Abstract
Cell-wall invertases (CWINVs), responsible for the hydrolysis of sucrose into glucose and fructose, play important roles in plant development. Maize (Zea mays L.) ear-related traits were suggested to be important components of kernel yield. However, the association and nucleotide polymorphisms of maize CWINV genes with ear-related traits remain largely unclear. Herein, the maize ZmCWINV3 gene was re-sequenced in 301 inbred lines, 71 landraces and 31 teosintes to mine excellent variations associated with ear-related traits. A total of 594 variants, including 498 single nucleotide polymorphisms (SNPs) and 96 insertions and deletions (InDels), were identified in maize ZmCWINV3 locus. Five polymorphism sites in ZmCWINV3 were found to be significantly associated with six ear-related traits. Among them, a non-synonymous mutation SNP3694 was found to be significantly associated with ear weight (EW), ear grain weight (EGW), ear length (EL) and kernel length (KL). Further evidence revealed that the excellent allele T of SNP3694 was not exist in teosintes, while its frequencies raised to 9.86% and 69.44% in landraces and inbred lines, respectively. These observations suggested that the excellent allelic variations in ZmCWINV3 might possess potential application values in the genetic improvement of ear-related traits in maize.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Maize (Zea mays L.), a high-yield staple crop, plays critical role in food production in China as well as in the world. In addition, maize is also one of the vital sources of fodder, industrial raw materials and economic crops. Improving maize kernel yield is an essential goal during maize domestication and genetic improvement (Li et al. 2011a). However, the formation of kernel yield is a very complicated process. Understanding the genetic basis of ear-related traits will contribute tremendously to maize breeding (Zhou et al. 2020). The ear-related traits, including ear weight (EW), ear grain weight (EGW), ear length (EL) and kernel length (KL), show important influence on maize yield. However, most of these traits belong to complex quantitative traits which are influenced by both genetic background and environmental factors (Liu et al. 2012). The detection and application of superior alleles of ear-related traits associated genes have contributed a lot to maize yield improvement in the past few years (Jia et al. 2020; Chen et al. 2020). Therefore, it is of great importance to identify the genes related to ear-related traits and to detect the superior alleles for maize breeding.
Invertase (β-D-fructofuranoside fructohydrolase), a key enzyme of sucrose metabolism in both source and sink tissues (Juarez-Colunga et al. 2018), plays a critical role in the hydrolysis of sucrose to glucose and fructose (Slewinski 2011). According to their subcellular localization and optimum pH values, invertases were divided into three types, including alkaline/neutral cytoplasmic invertase (CINV) and acid invertase, which was further classified into vacuolar invertase (VINV) and cell wall invertase (CWINV) (Sturm 1999). A total of 21 invertases have been identified in maize, including ten CINVs, three VINVs and eight CWINVs (Juarez-Colunga et al. 2018). The activity of invertase was suggested to be consistent with the number of endosperm cells during grain filling stage, indicating that the activity of invertase was associated with grain filling rate (Qin et al. 2016; Estruch and Beltrán 1991). Importantly, the positive association between CWINV activity and seed development has been illustrated in multiple plant species, including Litchi chinensis Sonn, Manihot esculenta Crantz, and Solanum lycopersicum L. (Zhang et al. 2018; Shen et al. 2019; Yan et al. 2019). Generally, CWINVs showed high activity in the meristem and fast-growing young tissues of plants, indicating that CWINVs might possess functions in regulating plant growth and organogenesis (French et al. 2014). Crop CWINVs were also suggested to play decisive roles in the transportation of assimilates to developing grains (Cho et al. 2005; Chourey et al. 2006). However, the genetic variations of ZmCWINVs in cultivated maize populations and its association with ear-related traits have not been revealed.
In the present research, we re-sequenced the gene ZmCWINV3 from 301 inbred lines, 71 landraces and 31 teosintes. The purposes of this study include: (1) to detect the nucleotide polymorphisms of this gene, (2) to identify the polymorphisms associated with maize ear-related traits, and (3) to estimate the differences of ear-related traits in different haplotypes.
Materials and methods
Plant materials and the analysis of phenotypic data
In this study, a total of 301 inbred lines, 71 landraces, and 31 teosintes have been selected. These lines had been planted in the field using a randomized complete block design with two replications at Sanya (18°23ʹ N, 109°44ʹ E) in 2015 and 2016, and Yangzhou (32°39ʹ N, 119°42ʹ E) in 2017. An inbred line was planted in a sequential row patterns with 15 plants, 3.5 m long and 0.4 m between adjacent rows. Then after postharvest drying, three well-developed ears have been chosen to measure ear-related traits, including ear weight (EW), ear grain weight (EGW), ear length (EL), ear diameter (ED), ear row number (ERN), kernel number per row (KNR), hundred kernel weight (HKW), kernel length (KL), kernel width (KW), and kernel thickness (KT).
ANOVA of the phenotypic data of all ear-related traits in the three different environments were performed using the ‘aov’ function, and correlation coefficient analysis was carried out by ‘psych’ package in R. The ‘lme4’ package was used to calculate the broad-sense heritability (h2) for ear-related traits (Bates et al. 2015). The phenotypic data for each ear trait was analyzed by the best linear unbiased predictor (BLUP) method by ‘lme4’ package.
DNA extraction and ZmCWINV3 resequencing
Fresh and young leaves were collected from each line at the seeding stage, and a modified cetyl trimethyl ammonium bromide (CTAB) method was used to extract genomic DNA. The gene ZmCWINV3 (GRMZM2G123633) was sequenced by BGI (Beijing Genomics Institute) Life Tech Co. China using targeted sequence capture technology on the NimbleGen platform (Choi et al. 2009).
Analysis of sequence data
The software Clustal X (Larkin et al. 2007) was used for multi-sequence alignment of the maize ZmCWINV3 and further edited manually. The single nucleotide polymorphisms (SNP), allelic diversities and haplotype diversity of all tested lines were analyzed using DNASP5.0 software (Librado and Rozas 2009). Nucleotide diversity (π) in the ZmCWINV3 gene was defined as the mean number of nucleotide differences per site between any two DNA sequences using R package ‘PopGenome’ and using sliding window method with a window size of 200 bp and a step length of 50 bp (Pfeifer et al. 2014).
Marker-trait association analysis in inbred lines
TASSEL5.0 software (Bradbury et al. 2007) was used to analyze the relationship between the candidate ZmCWINV3 and promoter region sequences of 301 inbred lines and the BLUP values of EW, EGW, EL, ED, ERN, KNR, HKW, KL, KW, and KT. Single nucleotide polymorphisms (SNPs) and insertions and deletions (InDels) were screened out by a small allele frequency < 5% (Supplementary table S1). TASSEL5.0 software was also used to calculate principal component analysis (PCA) and kinship. LD (Linkage disequilibrium) analysis was estimated between significantly associations within the sequenced region of ZmCWINV3. The LD heatmap and R2 were generated using R packages ‘LDheatmap’ and ‘pegas’, respectively (Vens and Ziegler 2017; Paradis 2010).
Results
Sequence polymorphisms of ZmCWINV3
To detect the nucleotide polymorphisms of the maize ZmCWINV3 gene, the full-length sequences of this locus were re-sequenced in 301 inbred lines, 71 landraces, and 31 teosintes (Supplementary dataset S1). After multiple sequence alignment, a whole of 4282 bp sequences were obtained, including a 1390 bp upstream and a 117 bp 5’UTR regions, a 1803 bp exon region constituting of six exons, a 922 bp intron region including five introns, and a 50 bp 3’UTR region (Table 1). Sequence polymorphisms, including SNPs and InDels (insertion and deletion), had been identified at ZmCWINV3. A total of 594 variations, including 498 SNPs and 96 InDels, were detected. On average, one SNP and InDel were detected every 8.60 bp and 44.60 bp, respectively. And the highest frequencies of SNPs and InDels were both detected in the promoter region (4.95 bp and 23.17 bp, respectively).
The nucleotide diversity (π) was calculated for the ZmCWINV3 gene. Results showed that the overall nucleotide diversity (π) in this locus was 0.017. We also noticed that the estimated π values showed large variations in different regions. Compared with coding regions, the estimated π values of non-coding regions were relatively higher. Among five defined regions of the ZmCWINV3, nucleotide diversity (π) was lowest in the 5’UTR region, followed by exon regions. While the highest frequency of polymorphism was observed in the promoter region.
Nucleotide diversity and selection of ZmCWINV3 in inbred lines, landraces and teosintes
The genetic diversity of ZmCWINV3 were further compared in 301 inbred lines, 71 landraces, and 31 teosintes (Table 2). We found all the estimated π values are highest in teosintes and lowest in inbred lines, suggesting that putative selection occurred in the full range of the gene sequence. When using a sliding window of 200 bp with a step length of 50 bp, we observed differential nucleotide diversity in 14 regions (promoter, 5’UTR, 6 exons, 5 introns and 3’UTR) of the ZmCWINV3 gene. The most obvious difference was observed between inbred lines and teosintes in the promoter region, which was also the region with the highest nucleotide diversity, while nucleotide diversity of other regions was relatively low (Fig. 1). And the value of π was lowest in exons (Table 2). This uneven distribution of polymorphisms might be mainly due to the coding region had lower frequency of variants.
The neutrality of ZmCWINV3 gene was tested by Tajima’s D, Fu and Li’s D*, and Fu and Li’s F* (Tables 1 and 2). The Tajima’s D values of the three different populations for the entire region didn’t achieve a significant level. Furthermore, we noticed that the estimates of Fu and Li’s D* and F*statistic for this gene were significantly higher than zero in inbred lines. These results suggest that there are many moderate frequency alleles in this population.
Phenotypic variations and association analysis
A total of ten ear-related traits, including EW, EGW, EL, ED, ERN, KNR, HKW, KL, KW and KT, were obtained for 301 maize inbred lines (Table 3). ANOVA analyses revealed that all these traits showed significant variations among inbred lines, suggesting that this population hold genetic characteristics for association analysis. The broad-sense heritability estimation revealed that most ear-related traits had high heritability (Table 3), indicating the data were suitable for further association analysis.
In order to explore the relationship between these ten ear-related traits, pairwise correlation analyses have been performed, and the Pearson correlation coefficients (r) between any two ear-related traits had been calculated. We noticed that most of the trait pairs showed significant positive correlations (p < 0.05) (Fig. 2). Notably, EGW, an important yield trait, showed significant positive correlations with most ear-related traits except KT. Among them, EW/EGW had the highest correlation (r = 0.975, p < 0.0001).
Pearson correlation coefficients (r) between any two ear-related traits in 301 inbred lines. Abbreviations for traits are as follows: EW, ear weight; EGW, ear grain weight; EL, ear length; ED, ear diameter; ERN, ear row number; KNR, kernel number per row; HKW, hundred kernel weight; KL, kernel length; KW, kernel width and KT, kernel thickness. Asterisks indicate significant differences as determined by Student’s t-test (***p < 0.001; **p < 0.01; *p < 0.05)
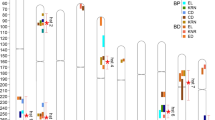
To detect the significant variations in ZmCWINV3 associated with ear-related traits, association analyses were performed in 301 inbred lines. Under the MLM (PCA + K) model, a total of five polymorphism sites (2 SNPs and 3 InDels) were found to be significantly related to six ear-related traits (EW, EGW, EL, ERN, KNR and KL) (Fig. 3). Two variation sites (SNP3577 and SNP3694) were found to be statistically associated with EW, EGW and EL, explaining 3.37–4.31% of the phenotype variations. SNP3694 was also found to be significantly associated with KL, explaining 3.21% of the phenotype variation. InDel3496 and SNP3577 were associated with KNR, explaining 3.37–4.63% of the variations of phenotype. In addition, two other InDels, including InDel1065 and InDel1072, were estimated to be statistically associated with ERN, explaining 3.27–4.18% of the phenotype variations (Fig. 4a, b). Among the SNP sites associated with ear-related traits, SNP3694 with a non-synonymous mutation of the C to T transition lead to arginine mutates to tryptophan.
LD analysis showed that InDel3496, SNP3577 and SNP3694 had relatively high linkage across inbred lines (Fig. 5a). Among them, both SNP3577 and SNP3694 were significant associated with EW, EGW and EL. Three major haplotypes were divided based on these two SNPs. The phenotypic differences in EW, EGW and EL among three main haplotypes were compared, and significant differences were tested by ANOVA among haplotypes. We found that hap-1 and hap-3 had significant higher values of EW, EGW and EL than hap-2 (Fig. 5b–d). In addition, we noticed that SNP3694 in both hap-1 and hap-3 is allele T, indicating that SNP3694-T was a superior allele. We further classified haplotypes based on the variation of SNP3694, and divided the inbred lines into two main groups. Significant differences between two haplotypes were observed for EW, EGW, EL and KL. The allele T group possessed significantly higher values of EW, EGW, EL, and KL than the allele C group (Fig. 5e–h). Two variants at sites InDel3496 and SNP3577 were significantly associated with KNR, and the tested inbred lines have been divided into two haplotypes based on SNP3577, and a significant distinction was observed between two haplotypes. Hap-1, carrying the increased allele A, had significant higher kernel number per row compared with hap-2, which carried the decreased allele G (Fig. 5j).
Comparisons of maize ear-related traits (EW, EGW, EL and KNR) among groups carrying different ZmCWINV3 alleles. The p values for Student’s t-test analysis and ANOVA comparing the groups carrying different alleles were indexed on the top. a Linkage disequilibrium (LD) heatmap for five significant variants associated with ear-related traits; b-d Comparisons of haplotypes in ZmCWINV3 among natural variations based on SNP3577 and SNP3694; e-i Comparisons of EW, EGW, EL, KL and KNR between two alleles of SNP3694 and SNP3577 (***, p < 0.001; **, p < 0.01; *, p < 0.05); j-k The allele frequencies of SNP3577-A and SNP3694-T in teosintes, landraces and inbred lines
Based on these results, combined with the correlation analysis of ear-related traits, we can infer that the allele T of SNP3694 positively affect EW through increasing KL, the allele A of SNP3577 positively affect KNR, eventually leading to the increase of EW and EGW. We further investigated the frequencies of superior alleles among 301 inbred lines, 71 landraces and 31 teosintes. Interestingly, we noticed both of the superior alleles of two polymorphism sites were not observed in teosintes. The frequencies of SNP3694-T and SNP3577-A in landraces were 9.86% and 5.63%, and increased to 69.44% and 61.46% in inbred lines, respectively (Fig. 5k, l).
Discussion
Association analysis is a widely used method in exploring the genetic basis of complex traits, and it is also an efficient method for confirming candidate genes or detecting the relationship of phenotype with new genes (Flint-Garcia et al. 2003). Association analysis takes advantage of linkage disequilibrium to link phenotypes to genotypes, and can explore all the recombination events and mutations in a given population in high resolution (Thornsberry et al. 2001). Maize is a monoecious cross-pollination crop with extensive morphological variation, high recombination frequency and genetic diversity (Jiao et al. 2012; Li et al. 2012; Whitt et al. 2002). Candidate gene association mapping has been widely used in the detection of functional SNPs or alleles related to agronomic traits in maize. Many functional genes were identified using this method, such as Zmisa2 (Yang et al. 2014) and ZmBT1 for starch properties (Xu et al. 2014), ZmYS1 for kernel mineral concentrations (Yang et al. 2015), ZmMADS60 (Li et al. 2020) for root morphology, ZmHKT1 (Li et al. 2019a) and ZmPGP1 (Li et al. 2019b) for plant architecture.
In the present research, results showed that the maize ZmCWINV3 gene possessed abundant nucleotide polymorphisms among the tested populations, and the association of nucleotide polymorphisms was established of ZmCWINV3 with ear-related traits. Our results revealed that five polymorphic sites possess significant associations with the phenotypes of six ear-related traits, including EW, EGW, EL, ERN, KNR and KL. Among them, SNP3577 and SNP3694 were found to be associated with EW, EGW and EL. In addition, we further noticed that these two sites are highly linked across inbred lines. The characteristic of high linkage was also detected between InDel1065 and InDel1072, which are found to both associated with KNR. The potential mechanism of this association might be owed to the correlation relationship among ear-related traits and the LD between these two sites.
Ear and kernel traits are the key factors affecting maize yield, and they were also the target traits of maize breeding (Li et al. 2011b). Identifying the natural variations of these traits is helpful to improve the efficiency of maize breeding. In this study, we identified significant association between the polymorphisms of ZmCWINV3 locus and ear-related traits. Among them, a non-synonymous mutation SNP3694 was found to be significantly associated with EW, EGW, EL, and KL. We also noticed that the excellent allele T of SNP3694 was not exist in teosintes, while its frequencies raised to 9.86% and 69.44% in landraces and inbred lines, respectively. These observations suggested that this site might have been applied in the practice of maize breeding. In addition, we also noticed that the estimates of Fu and Li’s D* and F* are significantly higher than zero in the population of inbred lines, suggestive many alleles with moderate frequency caused by the bottleneck effect (Wang et al. 1998).
In summary, we re-sequenced the maize ZmCWINV3 gene in 301 inbred lines, 71 landraces, and 31 teosintes. Our results revealed that a total of five variants were significantly related to six ear-related traits. Especially, the non-synonymous mutation SNP3694 was significantly associated with EW, EGW, EL, and KL. These results revealed that the superior allelic variations of ZmCWINV3 possess potential application values in maize molecular breeding.
Data availability
Data supporting the findings of this study are available in Supplementary Table S1 and Supplementary Dataset S1.
References
Bates D, Machler M, Bolker BM, Walker SC (2015) Fitting linear mixed-effects models using lme4. J Stat Softw 67(1):1–48. https://doi.org/10.18637/jss.v067.i01
Bradbury PJ, Zhang Z, Kroon DE, Casstevens TM, Ramdoss Y, Buckler ES (2007) TASSEL: software for association mapping of complex traits in diverse samples. Bioinformatics 23(19):2633–2635. https://doi.org/10.1093/bioinformatics/btm308
Chen L, Li YX, Li C, Shi Y, Song Y, Zhang D, Wang H, Li Y, Wang T (2020) The retromer protein ZmVPS29 regulates maize kernel morphology likely through an auxin-dependent process(es). Plant Biotechnol J 18(4):1004–1014. https://doi.org/10.1111/pbi.13267
Cho JI, Lee SK, Ko SH, Kim HK, Jun SH, Lee YH, Bhoo SH, Lee KW, An GH, Hahn TR, Jeon JS (2005) Molecular cloning and expression analysis of the cell-wall invertase gene family in rice (Oryza sativa L.). Plant Cell Rep 24(4):225–236. https://doi.org/10.1007/s00299-004-0910-z
Choi M, Scholl UI, Ji W, Liu T, Tikhonova IR, Zumbo P, Nayir A, Bakkaloğlu A, Ozen S, Sanjad S, Nelson-Williams C, Farhi A, Mane S, Lifton RP (2009) Genetic diagnosis by whole exome capture and massively parallel DNA sequencing. Proc Natl Acad Sci U S A 106(45):19096–19101. https://doi.org/10.1073/pnas.0910672106
Chourey PS, Jain M, Li QB, Carlson SJ (2006) Genetic control of cell wall invertases in developing endosperm of maize. Planta 223(2):159–167. https://doi.org/10.1007/s00425-005-0039-5
Estruch JJ, Beltrán JP (1991) Gibberellic acid stimulates acid invertase secretion in pea ovary protoplasts. FEBS Lett 279(2):303–306. https://doi.org/10.1016/0014-5793(91)80173-z
Flint-Garcia SA, Thornsberry JM, Buckler ES (2003) Structure of linkage disequilibrium in plants. Annu Rev Plant Biol 54(1):357–374. https://doi.org/10.1146/annurev.arplant.54.031902.134907
French SR, Abu-Zaitoon Y, Uddin MM, Bennett K, Nonhebel HM (2014) Auxin and cell wall invertase related signaling during rice grain development. Plants (basel) 3(1):95–112. https://doi.org/10.3390/plants3010095
Jia H, Li M, Li W, Liu L, Jian Y, Yang Z, Shen X, Ning Q, Du Y, Zhao R, Jackson D, Yang X, Zhang Z (2020) A serine/threonine protein kinase encoding gene KERNEL NUMBER PER ROW6 regulates maize grain yield. Nat Commun 11(1):988. https://doi.org/10.1038/s41467-020-14746-7
Jiao Y, Zhao H, Ren L, Song W, Zeng B, Guo J, Wang B, Liu Z, Chen J, Li W, Zhang M, Xie S, Lai J (2012) Genome-wide genetic changes during modern breeding of maize. Nat Genet 44(7):812-U124. https://doi.org/10.1038/ng.2312
Juarez-Colunga S, Lopez-Gonzalez C, Cecilia Morales-Elias N, Armando Massange-Sanchez J, Trachsel S, Tiessen A (2018) Genome-wide analysis of the invertase gene family from maize. Plant Mol Biol 97(4–5):385–406. https://doi.org/10.1007/s11103-018-0746-5
Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG (2007) Clustal W and clustal X version 2.0. Bioinformatics 23(21):2947–2948. https://doi.org/10.1093/bioinformatics/btm404
Li Y, Ma X, Wang T, Li Y, Liu C, Liu Z, Sun B, Shi Y, Song Y, Carlone M, Bubeck D, Bhardwaj H, Whitaker D, Wilson W, Jones E, Wright K, Sun S, Niebur W, Smith S (2011a) Increasing maize productivity in china by planting hybrids with germplasm that responds favorably to higher planting densities. Crop Sci 51(6):2391–2400. https://doi.org/10.2135/cropsci2011.03.0148
Li C, Li Q-G, Dunwell JM, Zhang Y-M (2012) Divergent evolutionary pattern of starch biosynthetic pathway genes in grasses and dicots. Mol Biol Evol 29(10):3227–3236. https://doi.org/10.1093/molbev/mss131
Li P, Pan T, Wang H, Wei J, Chen M, Hu X, Zhao Y, Yang X, Yin S, Xu Y, Fang H, Liu J, Xu C, Yang Z (2019a) Natural variation of ZmHKT1 affects root morphology in maize at the seedling stage. Planta 249(3):879–889. https://doi.org/10.1007/s00425-018-3043-2
Li P, Wei J, Wang H, Fang Y, Yin S, Xu Y, Liu J, Yang Z, Xu C (2019b) Natural variation and domestication selection of ZmPGP1 affects plant architecture and yield-related traits in maize. Genes (basel). https://doi.org/10.3390/genes10090664
Li P, Ge Z, Wang H, Wei J, Wang Y, Xu Y, Yang Z, Xu C (2020) Nucleotide diversity and association analysis of ZmMADS60 with root length in the maize seedling stage. Agronomy (basel). https://doi.org/10.3390/agronomy10030342
Librado P, Rozas J (2009) DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25(11):1451–1452. https://doi.org/10.1093/bioinformatics/btp187
Liu R, Jia H, Cao X, Huang J, Li F, Tao Y, Qiu F, Zheng Y, Zhang Z (2012) Fine mapping and candidate gene prediction of a pleiotropic quantitative trait locus for yield-related trait in zea mays. PLoS ONE. https://doi.org/10.1371/journal.pone.0049836
Paradis E (2010) pegas: an R package for population genetics with an integrated-modular approach. Bioinformatics 26(3):419–420. https://doi.org/10.1093/bioinformatics/btp696
Pfeifer B, Wittelsbuerger U, Ramos-Onsins SE, Lercher MJ (2014) PopGenome: an efficient swiss army knife for population genomic analyses in R. Mol Biol Evol 31(7):1929–1936. https://doi.org/10.1093/molbev/msu136
Qin G, Zhu Z, Wang W, Cai J, Chen Y, Li L, Tian S (2016) A tomato vacuolar invertase inhibitor mediates sucrose metabolism and influences fruit ripening. Plant Physiol 172(3):1596–1611. https://doi.org/10.1104/pp.16.01269
Shen S, Ma S, Liu Y, Liao S, Li J, Wu L, Kartika D, Mock HP, Ruan YL (2019) Cell wall invertase and sugar transporters are differentially activated in tomato styles and ovaries during pollination and fertilization. Front Plant Sci 10:506. https://doi.org/10.3389/fpls.2019.00506
Slewinski TL (2011) Diverse functional roles of monosaccharide transporters and their homologs in vascular plants: a physiological perspective. Mol Plant 4:641–662. https://doi.org/10.1093/mp/ssr051
Sturm A (1999) Invertases. Primary structures, functions, and roles in plant development and sucrose partitioning. Plant Physiol 121(1):1–8. https://doi.org/10.1104/pp.121.1.1
Thornsberry JM, Goodman MM, Doebley J, Kresovich S, Nielsen D, Buckler ES (2001) Dwarf8 polymorphisms associate with variation in flowering time. Nat Genet 28(3):286–289. https://doi.org/10.1038/90135
Vens M, Ziegler A (2017) Estimating disequilibrium coefficients. In: Elston RC (ed) Statistical human genetics: methods and protocols. Springer, NY, pp 117–132. https://doi.org/10.1007/978-1-4939-7274-6_7
Wang J, Caballero A, Hill WG (1998) The effect of linkage disequilibrium and deviation from Hardy-Weinberg proportions on the changes in genetic variance with bottlenecking. Heredity 81(2):174–186. https://doi.org/10.1046/j.1365-2540.1998.00390.x
Whitt SR, Wilson LM, Tenaillon MI, Gaut BS, Buckler ESt, (2002) Genetic diversity and selection in the maize starch pathway. Proc Natl Acad Sci U S A 99(20):12959–12962. https://doi.org/10.1073/pnas.202476999
Xu S, Yang Z, Zhang E, Jiang Y, Pan L, Chen Q, Xie Z, Xu C (2014) Nucleotide diversity of Maize ZmBT1 gene and association with starch physicochemical properties. PLoS ONE 9(8):e103627. https://doi.org/10.1371/journal.pone.0103627
Yan W, Wu X, Li Y, Liu G, Cui Z, Jiang T, Ma Q, Luo L, Zhang P (2019) Cell wall invertase 3 affects cassava productivity via regulating sugar allocation from source to sink. Front Plant Sci 10:541. https://doi.org/10.3389/fpls.2019.00541
Yang Z, Zhang E, Jiang Y, Xu S, Pan L, Chen Q, Xu C (2014) Sequence polymorphisms in Zmisa2 gene are significantly associated with starch pasting and gelatinization properties in maize (Zea mays L.). Mol Breeding 34(4):1833–1842. https://doi.org/10.1007/s11032-014-0142-z
Yang ZF, Ma SJ, Hu YY, Zhang EY, Xie ZW, Xu SH, Liu L, Deng LL, Xu CW, Huang JL (2015) Association analysis of the maize gene ZmYS1 with kernel mineral concentrations. Plant Mol Biol Rep 33(5):1327–1335. https://doi.org/10.1007/s11105-014-0836-8
Zhang J, Wu Z, Hu F, Liu L, Huang X, Zhao J, Wang H (2018) Aberrant seed development in Litchi chinensis is associated with the impaired expression of cell wall invertase genes. Hortic Res. https://doi.org/10.1038/s41438-018-0042-1
Zhou Z, Li G, Tan S, Li D, Weiss TM, Wang X, Chen S, Wuerschum T, Liu W (2020) A QTL atlas for grain yield and its component traits in maize (Zea mays). Plant Breeding 139(3):562–574. https://doi.org/10.1111/pbr.12809
Acknowledgements
This work was supported by the grant from the National Natural Science Foundation of China (32070558, 32061143030, 31801028, 31972487, 32100448), a project Funded by the Priority Academic Program Development of Jiangsu Higher Education Institutions (PAPD), and Natural Science Foundation of Jiangsu Province (BK20210799).
Funding
The authors have not disclosed any funding.
Author information
Authors and Affiliations
Contributions
M.Z., C.X. and Z.Y. conceived and designed the research. Y.L., R.C., M.Z., Z.Z. and Y.Z. performed the association analyses and relative analyses. T.T., Y.J., Z.Z., E.Z., D.H., P.L. and Y.X. collected and analyzed the phenotypic data. M.Z., C.X. and Z.Y. wrote the manuscript. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Consent for publication
All authors have given their consent for publication.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Zhu, M., Lu, Y., Chen, R. et al. Nucleotide polymorphisms of the maize ZmCWINV3 gene and their association with ear-related traits. Genet Resour Crop Evol 69, 2115–2124 (2022). https://doi.org/10.1007/s10722-022-01360-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10722-022-01360-3