Abstract
The flag leaf size of wheat is an “ideotypic” morphological trait that plays a critical role in plant architecture and grain yield by providing photosynthetic assimilates in wheat. Although many of the genomics research studies covered the flag leaf traits, including flag leaf length (FLL), width (FLW), area (FLA), thickness (FLT), and volume (FLV), for a better understanding, this research used a recombinant inbred line (RIL) population derived from a cross between DH118 and Jinmai 919 to evaluate the genetic regions across six environments, including BLUP under both drought stress (DS) and well-watered (WW) conditions and analyze their correlation with traits related to grain yield. A total of 40 (QTL) quantitative trait loci controlling the five traits were detected across all environments, with phenotypic variance explaining (PVE) 5.09%-15.26%. Among them, 12 QTL were identified as stable, including two QTL for FLL, two for FLW, three for FLA, two for FLT and three for FLV, in which nine QTL were found to be validated in more than three environments through a double haploid (DH) population Jinchun 7 × Jinmai 919. The Qflw.saw-2A, Qfla.saw-2A, Qflv.saw-2A, Qflt.saw-2B, and Qflt.saw-3B were stated as novel due to not being reported by any of the previous research studies related to flag leaf traits. In addition, traits related to flag-leaf and grain yield were significantly correlated in both water regimes. These results provide a better understanding of the genetic basis underlying flag leaf traits. Also, target regions for fine mapping and marker-assisted selection (MAS) were identified and will be valuable for breeding high-yielding bread wheat.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Wheat (Triticum aestivum L.) is one of the most important staple food crops worldwide, growing in a wide range of environmental conditions such as water regimes, climatic factors, and soil types. Due to the human food security issue in the current global climatic changes, especially under drought (Trenberth 2011), the increased production of wheat must meet the sustainable food demand for the growing human population. Specifically, terminal drought is responsible for substantial reductions in grain yield by setting poor grain and development in arid and semi-arid regions (Nawaz et al. 2013). Therefore, selecting wheat cultivars for dryland environments, improvements in high grain yield, and drought tolerance are crucial.
The grain yield of wheat is quantitatively inherited and determined by the number of grains per spike (GNS), tiller number (TN) and thousand-grain weight (TGW) (Cao et al. 2020), and significantly influenced by the environmental factors (Cui et al. 2014; Li et al. 2018). The uppermost leaf is regarded as flag leaf morphology (FLM) (length, width, area, thickness, and volume) significantly determines these three yield traits (Liu et al. 2021). For instance, ideotypic plant types, including relatively short, wide, and erect flag leaves of wheat (Donald 1968), can improve light capture for photosynthesis of lower leaves and increase the leaf area index (Sinclair and Sheehy 1999) thus increasing accumulation of nitrogen in leaves and increased the proteostasis, lipid re-modeling, and nitrogen re-mobilization of grains (Cohen et al. 2022) which finally enhancing grain yield (Pendleton et al. 1968; Duncan 1971; Stewart et al. 2003) and also sensing environmental signals for adaptation (Tian et al. 2015). Insufficient photoassimilates yielded unfruitful spikelet’s, reducing GNS and 10.70% grain yield per plant (Duwayri 1984). The flag leaf is also regarded as the "functional leaf" in wheat production, which contributes 45–58% of plant photosynthate and 41–43% of the carbohydrates during the grain-filling period (Duncan 1971; Xu et al. 1995; Sharma et al. 2003; Khaliq et al. 2008), while TGW will be incomplete due to insufficient photosynthetic products (Zahedi and Jenner 2003; Yang and Zhang 2006). Drought stress is a common phenomenon at this stage and yielded flag leaf senescence (Sawhney and Singh 2002), leading to an aborted grain (Farooq et al. 2014) due to a reduction in the accumulation of photoassimilates and lower fixation and CO2 assimilation (Yang et al., 2016). Leaf thickness is crucial in the amount of light absorbed by leaves and CO2 diffusion through the leaf tissue (Givnish 1979; Agustí et al. 1994; Syvertsen et al. 1995). Studies have shown that leaf thickness is related to photosynthesis and growth rate and can increase the number of chloroplasts, improve photosynthetic capacity, and enhance leaf water content (Araus et al. 1986; Devika et al. 2018; Wang et al. 2011). Increasing leaf mass area, thickness, and stomatal closure can reduce water loss and achieve higher yield under drought and high-temperature stress (Cellier et al. 2000; Peña-Rojas et al. 2005). Moreover, flag leaf traits have proven to have a significant relationship with yield-related traits, such as thousand-grain weight (TGW), grain number per spike (GNS), grain weight per spike (GWS), grain hardness, grain yield (Guitman et al. 1991; Sakamoto et al. 2006), specially FLW was crucial in determining GNS (Li et al., 2018). Wang et al. (2022) also revealed that FLW, FLA, and FLV had highly significant and positive relationships with SN, GT, GNS, and TGW under drought and irrigated conditions. Qian et al. (2009) also revealed that FLL and FLW were positively correlated with yield-related traits of wheat under drought conditions. Therefore, to better breed for high yield of wheat, it is essential to understand the genetic basis of FLM traits for drought tolerance.
Understanding the molecular genetic basis of wheat traits is crucial for developing molecular marker-assisted selection and exploring novel functional genes for improved drought tolerance. The flag leaf traits, including FLL, FLW, FLA, FLT, and FLV, are complex quantitative traits significantly influenced by multiple genes and environmental factors (Simón 1999; Coleman et al. 2001; Kobayashi et al. 2003). A large number of QTL associated with FLM flag leaf traits were identified in wheat with the improvement of a molecular marker-based genetic map (Wu et al. 2016; Xue et al. 2013; Fan et al. 2015; Tu et al. 2021; Ma et al. 2020, Chen et al., 2022b; Niu et al. 2023). A QTL for FLW on chromosome 5A was fine-mapped by Xue et al. (2013). Fan et al. (2015) revealed a total of 31 QTL associated with traits related to flag leaf by using a RIL population, including three major and stable QTL on chromosomes 2A (QFlw-2A), 3B (QFll-3B) and 4A (QFll-4A). Another study identified a total of 61 QTL associated with traits related to flag leaf in a RIL population that integrated with high-density simple sequence repeat (SSR) and single-nucleotide polymorphism (SNP) markers (Wu et al. 2016). A total of 23 putative QTL were identified to be associated with flag leaf traits, including 15 QTL detected in at least two environments (Liu et al. 2018). Hu et al. (2020) identified 43 QTL associated with FLM traits using four RIL populations. Tu et al. (2021), Ma et al. (2020), and Niu et al. (2023) detected a total of eight major, eight stable, and eight major QTL related to flag leaf in 10, 11, and eight environments, respectively. Genetic dissection of leaf thickness has been studied in rice. For example, Narawatthana et al. (2023) identified four loci related to leaf thickness in rice by using multi-model GWAS, and Chen et al. (2022a) confirmed that chromosome 9 contributed to the flag leaf thickness in rice by fine mapping. Although QTL for FLL, FLW, and FLA have been studied in previous studies, but the genetic basis of FLT and FLV in common wheat has not been studied in detail except Wang et al. (2022), who identified two QTL for FLT and two for FLV on chromosome 2B, 6A, 2A, and 7D by using a DH population.
The study emphasizes the need to identify and pyramid major, stably expressed loci for FLM traits from diverse wheat germplasm resources due to their significant impact on wheat yield. The present research developed and used a simple yet highly accurate FLT detection technique for calculating FLV. By using new and conventional techniques, a RIL population cross between the common wheat cultivars DH118 and Jinmai 919 under drought stress (DS) and well-watered (WW) conditions across five environments for five flag leaf traits to i) identify and validate the novel, major, and stable QTL and analyze their effects; ii) assess the relationship between flag leaf and yield-related traits under both of water regimes. The findings will provide a better understanding for the genetic mechanisms of flag leaf traits to increase wheat grain yield.
Materials and methods
Plant materials and field trials
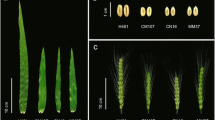
The RIL of F10 population of 165 lines used for QTL mapping in the present study was derived from the cross between two cultivars, such as DH118 and Jinmai 919, both of which were released by Shanxi Province. The DH118 was a high-yielding variety selected for irrigated conditions, and Jinmai 919 for drought resistance. Jinmai 919 has wider and longer leaves than DH118, and DH118 is thicker than Jinmai 919. Three flag leaves were collected 20 days after flowering from each of the five wheats. The 180-line double haploid (DH) population from Jinchun 7 × Jinmai 919 was used to validate QTL identified in the mapping population.
The RIL populations were planted at the Yaodu Experimental Station (YD) (36°08′N, 111°52′E, altitude 450 m), the Hancun Experimental Station (HC) (36°25′N, 111°67′E, altitude 450 m) in Shanxi Province in 2018–2019, 2019–2020, and 2020–2021 (Only Yaodu). Plants were grown under drought-stressed (DS) and well-watered (WW) conditions across five environments designated as E1 (DS, 2019-HC), E2 (WW, 2019-YD), E3 (DS, 2020-HC), E4 (WW, 2020-YD), and E5 (WW, 2021-YD). The DH population was planted under the environmental conditions E2, E4, E5, 2021-HC and 2020-YC (Here, YC—Yuncheng Experimental Station (34°35' N, 110°15' E, altitude 450 m on 2020–2021). The field design was a randomized complete block with three replications. Each plot consisted of two 1.5 m rows spaced 0.3 m apart at 21 seeds per row. Field management used standard wheat production practices in these regions.
Measurement of flag leaf traits
FLL, FLW, FLA, FLT, and FLV were measured on the main tiller of ten randomly selected plants. FLL (cm) was measured from the leaf collar to the tip. FLW (cm) was measured at the widest part of the leaf. The derived trait FLA (cm2) was calculated by FLA = FLL × FLW × 0.83 (Fan et al. 2015). FLV (mm3) was calculated as FLV = FLT × FLA (Wang et al. 2022). For calculating the thickness of the flag leaf, the formula h = S/W was used, where h is the flag leaf thickness (FLT), S is the cross-sectional area, and W is the leaf width at the cut. The thickness of each of the 20 segments was then compared with their average (Wang et al. 2022).
Statistical analysis
Basic statistics and Pearson’s correlation analysis were performed on the phenotypic data from each environment. Analysis of variance (ANOVA) was done using the SPSS (V22.0) statistical package (IBM SPSS, Armonk, NY, USA). SAS V8.0 (SAS Institute, Cary, North Carolina, USA; https://www.sas.com) was used to calculate the best linear unbiased predictions (BLUP) and broad-sense heritability (H2) (Smith et al. 1998; Qin et al. 2016).
High-density genetic linkage map construction and QTL mapping
A genetic map of the mapping population was generated with 3553 polymorphic SNPs from the wheat Illumina 90k SNP assay, representing all 21 hexaploid wheat chromosomes (Yang et al. 2022). QTL was detected using WinQTLCart version 2.5 (https://brcwebportal.cos.ncsu.edu/qtlcart/WQTLCart.htm) and composite interval mapping. The minimal LOD score to accept the presence of a QTL was set at 2.5. The QTL was detected in at least three environments (including the BLUP data set as an environment) and stated as stable. Major QTL were those that met these criteria in at least three environments (including the BLUP data set as an environment) with more than 10% of phenotypic variance explained in at least one environment. QTL that was either within 1 cM of one another or shared common flanking markers were considered identical. The naming of QTL followed the International Rules of Genetic Nomenclature (https://wheat.pw.usda.gov/ggpages/wgc/98/Intro.htm). To determine the physical position of identified QTL regions, a BLAST search in WheatOmics 1.0 (http://wheatomics.sdau.edu.cn/) was performed to align the QTL-associated peak and flanking SNP marker sequences (Ma et al. 2021).
Marker development and validation of stable QTLs
To develop kompetitive allele-specific PCR (KASP) tags from the peak marker SNP sequence of the major QTLs, two specific primers (F1/F2) and a universal primer (R) were designed for each SNP. An F1 tail that could bind to induce FAM fluorescence and an F2 tail that could bind to induce HEX fluorescence were added to the specific sequences. KASP primers were designed by Polymarker (http://www.polymarker.info/) and synthesized by Beijing Jiacheng Biotechnology Co. Ltd. The developed KASP markers were used in PCR to detect previously identified QTLs in the DH population as a means of validation. Following genotyping, the validation population was divided into two groups and differences in flag leaves between the groups were assessed by t-tests in SAS V8.0.
Prediction of candidate genes
Genes within the target region of major QTL were obtained using the genome browser (JBrowse) on the Triticeae Multi-omics website http://wheatomics.sdau.edu.cn/jbrowse.html). Functional annotation and enrichment analysis of genes in these regions were done using the GO (gene ontology) database and the R package cluster Profiler. Analysis of orthologs between wheat and rice used the Triticeae-Gene Tribe website (http://wheat.cau.edu.cn/TGT/).
Results
Phenotypic variation of flag leaf traits
The phenotypic variation of the RIL population and the parental lines was evaluated in five environments. Flag leaves of Jinmai 919 were larger than DH118 as measured by FLL, FLW, FLA and FLV (Table 1, Fig. 1A) across all environments, including the BLUP data. The flag-leaf-related traits of the RIL population ranged from 8.18 to 30.12 cm for FLL, 1.10 to 2.47 cm for FLW, 0.11 to 0.37 mm for FLT, 9.37 to 61.45 cm2 for FLA, and 112.26 to 1324.00 mm3 for FLV. The H2 of five flag leaf traits ranged from 0.68 to 0.84, indicating that genetic factors significantly affected these traits. Principal component analysis showed that environmental factors influenced phenotypic values considerably, and drought stress increases the phenotypic variation (Fig. 1B). The Pearson correlations among different environments were significant (Table S1). All traits were distributed normally in the RIL population (Fig. 2).
Correlation analysis for traits related to flag leaf and yield
The relationship between flag leaf traits within different environments and traits related to yield was analyzed under two different water regimes. All flag leaf traits were highly significant and positively correlated within different environments except for a few negative correlations with dependent trait FLT (Table S1). Furthermore, flag leaf traits, such as FLL, had a highly significant and positive correlation with FLW, FLA and FLV in both water regimes (Fig. 2). All four traits showed a significant and positive correlation with FLV. Interestingly, FLL and FLW showed highly significant and positive correlations with FLT under WW conditions but non-significant and positive correlations under DS conditions, indicating that FLT is performed well under irrigated conditions.
Subsequently, the relationship between flag-leaf and yield-related traits was also evaluated. Under both water regimes, FLW showed highly significant positive correlations with GL, GW and TGW, but FLL showed no correlations with yield-related traits. Furthermore, FLT had highly significant and positive correlations with GL and GW in WW conditions and only in TGW under DS conditions. FLA and FLV had highly significant and positive correlations with TGW under both water regimes, and FLW, FLA, and FLV had highly significant and positive correlations with SNS in DS conditions (Fig. 2). Correlation coefficients in DS conditions across environments were slightly lower than in WW conditions.
QTL mapping
A total of 40 QTL, including 6 for FLL, 7 for FLW, 9 for FLT, 10 for FLA, and 8 for FLV, were identified and located on the chromosomes of 1B, 2A, 2B, 3B, 3D, 4A, 5A, 5A, 5B, 6B, 6D, 7A and 7B (Table 2). These QTL phenotypic variances explained (PVE) about 0.80%-15.26% in different environments. Out of the 40 QTL, the DH118 alleles contributed 14, whereas Jinmai 919 contributed 26.
For FLL, six QTL were mapped to chromosomes 4A, 5A, 5B, 6D, and 7A, with PVE ranging from 5.54–8.85%. Among them, Qfll.saw-4A and Qfll.saw-5A were found to be stable QTL and detected in four environments, including BLUP, explaining PVE ranging from 6.32–8.28% and 5.72–8.85%, respectively. The additive effect values indicated that the positive alleles at Qfll.saw-4A were from DH118, and those at Qfll.saw-5A were from Jinmai 919.
For FLW, a total of seven QTL were identified to be distributed on chromosomes 1B, 2A, 3B, 3D, 4A, 5B, and 7B, with the PVE ranging from 6.15 to 12.80% in different environments. In which, Qflw.saw-2A was identified as novel and stable and Qflw.saw-5B as stable, as well as were detected in four environments including BLUP within the PVE ranging from 8.94–12.80% and 6.71–9.15%, respectively. Both of the QTL contributed to an increase in the favourable alleles for FLW by Jinmai 919.
For FLA, ten QTL were mapped to chromosomes 2A, 3B, 3D, 4A, 5A, 5B, 6D, and 7B, with individual QTL contributing 5.09–9.91% of the PVE. Among them, Qfla.saw-2A, Qfla.saw-4A, and Qfla.saw-5B were stably detected in three to four environments, including BLUP, with PVE ranging from 6.43–9.91%, 5.78–7.00% and 5.09–6.22%, respectively. Qfla.saw-2A and Qfla.saw-5B contributed to increased FLA through Jinmai 919 alleles, whereas Qfla.saw-4A by DH118.
For FLT, nine QTL were detected to be distributed on chromosomes 2B, 3B, 4A, 5A, 6B, 7A, and 7B, with the PVE ranging from 5.52 to 15.26%. The QTL Qflt.saw-2B and Qflt.saw-3B were observed in four environments, including BLUP and PVE, from 5.78 to 13.30% and 6.29 to 12.32%, respectively, and stated as major and stable QTL. The Qflt.saw-2B alleles that increased FLT were contributed by Jinmai 919 and Qflt.saw-3B by DH118.
For FLV, eight additive effects QTL were detected on chromosomes 2A, 2B, 3B, 4A, 5B, and 7B, and the PVE by individual QTL ranged from 0.80–13.70%. The QTL Qflv.saw-2A, Qflv.saw-4A.1, and Qflv.saw-5B were found to be stable and identified in three to four environments, including BLUP, with the PVE ranging from 0.80–12.13%, 7.81–9.08%, and 6.34–9.70%, respectively, and Qflv.saw-2A was stated as a major and stable QTL. The additive effect of Qflv.saw-2A and Qflv.saw-5B indicated that the allele contributing to the increase of FLV was from Jinmai 919, whereas DH118 contributed Qflv.saw-4A.1.
Remarkably, we found three overlapping regions underlying the QTL for flag leaf traits, suggesting these loci simultaneously control flag leaf morphology in multiple dimensions (Fig. 3). For example, two genomic regions containing the QTL Qflw.saw-2A/Qfla.saw-2A/Qflv.saw-2A and Qflw.saw-5B/Qfla.saw-5B/Qflv.saw-5B were found to be simultaneously control FLW, FLA, and FLV. In addition, there are three stable QTL Qfll.saw-4A, Qfla.saw-4A and Qflv.saw-4A.1 were also co-located in a region 40.8–49.2 cM (Fig. 3, Table 2).
Genetic map of the stable QTL Qflw.saw-2A/Qfla.saw-2A/Qflv.saw-2A, Qfll.saw-4A/Qfla.saw-4A/Qflv.saw-4A.1, and Qflw.saw-5B/Qfla.saw-5B/Qflv.saw-5B and their effects. A, C and E: Genetic map of 2A, 4A and 5B; B, D and F: Effect of QTL on FLL, FLW, FLA, and FLV shown as box plots calculated after grouping the DH118 × Jinmai 919 RIL population into two classes based on the allele of the flanking markers. *, **, and *** represent significance at P < 0.05, P < 0.01, and P < 0.001, respectively
Analyses of additive effects of the major QTL
Generally, the higher the number of positive alleles, the larger the leaf size. The linear relationship between phenotypic data and the number of positive alleles was analyzed. We detected 12 stable QTL, including two for FLL (Qfll.saw-4A and Qfll.saw-5A), two for FLW (Qflw.saw-2A, and Qflw.saw-5B), three for FLA (Qfla.saw-2A, Qfla.saw-4A, and Qfla.saw-5B), two for FLT (Qflt.saw-2B and Qflt.saw-3B), and three for FLV (Qflv.saw-2A, Qflv.saw-4A.1, and Qflv.saw-5B). The additive effects for each trait were further analyzed based on linked peak markers. The average trait value increased as the number of positive alleles increased (Fig. 4, Table S2). RIL lines with positive alleles at both QTL regions had an average FLL 1.27 cm greater than lines with contrasting alleles. Similarly, lines with more than two positive alleles showed significantly increased FLW. RIL lines with positive alleles at all two QTL regions had an average FLW 0.16 cm greater than lines with contrasting alleles. RIL lines with positive alleles at three QTL regions had an average FLA of 4.80 cm2 greater than lines with contrasting alleles. RIL lines with positive alleles at two QTL regions had an average FLT 0.01mm thicker than lines with contrasting alleles. RIL lines with positive alleles at three QTL regions had an average FLV of 203.84 mm3 greater than lines with contrasting alleles.
Linear regressions between the number of positive alleles and trait values for five flag leaf traits in the DH118 × Jinmai 919 RIL population. The number of lines carrying the corresponding number of positive alleles is shown in brackets. The letter above the bars indicates comparison results at the significant level of 0.05
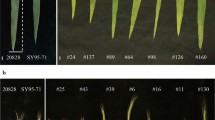
Quantitative trait loci (QTL) validation
To further validate the stable QTL, the KASP markers for each QTL were used to evaluate their effects in the Jinchun 7 × Jinmai 919 population (Table S3). The KASP markers for Qfll.saw-5A, Qflw.saw-5B, Qfla.saw-5B, and Qflv.saw-5B were not polymorphic between Jinchun 7 and Jinmai 919; thus, the effects could not be evaluated. The remaining eight QTL were polymorphic and evaluated. The effect of Qflt.saw-3B, Qfll.saw-4A, Qfla.saw-4A, and Qflv.saw-4A did not differ significantly between the two groups in the Jinchun 7 × Jinmai 919 population (Fig. 5). The effect of the other four QTL, Qflw.saw-2A, Qfla.saw-2A, Qflv.saw-2A, and Qflt.saw-2B were highly significant (P < 0.05) in more than three environments. According to the markers profile of four QTL, lines with homozygous alleles from Jinmai 919 had significantly higher (P < 0.05) values than those from DH118. The Qflw.saw-2A, Qfla.saw-2A, and Qflv.saw-2A lines were homozygous for the Jinmai 919 alleles had significantly higher phenotypic values than those with the Jinchun 7 alleles irrespective of QTL region, with the differences in FLW, FLA and FLV ranging from 3.17 to 8.06%, 4.45 to 10.04%, and 7.32 to 14.84%, respectively. The FLT of lines with the Qflt.saw-2B allele was significantly higher than for lines lacking the Qflt.saw-2B allele and the difference was between 2.54 and 12.47%.
QTL for FLT and FLV co-located with yield-related traits
Hundreds of yield-related QTL have been mapped on all chromosomes of wheat (Cao et al. 2020). We detected five stable QTL for FLT and FLV (Table 2). The Qflt.saw-2B was mapped between IACX8202 and IAAV1798, which is physically located between 765.06 and 788.52 Mb on chromosome 2B. It overlapped with loci TKW-IWB32380 for a thousand kernel weight on 2B, as reported by Sukumaran et al. (2018). The Qflt.saw-2B was near to SNS-wsnp_Ex_c41300_48154348 for SNS that was physically located at 786.23 Mb on chromosome 2B, suggesting they may be allelic (Tadesse et al. 2019).
Qflv.saw-2A mapped between Ku_c23118_149 and wsnp_Ex_rep_c103167_88182254 and was physically located between 33.04 and 50.91 Mb on chromosome 2A. It overlapped with QHGfd.bhu-2A (Tiwari et al. 2013) and QTgw.nfcri-2A (Guan et al. 2018) for TGW, and qSlt-2A (Zhao et al. 2019) for Spike-layer thickness, which was in the markers wmc177-gwm71, Tdurum_contig51537_275 and AX-110601484-AX-111567412, respectively. In addition, Qflv.saw-5B was mapped between TA014612-0744 and BobWhite_rep_c50066_63 and physically located between 21.43 and 27.83 Mb on chromosome 5B. Zhai et al. (2018 revealed that QGa.cau-5B.1 and QTgw.cau-5B.1 were also located in the same interval. The QSL-4A.1 for spike length mapped between Tdurum_contig1868_24 and Ku_c7594_1179 and was physically located between 11.71 and 16.96 Mb on chromosome 4A (Hu et al. 2020), which also overlapped with Qflv.saw-4A.1. Therefore, it can be suggested that FLT and FLV have pleiotropic effects and close connection with grain yield traits.
Putative candidate genes
Based on the result of local-blast browse through the IWGSCv1.1 reference sequence, no homologues of the above genes were found in the physical regions of 33.04 and 50.91Mb Mb for Qflw.saw-2A, Qfla.saw-2A, Qflv.saw-2A on 2AS, 765.06 and 788.52 Mb for Qflt.saw-2B on 2BL, and 54.75–66.18 Mb for Qflt.saw-3B on 3BS in wheat. It indicated that there might be novel genes related to flag leaf among the three regions. But Qflt.saw-3B were not validated in the DH population. Thus, these four QTL Qflw.saw-2A, Qfla.saw-2A, Qflv.saw-2A and Qflt.saw-2B were chosen for further analysis. A total of 495 genes were identified within the two regions, including 212 genes in Qflw.saw-2A, Qfla.saw-2A, Qflv.saw-2A (Table S4), and 283 genes in Qflt.saw-2B (Table S5). According to gene functional annotations in the Gene Ontology (GO) public database, 24 of these genes are involved in chloroplast stroma and auxin/cytokinin-activated signaling pathways. Five genes were related to the hormone signaling pathway. For instance, TraesCS2A02G076100, TraesCS2B02G591500 and TraesCS2B02G578500 were involved in auxin-activated signaling pathways, such as the series of molecular signals generated by the binding of the plant hormone auxin to a receptor, and ending with modulation of a downstream cellular process, e.g. transcription. In addition, TraesCS2A02G081900 and TraesCS2B02G582300 were involved in cytokinin-activated signaling pathways, and nineteen genes were related to chlorophyll photosynthesis.
Discussion
Multiple environment-based analyses of FLM traits and correlation with GY traits
In wheat, the flag leaf morphology can not only alter the plant architecture at the vegetative growth stage but also influence the capacity of light capture and photosynthate assimilation (upto 45–58%) (Sharma et al. 2003; Khaliq et al. 2008) and 41–43% of the carbohydrates at the grain filling stage (Duncan 1971; Xu et al. 1995). Yang et al. (2016) found a significant positive correlation with each of the FLM traits under both of the water regimes, in which correlations in DS (r = 0.31* to 0.93**) were higher than WW (r = 0.29* to 0.81**) conditions. Qian et al. (2009) also found that, under drought stress, FLW was positively correlated with yield component traits during grain-filling. A recent study revealed to be SN, GNS, GT, and TGW were significantly and positively associated with FLW, FLA and FLV (Wang et al. 2022). For instance, in this research, correlation results were also similar to Yang et al. (2016) under both water regimes and DS (0.31** to 0.86**) > WW (0.18* to 0.84**) condition, which suggested that phenotypic reduction could be a more efficient way to coordinate all flag leaf traits under DS components in order to endure unfavourable conditions. TGW was significantly correlated with FLW and FLV under both DS and WW conditions but non-significant with FLA in DS and FLT in WW condition, as well as FLA significantly and positively associated with SNS under DS, indicating that FLA contributed more to grain yield.
In previous research studies, identified genetic regions associated with flag leaf traits were from different genetic backgrounds (Ma et al. 2020; Tu et al. 2021), but the present research study used two important traits, FLT and FLV along with flag leaf traits and comprehensively investigated in six environments including BLUP. Interestingly, out of 12 stable QTLs in this research, nine were validated in all environments by Jinchun 7 × Jinmai 919 population, of which three were related to FLV. In contrast, other research studies detected few major QTLs (very few were validated) by using more environments and different genetic backgrounds (Ma et al. 2020; Tu et al. 2021). Therefore, the KASP markers created were helpful in wheat breeding, and these nine QTLs for traits linked to flag leaves were strongly recognized in most environments studied with PVE and higher LOD values.
Identification of four novel stable QTL related to flag leaf traits
A total of twelve stable QTLs for flag-leaf-related traits were identified in the present study. In order to compare the intervals of the previously identified QTL with those detected, QTL was mapped onto target chromosomes in Chinese Spring. The QTL viz. Qfll.saw-4A, Qfla.saw-4A and Qflv.saw-4A.1 were mapped between BS00043286_51 and BS00108852_51 and located within the physical distance of 7.40–11.31 Mb on chromosome 4A, which is overlapped with RAC875_c8121_1490 (3.87 Mb) (Muhammad et al. 2021). The QTL Qfll.saw-5A (chr 5A: 580.94–616.52 Mb) were mapped between wsnp_Ex_c62818_62296773 and wsnp_Ex_c3772_6866645, co-located by Yao et al., (2020) (chr 5A: 591.32 Mb). As well as Qflw.saw-5B, Qfla.saw-5B, and Qflv.saw-5B overlapped with QFlw.sicau-6A.2 as reported by Ma et al., (2020) and was also closer to the QFLW-6A (Yan et al. 2020). The QFLW-7A.1 overlapped with a genetic region reported by Li et al. (2023) and Zanella et al. (2022). Furthermore, comparing physical intervals suggested that none of the studies had yet been detected and co-located with the Qflw.saw-2A, Qfla.saw-2A, Qflv.saw-2A, Qflt.saw-2B, and Qflt.saw-3B, and stated as novel.
Leaf thickness is of great significance to grain yield
Flag leaf morphological characteristics contribute significantly to "the source" during the grain-filling stage and are crucial in determining crop yield and biomass. Leaf thickness is an essential leaf morphological trait that affects the utilization of light energy and wheat grain yield. In this research, FLT was identified to have a highly significant positive relationship with TGW (0.250**) under the DS condition, whereas, in the WW condition, there was a significant positive correlation with GL (0.164*) and GW (0.154*). The linkage mapping analysis results of this research study, out of two stable QTL (Qflt.saw-2B and Qflt.saw-3B), Qflt.saw-2B was co-located with the previously reported loci related to TKW (IWB32380) (Sukumaran et al. 2018) and SNS (wsnp_Ex_c41300_48154348) (Tadesse et al. 2019), indicating this locus have pleiotropic effects and close connection with grain yield traits. Moreover, another QTL (Qflt.saw-3B) was found to be novel compared to other studies. Improving grain yield is believed to be possible through breeding for the ideal size of the flag leaf. To restrict shade and the population of plant's susceptibility to disease, there is a limit on how much leaf area may be raised in order to maximize grain yield. Thus, breeding for increased leaf thickness provides a further means of boosting grain yield and photosynthetic potential. Furthermore, the SNP for FLT found in this work may be transformed into KASP markers and utilized in future molecular marker-assisted breeding.
Data availability
Data generated or analyzed during this study are included in this published article (and its Supplementary Material).
Code availability
Not applicable.
References
Agustí S, Enriquez S, Frostchristensen H et al (1994) Light harvesting among photosynthetic organisms. Funct Ecol 8:273–279. https://doi.org/10.2307/2389911
Araus JL, Alegre L, Tapia L et al (1986) Relationships between photosynthetic capacity and leaf structure in several shade plants. Am J Bot 73:1760–1770. https://doi.org/10.1002/j.1537-2197.1986.tb09708.x
Cao SH, Xu DG, Hanif M et al (2020) Genetic architecture underpinning yield component traits in wheat. Theor Appl Genet 133(6):1811–1823. https://doi.org/10.1007/s00122-020-03562-8
Cellier F, Conejero G, Casse F (2000) Dehydrin transcript fluctuations during a day/night cycle in drought-stressed sunflower. J Exp Bot 51:299–304. https://doi.org/10.1093/jexbot/51.343.299
Chen DG, Zhou XQ, Chen K et al (2022a) Fine-mapping and candidate gene analysis of a major locus controlling leaf thickness in rice (Oryza sativa L.). Mol Breeding 42:6. https://doi.org/10.1007/s11032-022-01275-y
Chen LG, Xu ZB, Fan XL et al (2022b) Genetic dissection of quantitative trait loci for flag leaf size in bread wheat (Triticum aestivum L.). Front Plant Sci 13:1047899. https://doi.org/10.3389/fpls.2022.1047899
Cohen M, Hertweck K, Itkin M et al (2022) Enhanced proteostasis, lipid remodeling, and nitrogen remobilization define barley flag leaf senescence. J Exp Bot 73(19):6816–6837. https://doi.org/10.1093/jxb/erac329
Coleman RK, Gill G, Rebetzke GJ (2001) Identification of quantitative trait loci for traits conferring weed competitiveness in wheat (Triticum aestivum L.). Aust J Agric Res 52:1235–1246. https://doi.org/10.1071/AR01055
Cui F, Zhao CH, Ding AM et al (2014) Construction of an integrative linkage map and QTL mapping of grain yield-related traits using three related wheat RIL populations. Theor Appl Genet 127:659–675. https://doi.org/10.1007/s00122-013-2249-8
Devika S, Ravichandran V, Boominathan P (2018) Physiological analyses of nitrogen use efficiency and yield traits of rice genotypes. Indian J Plant Physiol 23:100–110. https://doi.org/10.1007/s40502-018-0358-8
Donald CM (1968) The breeding of crop ideotypes. Euphytica 17:385–403. https://doi.org/10.1007/BF00056241
Duncan W (1971) Leaf angles, leaf area, and canopy photosynthesis. Crop Sci 11:482–485. https://doi.org/10.2135/cropsci1971.0011183X001100040006x
Duwayri M (1984) Effect of flag leaf and awn removal on grain yield and yield components of wheat grown under dryland conditions. Field Crop Res 8:307–313. https://doi.org/10.1016/0378-4290(84)90077-7
Fan XL, Cui F, Zhao CH et al (2015) QTLs for flag leaf size and their influence on yield-related traits in wheat (Triticum aestivum L.). Mol Breeding 35:24. https://doi.org/10.1007/s11032-015-0205-9
Farooq M, Hussain M, Siddique KHM (2014) Drought stress in wheat during flowering and grain-filling periods. Crit Rev Plant Sci 33:331–349. https://doi.org/10.1080/07352689.2014.87529
Givnish T (1979) On the Adaptive Significance of Leaf Form In: Solbrig, OT, Jain, S, Johnson, GB, Raven, PH (eds) Topics in Plant Population Biology. Palgrave, London. https://doi.org/10.1007/978-1-349-04627-0_17
Guan PF, Lu LH, Jia LJ et al (2018) Global QTL analysis identifies genomic regions on Chromosomes 4A and 4B harboring stable loci for yield-related traits across different environments in wheat (Triticum aestivum L.). Front Plant Sci 9:529. https://doi.org/10.3389/fpls.2018.00529
Guitman MR, Arnozis PA, Barneix AJ (1991) Effect of source-sink relations and nitrogen nutrition on senescence and N remobilization in the flag leaf of wheat. Physiol Plant 82:278–284. https://doi.org/10.1111/j.1399-3054.1991.tb00094.x
Hu JM, Wang XQ, Zhang GX et al (2020) QTL mapping for yield-related traits in wheat based on four RIL populations. Theor Appl Genet 133:917–933. https://doi.org/10.1007/s00122-019-03515-w
Khaliq I, Irshad A, Ahsan M (2008) Awns and flag leaf contribution towards grain yield in spring wheat (Triticum aestivum L.). Cereal Res Commun 36:65–76. https://doi.org/10.1556/CRC.36.2008.1.7
Kobayashi S, Fukuta Y, Morita S et al (2003) Quantitative trait loci affecting flag leaf development in rice (Oryza sativa L.). Breed Sci 53:255–262. https://doi.org/10.1270/jsbbs.53.255
Li FJ, Wen WE, He ZH et al (2018) Genome-wide linkage mapping of yield-related traits in three chinese bread wheat populations using high-density SNP markers. Theor Appl Genet 131:1903–1924. https://doi.org/10.1007/s00122-018-3122-6
Li YB, Tao FL, Hao YF et al (2023) Variations in phenological, physiological, plant architectural and yield-related traits, their associations with grain yield and genetic basis. Ann Bot 131(3):503–519. https://doi.org/10.1093/aob/mcad003
Liu KY, Xu H, Liu G et al (2018) QTL mapping of flag leaf-related traits in wheat (Triticum aestivum L.). Theor Appl Genet 131:839–849. https://doi.org/10.1007/s00122-017-3040-z
Liu XJ, Yin BZ, Hu ZH et al (2021) Physiological response of flag leaf and yield formation of winter wheat under different spring restrictive irrigation regimes in the Haihe Plain, China. J Integr Agric 20:2343–2359. https://doi.org/10.1016/S2095-3119(20)63352-4
Ma J, Tu Y, Zhu J et al (2020) Flag leaf size and posture of bread wheat: genetic dissection, QTL validation and their relationships with yield-related traits. Theor Appl Genet 133:297–315. https://doi.org/10.1007/s00122-019-03458-2
Ma SW, Wang M, Wu JH, Guo WL et al (2021) WheatOmics: a platform combining multiple omics data to accelerate functional genomics studies in wheat. Mol Plant 14:1965–1968. https://doi.org/10.1016/j.molp.2021.10.006
Muhammad A, Li JG, Hu WC et al (2021) Uncovering genomic regions controlling plant architectural traits in hexaploid wheat using different GWAS models. Sci Rep 11:6767. https://doi.org/10.1038/s41598-021-86127-z
Narawatthana S, Phansenee Y, Thammasamisorn B-OC (2023) Multi-model genome-wide association studies of leaf anatomical traits and vein architecture in rice. Front Plant Sci 14:1107718. https://doi.org/10.3389/fpls.2023.1107718
Nawaz A, Farooq M, Cheema SA et al (2013) Stay green character at grain filling ensures resistance against terminal drought in wheat. Int J Agricul Biol. 15(6):1272–1276
Niu JQ, Si YQ, Tian SQ et al (2023) A Wheat 660 K SNP array-based high-density genetic map facilitates QTL mapping of flag leaf-related traits in wheat. Theor Appl Genet 136(3):51. https://doi.org/10.1007/s00122-023-04248-7
Peña-Rojas K, Aranda X, Joffre R et al (2005) Leaf morphology, photochemistry and water status changes in resprouting Quercus ilex during drought. Funct Plant Biol 32:117–130. https://doi.org/10.1071/fp04137
Pendleton JW, Smith GE, Winter SR et al (1968) Field investigations of the relationships of leaf angle in corn (Zea mays L.) to grain yield and apparent photosynthesis. Agron J 60:422–424. https://doi.org/10.2134/agronj1968.00021962006000040027x
Qian XY, Jing RL, Wang H et al (2009) Genetic characteristics of wheat functional leaves at filling stage under different water regimes. Ying Yong Sheng Tai Xue Bao 20(12):2957–2963
Qin P, Lin Y, Hu YD, Liu K, Mao SS, Li ZY, Wang JR, Liu YX, Wei YM, Zheng YL (2016) Genome-wide association study of drought-related resistance traits in aegilops tauschii. Genet Mol Biol 39:398–407. https://doi.org/10.1590/1678-4685-GMB-2015-0232
Sakamoto T, Morinaka Y, Ohnishi T et al (2006) Erect leaves caused by brassinosteroid deficiency increase biomass production and grain yield in rice. Nat Biotechnol 24(1):105–109. https://doi.org/10.1038/nbt1173
Sawhney V, Singh DP (2002) Effect of chemical desiccation at the post-anthesis stage on some physiological and biochemical changes in the flag leaves of contrasting wheat genotypes. Field Crop Res 77:1–6. https://doi.org/10.1016/S0378-4290(01)00192-7
Sharma SN, Sain RS, Sharma PK (2003) The genetic control of flag leaf length in normal and late sown durum wheat. J Agric Sci 141:323–331. https://doi.org/10.1017/S0021859603003642
Simón MR (1999) Inheritance of flag-leaf angle, flag-leaf area and flag-leaf area duration in four wheat crosses. Theor Appl Genet 98:310–314. https://doi.org/10.1007/s001220051074
Sinclair TR, Sheehy JE (1999) Erect leaves and photosynthesis in rice. Science 283:1455–1455. https://doi.org/10.1126/science.283.5407.1455c
Smith SE, Kuehl RO, Ray IM, Hui R, Soleri D (1998) Evaluation of simple methods for estimating broad-sense heritability in stands of randomly planted genotypes. Crop Sci 38:1125–1129. https://doi.org/10.2135/cropsci1998.0011183X003800050003x
Stewart DW, Costa C, Dwyer LM et al (2003) Canopy structure, light interception, and photosynthesis in maize. Agron J 95:1465–1474. https://doi.org/10.2134/agronj2003.1465
Sukumaran S, Lopes M, Dreisigacker S et al (2018) Genetic analysis of multi-environmental spring wheat trials identifies genomic regions for locus-specific trade-offs for grain weight and grain number. Theor Appl Genet 131:985–998. https://doi.org/10.1007/s00122-017-3037-7
Syvertsen JP, Lloyd J, McConchie C et al (1995) On the relationship between leaf anatomy and CO2 diffusion through the mesophyll of hypostomatous leaves. Plant, Cell Environ 18:149–157. https://doi.org/10.1111/j.1365-3040.1995.tb00348.x
Tadesse W, Suleiman S, Tahir I et al (2019) Heat-tolerant QTLs associated with grain yield and its components in spring bread wheat under heat-stressed environments of sudan and egypt. Crop Sci 59:199–211. https://doi.org/10.2135/cropsci2018.06.0389
Tian YH, Zhang HY, Xu PZ et al (2015) Genetic mapping of a QTL controlling leaf width and grain number in rice. Euphytica 202(1):1–11. https://doi.org/10.1007/s10681-014-1263-5
Tiwari C, Wallwork H, Kumar U et al (2013) Molecular mapping of high temperature tolerance in bread wheat adapted to the eastern gangetic plain region of india. Field Crop Res 154:201–210. https://doi.org/10.1016/j.fcr.2013.08.004
Trenberth KE (2011) Changes in precipitation with climate change research. Clim Res 47:123–138. https://doi.org/10.3354/cr00953
Tu Y, Liu H, Liu JJ et al (2021) QTL mapping and validation of bread wheat flag leaf morphology across multiple environments in different genetic backgrounds. Theor Appl Genet 134:261–278. https://doi.org/10.1007/s00122-020-03695-w
Wang RZ, Huang WW, Chen L et al (2011) Anatomical and physiological plasticity in Leymus chinensis (Poaceae) along large-scale longitudinal gradient in northeast China. PLoS ONE 6:e26209. https://doi.org/10.1371/journal.pone.0026209
Wang Y, Qiao L, Yang CK et al (2022) Identification of genetic loci for flag-leaf-related traits in wheat (Triticum aestivum L.) and their effects on grain yield. Front Plant Sci 13:990287. https://doi.org/10.3389/fpls.2022.990287
Wu QH, Chen YX, Lin F et al (2016) QTL mapping of flag leaf traits in common wheat using an integrated high-density SSR and SNP genetic linkage map. Euphytica 208:337–351. https://doi.org/10.1007/s10681-015-1603-0
Xu H, Zhao J (1995) Canopy photosynthesis capacity and the contribution from different organs in high-yielding winter wheat. Acta Agro Sin 21:204–209
Xue SL, Xu F, Li GQ et al (2013) Fine mapping TaFLW1, a major QTL controlling flag leaf width in bread wheat (Triticum aestivum L.). Theor Appl Genet 126(8):1941–1949. https://doi.org/10.1007/s00122-013-2108-7
Yan XF, Zhao L, Ren Y et al (2020) Identification of genetic loci and a candidate gene related to flag leaf traits in common wheat by genome-wide association study and linkage mapping. Mol Breed 40:58. https://doi.org/10.1007/s11032-020-01135-7
Yang JC, Zhang JH (2006) Grain filling of cereals under soil drying. New Phytol 169:223–236. https://doi.org/10.1111/j.1469-8137.2005.01597.x
Yang DL, Liu Y, Cheng HB et al (2016) Genetic dissection of flag leaf morphology in wheat (Triticum aestivum L.) under diverse water regimes. BMC Genet 17:94. https://doi.org/10.1186/s12863-016-0399-9
Yang B, Wen X, Wen H, Feng Y, Zhao J, Wu B, Zheng X, Yang C, Yang S, Qiao L, Zheng J (2022) Identification of genetic loci affecting flag leaf chlorophyll in wheat grown under different water regimes. Front Genet 13:832898. https://doi.org/10.3389/fgene.2022.832898
Yao JX, Zhang CL, Song XP et al (2020) QTL analysis of wheat spike length and flag length based on 90k SNP assay. J Triticeae Crop 40(11):1283–1289. https://doi.org/10.7606/j.issn.1009-1041.2020.11.01
Zahedi M, Jenner CF (2003) Analysis of effects in wheat of high temperature on grain filling attributes estimated from mathematical models of grain filling. J Agric Sci 141:203–212. https://doi.org/10.1017/S0021859603003411
Zanella CM, Marilena R, Greg M et al (2022) Longer epidermal cells underlie a quantitative source of variation in wheat flag leaf size. New Phytol 237(5):1558–1573. https://doi.org/10.1111/nph.18676
Zhai HJ, Feng ZY, Du XF et al (2018) A novel allele of TaGW2-A1is located in a finely mapped QTL that increases grain weight but decreases grain number in wheat (Triticum aestivum L.). Theor Appl Genet 131:539–553. https://doi.org/10.1007/s00122-017-3017-y
Zhao CH, Zhang N, Wu YZ et al (2019) QTL for spike-layer uniformity and their influence on yield-related traits in wheat. BMC Genet 20:23. https://doi.org/10.1186/s12863-019-0730-3
Acknowledgements
We would like to thank Dr. Mike Bonman, University of Idaho in the United States, for editing our manuscript.
Funding
This work was supported by Research Program of Shanxi Province (202102140601001–1 and 202201140601025–2-01), the fellowship of China Postdoctoral Science Foundation (2020M670701).
Author information
Authors and Affiliations
Contributions
Conceptualization and design—MUH and JZ; Data curation and Methodology—YX, JZ, MK and BW; Writing—original draft—YX and JZ; Writing – review & editing—MUH and JZ. All authors commented on previous versions of the manuscript. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Ethics approval
Not applicable.
Consent to participate
Not applicable.
Consent for publication
Not applicable.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Xu, Y., Zhao, J., Kai, M. et al. Identification of genetic loci for flag leaf traits in wheat (Triticum aestivum L.). Euphytica 220, 50 (2024). https://doi.org/10.1007/s10681-024-03302-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s10681-024-03302-9