculate the relative expression of ANAC069 in other tissues. Error bars were obtained from three replicates analyzed by real time RT-PCR. C GUS staining analysis of ANAC069 expression in response to ABA and mannitol treatment. D Real-time RT-PCR analysis of ANAC069 expression in response to ABA, salt and osmotic. The relative expression level values were log2 transformed. Data are mean ± SD from three independent experiments
Previous study showed that the expression of ANAC069 is induced by salt treatment (Park et al. 2011). To further examine the expression of ANAC069 in response to ABA and osmotic stress, 4-day-old transgenic plants were treated with ABA or mannitol for 24 h before staining. Compared with control plants (untreated), significantly elevated GUS activity was observed in the roots of ABA-treated seedlings, meanwhile the difference in GUS activity was more pronounced in the whole seedlings of mannitol-treated seedlings, including a region of stem that did not show ANAC069 expression in the absence of stress (Fig. 1C). Consistently, real-time RT-PCR showed that the expression of ANAC069 was dramatically induced after salt treatment during the studied period in leaves. In roots, ANAC069 was highly induced by osmotic stress and ABA for 24 h. These findings are consistent with GUS staining results, demonstrating that ANAC069 is induced by ABA and osmotic stress (Fig. 1D). These results suggested that ANAC069 is involved in abiotic stress responses.
Generation of Arabidopsis plants overexpressing and lacking ANAC069
To investigate the function of ANAC069 using gain-and-loss-of function method, transgenic Arabidopsis plants overexpressing ANAC069 were generated using the floral-dip method. We randomly selected three transgenic lines to analyze the expression of ANAC069. The results showed that the expression of ANAC069 increased greatly in these three transgenic lines compared with WT plants (Fig. S1b), indicating that ANAC069 had been transformed and expressed in these lines. The OE-2 and OE-3 lines have the medium and highest ANAC069 expression levels, respectively, and were selected for further study. The transcript levels of ANAC069 were reduced by 91% and 95%, respectively, in SALK_082353 and 095231C lines (Fig. S1b), indicating that ANAC069 was knocked down in these two lines. Therefore, the SALK-082353 and 095231C lines were selected for study, and were termed as KO-1 and KO-2 respectively. Both of the T-DNA insertion sites are located in the promoter region of ANAC069 (Fig. S1a).
ANAC069 modulates abiotic stress tolerance negatively in plants
No difference was found in germination rates, root lengths and fresh weights when the WT and mutant plants were compared under normal growth conditions. However, after salt and mannitol treatments, both WT and KO lines displayed higher germination rates compared with OE lines (Fig. S1c), with the highest root lengths and fresh weights being recorded in the KO lines (Fig. 2a–c). Interestingly, under ABA treatment, KO-1, KO-2 and WT plants showed similar germination rates, root lengths and fresh weights, with values significantly higher than that of OE-2 and OE-3 plants.
Effects of ABA, salt and osmotic stress on plant root lengths, fresh weights, growth and survival rates. a Root length assay. Images were taken after transfer of 5-day-old seedlings from 1/2 MS medium to plates containing 50 μM ABA, 150 mM NaCl or 400 mM mannitol for 10 days. b, c Root length and fresh weight statistical analysis. Seedlings of 5-day-old were subjected with ABA, NaCl or mannitol, respectively. After 10 days of growth, root lengths and fresh weights of seedlings were measured. Three independent experiments were averaged; bars indicate standard deviation. Asterisk indicates significant differences between the wild type and the KO or OE lines under the same conditions (P < 0.05). d Plant growth in soil under treatments of water (used as control), ABA, salt and osmotic stress conditions. e Survival rates of WT, OE and KO after recovery from stress treatments for 4 day. Each data point represents the mean of triplicate tests and 40–50 plants were evaluated in each test. Bars indicate SD. (*P < 0.05, **P < 0.01). WT wild type plants, OE overexpressing ANAC069 plants, KO ANAC069 knockdown plants
Soil-grown plants were also subjected to treatment with abiotic stresses and ABA. Consistent with above data, there was no difference in growth among the WT, OE-3 and KO-2 lines under normal growth conditions (Fig. 2d). However, OE-3 showed the most severe growth retardation and the lowest survival rate under salt, osmotic stress and ABA treatment conditions. Meanwhile, KO-2 and WT plants showed no significant difference under ABA treatment conditions. However, KO-2 showed higher tolerance to salt and osmotic stress compared with WT plants (Fig. 2d, e). Taken together, these findings indicated that ANAC069 plays a detrimental role in the salt and osmotic response of plants.
ANAC069 modulates ROS scavenging systems negatively
We examined O −2 and H2O2 cellular levels by DAB or NBT in situ staining. As shown in Fig. 3a, b, the steady state levels of both H2O2 and O −2 were largely reduced in KO lines compared with OE and WT plants after salt and mannitol treatments. We then evaluated the cellular levels of H2O2 in guard cells under abiotic stress, using H2DCF-DA staining. As shown in Fig. 3c, the H2O2 levels of KO lines in guard cells were much lower than in the WT or OE lines under salt and mannitol stress, whereas OE lines displayed the highest O −2 and H2O2 levels under abiotic stress conditions. These results demonstrated a positive correlation between the transcript level of ANAC069 and ROS levels in response to salt and osmotic stress.
Detection of ROS levels and physiological roles of ANAC069. a, b Analyses of ROS levels by histochemical staining in situ. Histochemical staining by DAB and NBT to reveal accumulation of O −2 and H2O2. Rosette leaves sampled from 4-week-old Arabidopsis plantlets treated with water (control), 50 μM ABA, 200 mM NaCl or 300 mM mannitol for 2 h were used for histochemical staining assay. c Representative microscopy images of ROS production in intact guard cells indicated by fluorescent dye H2DCF-DA. Rosette leaves sampled from plantlets treated as above were used to obtain epidermal peels. d Physiological roles of ANAC069. 4-week-old Arabidopsis plantlets were treated with water, ABA, NaCl, or mannitol for 3 day for physiological change evaluation. Bars indicate SD and data are mean ± SD from three independent experiments. Asterisk indicates significant differences between the wild type and the KO or OE lines under the same conditions (*P < 0.05)
We further assessed GST, SOD and POD activities, which were all significantly reduced in the OE-2 and OE-3 lines compared with the KO-1 and KO-2 lines under either normal growth or salt and mannitol-induced stress conditions (Fig. 3d). Under normal or ABA treatment conditions, both SOD and POD activities were lower in the OE lines than in the KO lines and WT plants; however, the KO lines and WT plants showed similar SOD and POD activities. Under salt and mannitol-induced stress conditions, the KO lines showed the highest SOD and POD activities, followed by WT plants; the OE lines displayed the lowest SOD and POD activities (Fig. 3d). Under normal growth and stress conditions, GST activity was significantly higher in the KO lines compared with the WT and OE lines. GST activity in the WT was similar to that of the OE lines under normal growth conditions. However, under abiotic and ABA treatment conditions, GST activity in both the KO lines were highest, followed by that in the WT; the OE lines had the lowest GST activities (Fig. 3d). The MDA content assay showed that the KO lines had the lowest MDA levels, followed by WT plants; the OE lines showing highest MDA levels under all studied conditions (Fig. 3d).
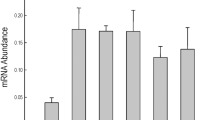
As GST, POD and SOD activities were significantly altered among the KO, WT and OE lines, we further assessed the expression of these genes in various plant lines. Nine SOD, ten POD and seven GST genes from Arabidopsis, which were previously confirmed to express proteins with POD, SOD and GST activities, were selected for analyses. Under salt or osmotic conditions, their expressions were more strongly induced in the KO lines and were highly decreased in the OE lines compared with the WT. In particular, in the KO lines, many genes showed the highest increases (Fig. 4). These results suggested that ANAC069 regulates the expression of SOD, POD, and GST genes negatively, especially under salt and osmotic stress.
Analysis of the expressions of SOD, POD and GST genes in WT, OE and KO plants. 4-week-old WT, OE and KO plants subjected with ABA (100 μM), NaCl (200 mM), or mannitol (300 mM) for 24 h were harvested for analysis. The transcript levels of the target Genes in WT plants under the same condition were used as a calibrator 1 to calculate their expression in other kinds of plants. The error bars were calculated from three replicates of the real-time RT-PCR
Expression of ANAC069 increases cell death and water loss rate
No difference was observed in electrolyte leakage rate among the KO, OE and WT lines under normal growth conditions. However, under ABA, salt and mannitol-induced stress conditions, the OE lines showed the highest electrolyte leakage rates, followed by the WT and KO lines, in that order (Fig. 5a). Consistent with these findings, Evans Blue staining showed decreased cell death in the KO lines compared with the WT and OE lines; the OE lines displaying the most cell damage under abiotic stress or ABA conditions (Fig. 5b). These results support the view that ANAC069 regulates the abiotic stress response negatively in Arabidopsis. The KO lines showed an enhanced capacity to conserve water compared with the WT. However, the water loss rates were higher in the OE lines compared with the WT under dehydration conditions (Fig. 5c). These data indicated a detrimental role of ANAC069 in the conservation of water, and the increased water loss rate at contributed, least in part, to the reduced salt and osmotic resistance observed in the OE lines.
Analysis of cell death and water loss rates. a Assay of electrolyte leakage. Relative electrolyte leakage was assessed after exposure to water (control), ABA (100 μM), NaCl (200 mM), or mannitol (300 mM) for 3 day. Three replicates were performed. Bars indicate SD from three replicates (*P < 0.05). b Analysis of cell death by Evans Blue staining. Rosette leaves sampled from seedlings treated with water, ABA, NaCl or mannitol for 2 h were used for Evans Blue staining. c Water loss rate assays. Rosette leaves were dried and weighed at the indicated time points. Leaves from ten plants were measured and averaged in each experiment, done in triplicates. Bars indicate SD
Proline biosynthesis is regulated negatively by ANAC069
We found no difference in proline contents among the OE, KO and WT plants under normal growth conditions (Fig. 6a). After ABA treatments, the WT and KO lines showed similar proline levels, while the OE lines displayed lower proline levels. Under salt and osmotic stress conditions, the KO lines exhibited the highest proline levels, followed by the WT and OE lines, in that order (Fig. 6a).
Proline metabolism in WT, OE and KO plants. a Proline content assays. Proline contents were measured after treated with water, ABA, NaCl, or mannitol for 3 day. Three independent assays were performed. (*P < 0.05, one-way ANOVA test). b Comparison of the expression of P5CS genes among WT, OE and KO plants. Four-week-old plants were treated as above and the expression of proline-related genes P5CS1 and P5CS2 were analyzed. The relative gene expression was calculated and normalized with the reference gene ACT7 and TUB2. Error bars represent the standard deviations from three independent replicates
Finally, we evaluated the expression of two known biosynthesis genes, P5CS1 and P5CS2, in the OE, WT and KO lines. We found that P5CS2 (AT3G55610) was highly expressed in the KO lines, followed by the WT plants and OE lines after exposure to salt, osmotic stress and ABA (Fig. 6b), which is fully consistent with the proline level patterns (Fig. 6a). However, P5CS1 (AT2G39800) was expressed at its highest levels in the WT plants, and the OE lines had the lowest P5CS1 level under all conditions, which also indicated that proline biosynthesis is highly inhibited in the OE lines.
Identification of genes affected by ANAC069 using a gene chip
To identify the target genes regulated by ANAC069, the expression profiles of the OE and KO lines under salt stress conditions were compared using Affymetrix Arabidopsis gene chips. A total of 339 and 226 genes with p values <0.05 and a fold change ≥2 were upregulated and downregulated in OE lines relative to the KO lines, respectively (Table S1). Ten differentially expressed genes identified in the gene chip data were selected randomly for real-time RT-PCR analyses to validate the gene chip results, and their locus number and primer sequences are shown as Table S3. The results showed a high correlation between the real-time RT-PCR and gene chip data (R2 = 0.9866, P < 0.05) (Fig. S2), demonstrating the reliability of the gene chip results. These gene chip data were submitted to the GEO database of the series accession number GSE82274.
Gene ontology (GO) enrichment analysis highlighted 52 (List Hits ≥4) significant GO terms classified as biological process, molecular function or cellular component (Fig. S3). Many genes regulated by ANAC069 were related with the stress response. For instance, in the biological process category, 11 GO sub-terms related to stress response were highly enriched (>5%), including response to chitin, response to wounding, and response to abscisic acid stimulus. This result suggested that ANAC069 alters the expression of stress response genes, resulting in abiotic stress sensitivity in plants. Therefore, these gene chip data will be helpful to reveal the regulatory mechanism of ANAC069 in response to salt stress.
ANAC069 recognizes NACRS and ORS1S11 motif
Previous studies showed that the NAC proteins could bind to NACRS and ORS1S11 motif (Tran et al. 2004; Matallana-Ramirez et al. 2013). To determine whether ANAC069 also recognizes the NACRS and ORS1S11 motif, yeast one-hybrid (Y1H) analysis was performed. The results showed that ANAC069 recognizes the NACRS motif (AGCTCTTCTTCTGTAACACGCATGT) and mutants of non-core sequences, Ma (in the sequence of “AAAAAATC AAAAAAAA CACGCATGT”, the mutated sequences are italics), or Mg “AGGGGG TC GGGGGGAACACGCATGT”, but is not able to recognize to the mutated core sequence Mcacg (in the sequence, “CACG” is underlined: “AGCTCTTCTTCTGTAACACGCATGT” and was mutated to “TTTT”) (Fig. 7b). To confirm the interactions identified in the Y1H assays, the effector (pROKII–ANAC069ΔC) and reporter constructs (pCAM-NACRS, -Ma, -Mg, and -Mcacg) were coexpressed into tobacco leaves. In accordance with the Y1H results, ANAC069 induced the expression of the GUS gene driven by the NACRS motifs and NACRS motifs with mutated non-core sequences (Ma and Mg; Fig. 7d), but failed to activate the GUS gene driven by the mutated core sequence Mcacg. These findings together demonstrated that ANAC069 can recognize the NACRS motif, and the core NACRS sequence, CACG, is crucial for ANAC069 protein recognition. As shown in Fig. 7f, ANAC069 could also recognize the ORS1S11 motif with the sequence of CGGGGTTACGTACGGCACACGCAACCGTGC (the core sequences for NAC binding are underlined), but failed in recognize to all-nucleotide mutations, both within the first motif (CGGGGTTAAAAACGGCACACGCAACCGTGC) and within the second binding motif (CGGGGTTACGTACGGCAAAAACAACCGTGC). The data indicated that ANAC069 can recognize the ORS1S11 motif and that both the core sequences ACGTA and CACG are essential for ANAC069 protein recognition.
Analyses of the recognition of ANAC069 to NACRS and ORS1S11 motif. a The sequence of NACRS and its mutants. b Y1H analyses of the recognition of ANAC069 to NACRS. pHIS2-NACRS, -Ma, -Mg, -Mcacg: One copy of NACRS or its three mutant types (-Ma, -Mg and -Mcacg) was respectively cloned into the pHIS2 upstream of the HIS3 gene as the reporter construct. pGADT7-ANAC069: pGADT7-Rec2 that encodes ANAC069 fused with GAL4AD as the effector construct. c Schematic diagram of the effector and reporter constructs used in plant transient expression assays. d Analyses of recognition of ANAC069 to NACRS in tobacco leaves by transient expression assays. The reporter constructs pCAM-NACRS, pCAM-Ma, pCAM-Mg and pCAM-Mcacg (one copy of the NACRS and mutant sequences Ma, Mg and Mcacg were fused to the minimal 35S promoter to drive GUS) were respectively cotransformed with the effector construct pROK II-ANAC069ΔC (consisting of residues 1–287 of ANAC069into pROK II driven by the 35S promoter) into tobacco leaves using the particle bombardment method. The 35S:LUC was cotransformed to normalize the transgenic efficiencies. Transformation of the empty pCAMBIA1301(35S:GUS) was used as positive control. Cotransformation of empty pROK II and pCAM-NACRS were used as negative control. Data represent mean values of three independent experiments. Bars indicate SD. e The sequence of ORS1S11 and its mutants. f Y1H analyses of the recognition of ANAC069 to ORS1S11. pHIS2-ORS1S11, -ORS1S11M1, -ORS1S11M2: one copy of ORS1S11 or its two mutant types was respectively cloned into the pHIS2 upstream of the HIS3 gene as the reporter construct. p53HIS2/pGADT7-p53 and p53HIS2/pGADT7-ANAC069 were used as positive and negative controls, respectively. Positive transformants were further identified by spotting serial dilutions (1, 10−1, 10−2 and10−3) of yeast onto SD/-His/-Leu/-Trp (TDO) with 40 mM 3-AT. The transformants grown at SD/-Leu/-Trp (DDO) were used as positive controls for transformant growth
Identification of the DNA sequence recognized by ANAC069
We randomly selected 59 genes that are highly differentially regulated by ANAC069 to study the conserved sequences in their promoter regions. Although both the NACRS (containing both CATGTG and CACG) and the ORS1S11 motifs are recognized by ANAC069, many genes regulated by ANAC069 do not contain either an NACRS or an ORS1S11 motif in their promoter regions (Table S2). This phenomenon prompted us to study whether there are other DNA sequences that can be recognized by ANAC069. Promoter sequences (from −1 to −1000) of these genes were searched using MEME. The conserved sequences present in the promoters of genes affected by ANAC069 were displayed using WebLogo, where the character size represents the frequency of occurrence. The motif detected by MEME was “[G/C/A][C/A]C[A/G]CG[T/G]” (Fig. 8a). The [G/C/A] and [C/A] both had low frequencies in the detected motif, indicating that they might not be important for recognition by ANAC069; therefore, the sequence of C[A/G]CG[T/G] was further studied. The recognition by ANAC069 of CACGT (C1), CACGG (C2), CGCGT (C3), CGCGG (C4) and mutants AACGT (M1), CAAGT (M2), CACTT (M3) and AAATT (M4) were studied using a Y1H assay. The results confirmed that ANAC069 could recognize to all these sequences, but failed to recognize the mutants (Fig. 8b). The recognition by ANAC069 of the C[A/G]CG[T/G] sequences was further studied in plants. Reporters containing sequences C1-4 or mutant sequences M1-4 fused with the 35S minimal promoter to drive the GUS gene were cotransformed with effector (35S:ANAC069ΔC) and 35S:LUC into tobacco plants. GUS activity was normalized using the LUC activity to normalize the transformation efficiencies. GUS activity measurements indicated that these sequences could be bound by ANAC069; however, the four mutants all failed to be recognized by ANAC069 (Fig. 8d). Taken together, these results suggested that ANAC069 could activate the GUS reporter gene when any of the four sequence types of C[A/G]CG[T/G] sequence were present.
Identification of the sequences of C[A/G]CG[T/G] recognized by ANAC069. a Analyses of the putative ANAC069-recognizing sequences using MEME. The size of the characters represents the frequency of occurrence. b Y1H analysis of the recognitions of ANAC069 to the sequences of C[A/G]CG[T/G]. C1-4: the different types of the sequences of “C[A/G]CG[T/G]” used for further study; M1-4: four types of mutant sequences. pHIS2-C1, C2, C3, C4, M1, M2, M3, M4: three tandem copies of these sequences were respectively cloned into the pHIS2 upstream of the HIS3 gene as the reporter constructs. pGADT7-ANAC069: pGADT7-Rec2 that encodes ANAC069 fused with GAL4AD as the effector construct. c Schematic diagram of the effector and reporter constructs used in plant transient expression assays. d Determination of the recognition of C1-4 to ANAC069 by in vivo study of plants. Three tandem copies of C1-4 and M1-4 were respectively fused to 35S minimal promoter to drive GUS gene as reporters. The effector vector was constructed by cloning the truncated ANAC069ΔC form into pROKII driven by the 35S promoter (pROK II-ANAC069ΔC). Each of these reporters was cotransformed with effector (pROK II-ANAC069ΔC) into tobacco leaves using the particle bombardment method. The 35S:LUC was also cotransformed to normalize the transgenic efficiencies. Empty pCAMBIA1301 was used as positive control (35S:GUS). pROK II-ANAC069ΔC plasmid was used as a negative control. The data represent mean values of three independent experiments. Error bars indicate SD. e ChIP analysis of the regulation of the binding of ANAC069 to “C[A/G]CG[T/G]” motifs present in promoter. PCR products amplified from the promoter of AT3G02840 was size fractionated by gel electrophoresis. Grey lines represent the promoter fragments and black shading represents the “C[A/G]CG[T/G]” motifs. M marker DL2000. Input, PCR amplification of sonicated chromatin. ChIP+, immunoprecipitated chromatin incubated with anti-GFP antibody; ChIP−, immunoprecipitated chromatin incubated with anti-hemagglutinin (HA) antibody. 1, 2 and 3, PCR product of the promoter fragments using three pairs of primers, respectively. f Binding of ANAC069ΔC to the promoters of NACs in vivo. The truncated promoters of ANAC019, AtNAP, ANAC055 and ATAF1 were PCR amplified from Input, ChIP+ and ChIP−. Black shading represents the C[A/G]CG[T/G] motifs
ChIP analysis of ANAC069 binding to the promoters of target genes
To further study whether the binding of ANAC069 to C[A/G]CG[T/G] sequences actually occurs in Arabidopsis plants, ChIP analysis was performed. The transgenic Arabidopsis plants expressing GFP-tagged ANAC069ΔC was generated under the control of CaMV35S promoter. AT3G02840, which was upregulated by ANAC069 in that microarray data and contained three C[A/G]CG[T/G] sequences and no other NAC binding motifs in its promoter region, was selected for ChIP analysis. The results showed that the truncated promoter of AT3G02840 was detected only from a chromatin immunoprecipitated fragment when precipitated using an anti-GFP antibody (ChIP+); whereas this promoter fragment was not detected when precipitated with the anti-HA antibody (ChIP−) (Fig. 8e), suggesting that ANAC069 binds to the C[A/G]CG[T/G]sequences in vivo.
Some NAC genes that were highly induced by ANAC069 according to the microarray contained C[A/G]CG[T/G] sequences. To study whether they could be regulated directly by ANAC069, ChIP analysis was performed. The results showed that the truncated promoters of AtANP, ATAF1, ANAC055 and ANAC019 could be amplified from DNA immunoprecipitated with the anti-GFP antibody (Fig. 8f), further indicating that ANAC069 could bind to C[A/G]CG[T/G] sequences to regulate the expressions of its target genes.
The C[A/G]CG[T/G] sequences exist commonly in the promoters of genes regulated by ANAC069
Seventy-seven genes that were highly upregulated by ANAC069 identified by the gene chip in this study were selected for study. The promoter regions (−1000 to −1) of these genes were screened for the C[A/G]CG[T/G] sequences, NACRS (containing both the discontinuous sequences of CACG and CATGTG) motifs and ORS1S11 motifs (containing both the discontinuous sequences of ACGTA and CACG). The results showed that 73 genes (94.8%) contain the C[A/G]CG[T/G] sequences in their promoter regions, six genes have NACRS motifs and two genes have ORS1S11 motifs (Table S2). These results indicated that the C[A/G]CG[T/G] sequences exist commonly in the promoter regions of genes regulated by ANAC069, and should play an important role in gene expression regulation mediated by ANAC069.
Discussion
NAC proteins play essential roles in plant development and stress responses. NACs with a transmembrane (TM) motif1-like (NTL) are TFs and have regions including the N-terminal NAC domain, the middle regulation region, and the C-terminal TM domain. In the present study, we studied NAC TF ANAC069. Previously, we had found that ANAC069 is regulated by AtDOF5.8 (He et al. 2015). Parket al. (2011) and Jung and Park (2011) showed that ANAC069 mediated salt signaling by crosstalk with auxin to delay seed germination under salinity stress conditions. These findings prompted further studies to reveal the mechanism by which ANAC069 is involved in abiotic stress regulation. Additionally, the physiological responses to abiotic stress mediated by ANAC069 remain mostly unknown. In the present study, we proposed a working model of ANAC069 in response to abiotic stress.
The regulatory mechanism of ANAC069 in response to abiotic stress
NAC proteins can bind the discontinuous motif NACRS (containing discontinuous core sequence of CATGTG and CACG), continuous core sequence TTNCGT[G/A] and CGT[G/A] to regulate gene expression (Zhouet al. 2013; Hu et al. 2006; Tran et al. 2004; Jensen et al. 2010; Xu et al. 2013; Lindemose et al. 2014). NAC proteins can also bind to the bi-partite sequence ORS1S11 (Matallana-Ramirez et al. 2013). Additionally, the ANAC069 protein was found to bind to the IAA30-BS sequence that contains the ACGT conserved sequence (Park et al. 2011). In the present study, ANAC069 was also found to bind to NACRS and ORS1S11 motifs (Fig. 7). In addition, MEME analysis showed that most of the genes regulated by ANAC069 have a C[A/G]CG[T/G] sequence in their promoters (Fig. 8a). Both Y1H assay and in vivo interaction between ANAC069 and the sequences of C[A/G]CG[T/G] in plants showed that the C[A/G]CG[T/G] sequence can be recognized specifically by ANAC069 (Fig. 8b, d–f). Therefore, besides recognition to discontinuous motifs such as NACRS and ORS1S11, ANAC069 can also recognize continuous the DNA sequences C[A/G]CG[T/G]. Furthermore, ANAC069 showed different transactivation activities when interacting with four sequence types of the sequences of C[A/G]CG[T/G]: CACGT showed the highest transactivation activity, followed by CGCGT and CACGG; CGCGG showed the lowest transactivation activity. These results suggested that the last nucleic acid “T” in the C[A/G]CG[T/G] sequences is important for ANAC069 recognition (Fig. 8d).
Some NAC proteins could recognize the DNA motif TTNCGT[G/A] (Jensen et al. 2010), or to the core sequence of CGT[G/A] (Xu et al. 2013; Lindemose et al. 2014). In the present study, we identified that ANAC069 also recognizes the sequences C[A/G]CG[T/G], which are similar to CGT[G/A] (reverse complementary sequences of CGT[G/A] contain CACG, which is a partial sequence of C[A/G]CG[T/G]). However, there were some differences between these motifs, and these differences might be useful to reveal the requirements of NAC proteins to recognize different DNA sequences. For instance, CGCGT (C3) and CGCGG (C4) are both recognized by ANAC069 (Fig. 8b, d), which are not similar to CGT[G/A]. These results indicated that NAC could recognize diverse DNA motifs. Additionally, ANAC069 could recognize the CACGT (C1), and CACGG (C2) motifs, but failed to interact with the mutated motifs AACGT (M1), CAAGT (M2), CACTT (M3) and AAATT (M4) (Fig. 2a), indicating that the CxCG sequence is important for NAC protein recognition.
Considering that the genes regulated differentially by ANAC069 also include that are not regulated directly by ANAC069 (i.e. they are regulated by genes that are regulated by ANAC069), the fact that 94.8% of the genes regulated by ANAC069 contain the sequences of C[A/G]CG[T/G] in their promoters (Table S2) suggested that most gene that are regulated directly by ANAC069 should contain the sequences of C[A/G]CG[T/G] in their promoters. Therefore, these results indicated that ANAC069 regulates the expression of genes mainly by recognizing the C[A/G]CG[T/G] sequences.
Overexpression of ANAC069 decreases tolerance to abiotic stresses and ABA
The ANAC069 (NTM2) protein consists of 457 residues, and is a membrane binding protein. However, the ΔC construct that contained only residues 1–287 was targeted to the nucleus (Park et al. 2011). Transgenic plants overexpressing the full CDS of ANAC069 were phenotypically indistinguishable from control plants, while plants overexpressing the ΔC form exhibited a dwarfed phenotype, with small, curled leaves, indicating that membrane release of the ANAC069 protein is essential for its activity (Park et al. 2011). In the present study, we found that plants overexpressing the full CDS of ANAC069 showed increased sensitivity to salt, osmotic and ABA stimuli, compared with the WT and ANAC069 knockdown plants, indicating that the full CDS of ANAC069 also works well in plants in response to salt, osmotic and ABA stimuli (Figs. 2, 3, 4, 5, 6). These results suggested that membrane release of the ANAC069 protein occurs when plants are exposed to salt, osmotic and ABA stimuli. Consistently, previous studies showed that most NTLs are localized to the endoplasmic reticulum, and are restricted to the nucleus. Arabidopsis NTL13, NTL10 and NTL8 contain certain inhibitory regions that mask the nuclear localization signal sequence in the N-terminus of the NAC domain and permit their diffusion between the cytoplasm and nucleus. The N-terminus of the NAC domain and the middle regulation region of NTL are translocated into the nucleus when activated by proteolytic cleavage events stimulated by internal and external environmental signals (Liang et al. 2015). Therefore, stress response studies could be carried out with the full CDS of ANAC069. Notably, the full CDS of ANAC069 inserted into pGADT7-rec2 in Y1H assays also confirmed the ability of ANAC069 to recognize NACRS and ORS1S11 motif and t C[A/G]CG[T/G] sequences in yeasts, suggesting that ANAC069 in yeast cells should be only partly membrane bound or not membrane bound. In addition, the WT and KO lines showed similar tolerances to ABA treatment. However, the OE lines were more sensitive to ABA, indicating that a certain level of ANAC069 in plants does not affect ABA tolerance. Nevertheless, high ANAC069 expression level result in sensitivity to ABA.
ANAC069 inhibits the expression of antioxidant genes, leading to reduced ROS scavenging ability
ROS scavenging is crucial in plant abiotic stress tolerance. As ANAC069 is sensitive to abiotic stress, we further investigated ROS accumulation in plants with different ANAC069 expressions. The results showed that ANAC069 affects ROS accumulation negatively (Fig. 3). Additionally, we found that the expression of Redox Responsive Transcription Factor1 (RRTF1) is highly induced by ANAC069 (Table S1). RRTF1 was upregulated rapidly and transiently by H2O2, as well as by biotic- and abiotic-induced redox signals, and increased expression of RRTF1 could induce ROS accumulation in Arabidopsis (Matsuo et al. 2015). Therefore, the accumulated ROS might represent the reason for the high induction of RRTF1 by ANAC069. Conversely, the induced RRTF1 might lead to enhanced ROS accumulation. Furthermore, ANAC069 expression is correlated negatively with GST, POD and SOD activities (Fig. 3d), and with the transcript levels of most SOD, POD and GST genes (Fig. 4). The fact that not all genes are correlated negatively with ANAC069 expression is reasonable, because these genes may be involved in different regulatory networks that do not involve ANAC069. Taken together, these data suggested that ANAC069 affects the expression of POD, SOD and GST genes negatively to modulate their activities, resulting in decreased ROS scavenging capability and sensitivity to salt and osmotic stress.
ANAC069 decreases the expression of P5CS genes to reduce proline accumulation
In plants, proline protects cells from damage by acting as both a radical scavenger and an osmotic agent. Once the stress is relieved, stored proline is degraded to provide a supply of energy for growth. In addition, proline maintains sustainable growth under long-term stress, and its homeostasis is important for actively dividing cells (Kavi and Sreenivasulu 2014). In Arabidopsis, two P5CS isoforms have been identified, P5CS1 (AT2G39800) and P5CS2 (AT3G55610), with different expression patterns and specific functions in primary metabolism and stress defense (Funcket al. 2012). We showed that both P5CS1 and P5CS2 displayed their lowest expression levels in the OE lines (Fig. 6b). Correspondingly, proline levels were also lower in the OE lines compared with the KO and WT plants (Fig. 6a). These data indicated that proline biosynthesis is highly decreased in the OE lines because of the downregulation of P5CS genes, resulting in sensitivity to salt, osmotic and ABA stress conditions. Furthermore, the expression of P5CS2 was higher in the KO than WT plants, but no difference in the expression level of P5CS1 was observed between the KO and WT plants; meanwhile, the proline content was significantly higher in the KO lines compared with the WT plants (Fig. 6a), which suggested that expression of P5CS2 is closely related to proline levels, probably playing a more critical role in proline biosynthesis mediated by ANAC069 when exposed to abiotic stressors. Therefore, high expression of P5CS2 results in elevated proline levels in the KO lines and the enhanced proline levels contribute to the improvement in salt and osmotic stress resistance in these lines.
Changing the expression of ANAC069 appears to have a major effect on other NAC genes
Our microarray results showed that many NAC genes were upregulated by ANAC069 (Table S1). For instance, the NAC genes such as ANAC019, AtNAP, ANAC055, ATAF1 and ANAC102 were significantly upregulated by ANAC069. Additionally, many of these NACs have the C[A/G]CG[T/G] sequences in their promoter regions, and some of them were confirmed to be regulated directly by ANAC069, according to the ChIP analysis (Fig. 8). These results suggested that altering ANAC069 expression in transgenic plants could have a major effect on other NACs and their target genes.
Working model of ANAC069 in response to abiotic stress
Our data suggest a working model for the function of ANAC069 in the abiotic stress response. Abiotic stresses, such as salt or osmotic stress conditions, induce stress signals to trigger the expression of ANAC069. Meanwhile, stress signals also induce the release of expressed ANAC069 from the membrane (Park et al. 2011). The activated ANAC069 then recognizes motifs such as C[A/G]CG[T/G] to regulate its target genes. Finally, the altered expressions of the downstream genes of ANAC069 lead to physiological changes that modulate the plant stress response, including decreased ROS scavenging leading to ROS accumulation, inhibition of proline biosynthesis to decrease osmotic potential and increased water loss rates, among others (Fig. 9).
Model of ANAC069 regulatory network involved in abiotic stress responses. Abiotic stresses such as salt or osmotic stress induce stress signals to trigger the expression of ANAC069. The activated ANAC069 in turn binds to the NAC binding motifs, such as CACGT, NACRS or ORS1S11, to regulate the expression of genes. The altered expression of genes finally leads to physiological changes that modulate plant stress response, including decreased ROS scavenging capability, reduced proline biosynthesis, and increased water loss rates, among others
References
Aida M, Ishida T, Fukaki H, Fujisawa H, Tasaka M (1997) Gene involved in organ separation in Arabidopsis: an analysis of the cup-shaped cotyledon mutant. Plant Cell 9:841–857
Balazadeh S, Siddiqui H, Allu AD, Matallana-Ramirez LP, Caldana C, Mehrnia M, Zanor MI, Kohler B, Mueller-Roeber B (2010) A gene regulatory network controlled by the NAC transcription factorANAC092/AtNAC2/ORE1 during salt-promoted senescence. Plant J 62:250–264
Balazadeh S, Kwasniewski M, Caldan C, Mehrni M, Zanor MI, Xue GP, Mueller-Roeber B (2011) ORS1, an H2O2-responsive NAC transcription factor, controls senescence in Arabidopsis thaliana. Mol Plant 4:346–360
Bates LS, Waldren RP, Teare JD (1973) Rapid determination of free proline for water stress studies. Plant Soil 39:205–207
Duval I, Lachance D, Giguère I, Bomal C, Morency MJ, Pelletier G, Boyle B, MacKay JJ, Séguin A (2014) Large-scale screening of transcription factor-promoter interactions in spruce reveals a transcriptional network involved in vascular development. J Exp Bot 65:2319–2333
Fang Y, You J, Xie K, Xie W, Xiong L (2008) Systematic sequence analysis and identification of tissue-specific or stress-responsive genes of NAC transcription factor family in rice. Mol Genet Genomics 280:547–563
Fang Y, Xie K, Xiong L (2014) Conserved miR164-targeted NAC genes negatively regulate drought resistance in rice. J Exp Bot 65(8):2119–2135
Funck D, Winter G, Baumgarten L, Forlani G (2012) Requirement of proline synthesis during Arabidopsis reproductive development. BMC Plant Biol 12:191
Guan Q, Yue X, Zeng H, Zhu J (2014) The protein phosphatase RCF2 and its interacting partner NAC019 are critical for heat stress-responsive gene regulation and thermotolerance in Arabidopsis. Plant Cell 26:438–453
Hao YJ, Wei W, Song QX, Chen HW, Zhang YQ, Wang F, Zou HF, Lei G, Tian AG, Zhang WK, Ma B, Zhang JS, Chen SY (2011) Soybean NAC transcription factors promote abiotic stress tolerance and lateral root formation in transgenic plants. Plant J 68:302–313
Haring M, Offermann S, Danker T, Horst I, Peterhansel C, Stam M (2007) Chromatin immunoprecipitation: optimization, quantitative analysis and data normalization. Plant Methods 3:11
He L, Su C, Wang Y, Wei Z (2015) ATDOF5.8 protein is the upstream regulator of ANAC069 and is responsive to abiotic stress. Biochimie 110:17–24
Hibara K, Karim MR, Takada S, Taoka K, Furutani M, Aida M, Tasaka M (2006) Arabidopsis CUP-SHAPED COTYLEDON3 regulates postembryonic shoot meristem and organ boundary formation. Plant Cell 18:2946–2957
Hu H, Dai M, Yao J, Xiao B, Li X, Zhang Q, Xiong L (2006) Overexpressing a NAM, ATAF, and CUC (NAC) transcription factor enhances drought resistance and salt tolerance in rice. Proc Natl Acad Sci USA 103:12987–12992
Hu R, Qi G, Kong Y, Kong D, Gao Q, Zhou G (2010) Comprehensive analysis of NAC domain transcription factor gene family in Populus trichocarpa. BMC Plant Biol 10:145
Jefferson R (1989) The GUS reporter gene system. Nature 342(6251):837
Jefferson RA, Kavanagh TA, Bevan MW (1987) GUS fusions: beta glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J 6:3901–3907
Jensen MK, Kjaersgaard T, Nielsen MM, Galberg P, Petersen K, O’Shea C, Skriver K (2010) The Arabidopsis thaliana NAC transcription factor family: structure–function relationships and determinants of ANAC019 stress signalling. Biochem J 426:183–196
Jin HX, Huang F, Cheng H, Song HN, Yu DY (2013) Overexpression of the GmNAC2gene, an NAC transcription factor, reduces abiotic stress tolerance in tobacco. Plant Mol Biol Rep 31:435–442
Jung JH, Park CM (2011) Auxin modulation of salt stress signaling in Arabidopsis seed germination. Plant Signal Behav 6(8):1198–1200
Kavi Kishor PB, Sreenivasulu N (2014) Is proline accumulation per se correlated with stress tolerance or is proline homeostasis a more critical issue? Plant Cell Environ 37:300–311
Kim M, Ahn JW, Jin UH, Choi D, Paek KH, Pai HS (2003) Activation of the programmed cell death pathway by inhibition of proteasome function in plants. J Biol Chem 278:19406–19415
Kim SG, Kim SY, Park CM (2007) A membrane-associated NAC transcription factor regulates salt-responsive flowering via FLOWERING LOCUS T in Arabidopsis. Planta 226:647–654
Kim MJ, Park MJ, Seo PJ, Song JS, Kim HJ, Park CM (2012) Controlled nuclear import of the transcription factorNTL6 reveals a cytoplasmic role of SnRK2.8 in the drought-stress response. Biochem J 448:353–363
Le DT, Nishiyama R, Watanabe Y, Mochida K, Yamaguchi-Shinozaki K, Shinozaki K, Tran LS (2011) Genomewide survey and expression analysis of the plant-specific NAC transcription factor family in soybean during development and dehydration stress. DNA Res 18:263–276
Lee S, Park CM (2012) Regulation of reactive oxygen species generation under drought conditions in Arabidopsis. Plant Signal Behav 7:599–601
Lee S, Lee HJ, Huh SU, Paek KH, Ha JH, Park CM (2014) The Arabidopsis NAC transcription factor NTL4 participates in a positive feedback loop that induces programmed cell death under heat stress conditions. Plant Sci 227:76–83
Liang C, Wang Y, Zhu Y, Tang J, Hu B, Liu L, Ou S, Wu H, Sun X, Chu J, Chu C (2014) OsNAP connects abscisic acid and leaf senescence by fine-tuning abscisic acid biosynthesis and directly targeting senescence-associated genes in rice. Proc Natl Acad Sci USA 111:10013–10018
Liang M, Li H, Zhou F, Li H, Liu J, Hao Y, Wang Y, Zhao H, Han S (2015) Subcellular distribution of NTL transcription factors in Arabidopsis thaliana. Traffic 16(10):1062–1074
Lindemose S, Jensen MK, Van de Velde J, O’Shea C, Heyndrickx KS, Workman CT, Vandepoele K, Skriver K, De Masi F (2014) A DNA-binding-site landscape and regulatory network analysis for NAC transcription factors in Arabidopsis thaliana. Nucleic Acids Res 42(12):7681–7693
Lu M, Sun QP, Zhang DF, Wang TY, Pan JB (2015) Identification of 7 stress-related NAC transcription factor members in maize (Zea mays L.) and characterization of the expression pattern of these genes. Biochem Biophys Res Commun 462(2):144–150
Matallana-Ramirez LP, Rauf M, Farage-Barhom S, Dortay H, Xue GP, Dröge-Laser W, Lers A, Balazadeh S, Mueller-Roeber B (2013) NAC transcription factor ORE1 and senescence-induced BIFUNCTIONAL NUCLEASE1 (BFN1) constitute a regulatory cascade in Arabidopsis. Mol Plant 6(5):1438–1452
Matsuo M, Johnson JM, Hieno A, Tokizawa M, Nomoto M, Tada Y, Godfrey R, Obokata J, Sherameti I, Yamamoto YY, Böhmer FD, Oelmüller R (2015) High REDOX RESPONSIVE TRANSCRIPTION FACTOR1 levels result in accumulation of reactive oxygen species in Arabidopsis thaliana shoots and roots. Mol Plant 8(8):1253–1273
Nakashima K, Tran LA, Van Nguyen S, Fujita M, Maruyama K, Todaka S, Ito Y, Hayashi N, Shinozaki K, Yamaguchi-Shinozaki K (2007) Functional analysis of a NAC-type transcription factor OsNAC6 involved in abiotic and biotic stress-responsive gene expression in rice. Plant J 51:617–630
Ohashi-Ito K, Oda Y, Fukuda H (2010) Arabidopsis VASCULAR-RELATED NACDOMAIN6 directly regulates the genes that govern programmed cell death and secondary wall formation during xylem differentiation. Plant Cell 22:3461–3473
Ooka H, Satoh K, Doi K, Nagata T, Otomo Y, Murakami K, Matsubara K, Osato N, Kawai J, Carninci P, Hayashizaki Y, Suzuki K, Kojima K, Takahara Y, Yamamoto K, Kikuchi S (2003) Comprehensive Analysis of NAC Family Genes in Oryza sativa and Arabidopsis thaliana. DNA Res 10:239–247
Park J, Kim YS, Kim SG, Jung JH, Woo JC, Park CM (2011) Integration of auxin and salt signals by the NAC transcription factor NTM2 during seed germination in Arabidopsis. Plant Physiol 156:537–549
Pfaffl MW, Horgan GW, Dempfle L (2002) Relative expression software tool (REST©) for group-wise comparison and statistical analysis of relative expression results in real-time RT-PCR. Nucleic Acids Res 30:e36
Rezaeia MK, Shobbar ZS, Shahbazia M, Abedinia R, Zare S (2013) Glutathione S-transferase (GST) family in barley: identification of members, enzyme activity, and gene expression pattern. J Plant Physiol 170:1277–1284
Sakuma Y, Maruyama K, Osakabe Y, Qin F, Seki M, Shinozaki K, Yamaguchi-Shinozaki K (2006a) Functional analysis of an Arabidopsis transcription factor, DREB2A, involved in drought-responsive gene expression. Plant Cell 18:1292–1309
Sakuma Y, Maruyama K, Qin F, Osakabe Y, Shinozaki K, Yamaguchi-Shinozaki K (2006b) Dual function of an Arabidopsis transcription factor DREB2A in water-stress-responsive and heat stress-responsive gene expression. Proc Natl Acad Sci USA 103:18822–18827
Seo PJ, Kim MJ, Park JY, Kim SY, Jeon J, Lee YH, Kim J, Park CM (2010) Cold activation of a plasma membrane tethered NAC transcription factor induces a pathogen resistance response in Arabidopsis. Plant J 61:661–671
Shahnejat-Bushehri S, Mueller-Roeber B, Balazadeh S (2012) Arabidopsis NAC transcription factor JUNGBRUNNEN1 affects thermomemory-associated genes and enhances heat stress tolerance in primed and unprimed conditions. Plant Signal Behav 7:1518–1521
Shan W, Kuang JF, Lu WJ, Chen JY (2014) A banana fruit NAC transcription factor MaNAC1 is a direct target of MaICE1 and involved in cold stress through interacting with MaCBF1. Plant Cell Environ 37:2116–2127
Song SY, Chen Y, Chen J, Dai XY, Zhang WH (2011) Physiological mechanisms underlying OsNAC5-dependent tolerance of rice plants to abiotic stress. Planta 234:331–345
Souer E, van Houwelingen A, Kloos D, Mol J, Koes R (1996) The No Apical Meristem gene of Petunia is required for pattern formation in embryos and flowers and is expressed at meristem and primordia boundaries. Cell 85:159–170
Takasaki H, Maruyama K, Kidokoro S, Ito Y, Fujita Y, Shinozaki K, Yamaguchi-Shinozaki K, Nakashima K (2010) The abiotic stress-responsive NAC-type transcription factor OsNAC5 regulates stress-inducible genes and stress tolerance in rice. Mol Genet Genomics 284:173–183
Takasaki H, Maruyama K, Takahashi F, Fujita M, Yoshida T, Nakashima K, Myouga F, Toyooka K, Yamaguchi-Shinozaki K, Shinozaki K (2015) SNAC-As, stress-responsive NAC transcription factors, mediate ABA-inducible leaf senescence. Plant J 84(6):1114–1123
Tran LS, Nakashima K, Sakuma Y, Simpson SD, Fujita Y, Maruyama K, Fujita M, Seki M, Shinozaki K, Yamaguchi-Shinozaki K (2004) Isolation and functional analysis of Arabidopsis stress-inducible NAC transcription factors that bind to a osmotic-responsive cis-element in the early responsive to dehydration stress 1 Promoter. Plant Cell 16:2481–2498
Ueki S, Lacroix B, Krichevsky A, Lazarowitz SG, Citovsky V (2009) Functional transient genetic transformation of Arabidopsis leaves by biolistic bombardment. Nat Protoc 4:71–77
Wang L, Xu C, Wang C, Wang Y (2012) Characterization of a eukaryotic translation initiation factor 5 A homolog from Tamarix and rossowii involved in plant abiotic stress tolerance. BMC Plant Biol 12:118
Welner DH, Lindemose S, Grossmann JG, Mollegaard NE, Olsen AN, Helgstrand C, Skriver K, Lo LL (2012) DNA binding by the plant-specific NAC transcription factors in crystal and solution: a firm link to WRKY and GCM transcription factors. Biochem J 444:395–404
Wu A, Allu AD, Garapati P, Siddiqui H, Dortay H, Zanor MI, Asensi-Fabado MA, Munné-Bosch S, Antonio C, Tohge T, Fernie AR, Kaufmann K, Xue GP, Mueller-Roeber B, Balazadeh S (2012) JUNGBRUNNEN1, a reactive oxygen species-responsive NAC transcription factor, regulates longevity in Arabidopsis. Plant Cell 24:482–506
Xie Q, Frugis G, Colgan D, Chua NH (2000) Arabidopsis NAC1 transduces auxin signal downstream of TIR1 to promote lateral root development. Gene Dev 14:3024–3036
Xu ZY, Kim SY, Hyeon DY, Kim DH, Dong T, Park Y, Jin JB, Joo SH, Kim SK, Hong JC, Hwang D, Hwang I (2013) The Arabidopsis NAC transcription factor ANAC096 cooperates with bZIP-type transcription factors in dehydration and osmotic stress responses. Plant Cell 25:4708–4724
Yao D, Wei Q, Xu W, Syrenne RD, Yuan JS, Su Z (2012) Comparative genomic analysis of NAC transcriptional factors to dissect the regulatory mechanisms for cell wall biosynthesis. BMC Bioinform 13:S10
Zhang X, Wang L, Meng H, Wen H, Fan Y, Zhao J (2011) Maize ABP9 enhances tolerance to multiple stresses in transgenic Arabidopsis by modulating ABA signaling and cellular levels of reactive oxygen species. Plant Mol Biol 75:365–378
Zhou Y, Huang W, Liu L, Chen T, Zhou F, Lin Y (2013) Identification and functional characterization of a rice NAC gene involved in the regulation of leaf senescence. BMC Plant Biol 13:132
Acknowledgements
This work was supported by the National Natural Science Foundation of China (No. 31270703).
Author contributions
YW conceived and designed the experiments; LH carried out the experiments; LH, XS, HYX, YW, KY and YG analyzed the experimental data. LH and YW wrote the manuscript. All authors read and approved the final manuscript.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
He, L., Shi, X., Wang, Y. et al. Arabidopsis ANAC069 binds to C[A/G]CG[T/G] sequences to negatively regulate salt and osmotic stress tolerance. Plant Mol Biol 93, 369–387 (2017). https://doi.org/10.1007/s11103-016-0567-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11103-016-0567-3