Abstract
Two new species in the Fusarium solani species complex (FSSC) are described and introduced. The new taxa are represented by German isolates CBS 142481 and CBS 142480 collected from commercial yard waste compost and vascular tissue of a wilting branch of hibiscus, respectively. The phylogenetic relationships of the collected strains to one another and within the FSSC were evaluated based on DNA sequences of 6 gene loci. Due to the limited sequence data available for reference strains in GenBank, however, a multi-gene phylogenetic analysis included partial sequences for the internal transcribed spacer region and intervening 5.8S nrRNA gene (ITS), translation elongation factor 1-alpha (tef1) and the RNA polymerase II second largest subunit (rpb2). Morphological and molecular phylogenetic data independently showed that these strains are distinct populations of the FSSC, nested within Clade 3. Thus, we introduce Fusarium stercicola and Fusarium witzenhausenense as novel species in the complex. In addition, 19 plant species of 7 legume genera were evaluated for their potential to host the newly described taxa. Eighteen plant species were successfully colonized, with 6 and 9 of these being symptomatic hosts for F. stercicola and F. witzenhausenense, respectively. As plants of the family Fabaceae are very distant to the originally sourced material from which the new taxa were recovered, our results suggest that F. stercicola and F. witzenhausenense are not host-specific and are ecologically fit to sustain stable populations in variety of habitats.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The Fusarium solani species complex (FSSC) is one of the most widespread groups of filamentous fungi that are intensively studied due to their ability to cause disease both in plants and humans (Zhang et al. 2006). This species complex causes vascular wilts and root rots in over 100 agricultural crops (Kolattukudy and Gamble 1995). Members of the complex are also emerging as clinically important fungal pathogens and account for approximately 60% of all human and animal fusarioses worldwide (Al-Hatmi et al. 2016; O’Donnell et al. 2008). The FSSC, however, are primarily soil and plant associated fungi characterised by considerable ecological plasticity that, in addition to pathogenic lifestyle, typically occur as saprotrophs and endophytes in agro-ecosystems (Leslie and Summerell 2006).
First described by von Martius (1842) as Fusisporium solani, later transferred to Fusarium by Appel and Wollenweber (1910), and recently epitypified as Fusisporium (Fusarium) solani (Schroers et al. 2016), the taxonomy and nomenclature of F. solani sensu lato (name used in traditional broad sense) still remains controversial. The traditional taxonomic concepts applied to F. solani by Snyder and Hansen (1941) and Matuo and Snyder (1973) were based on the morphology of asexual reproductive structures (particularly the shape and dimensions of macroconidia) and the ability of different isolates to reproduce sexually (the biological species concept). The system of Snyder and Hansen (1941), which was later adopted by Nelson et al. (1983), combined Wollenweber and Reinking (1935) sections Martiella and Ventricosum that initially recognized 7 species, 12 varieties and 6 forms into a single species F. solani accommodated within the section Martiella. This widely accepted morphological concept was generally characterised by the production of straight to slightly curved (usually 3 septate) macroconidia with a rounded apical cell and poorly developed or distinct foot-shaped basal cell found abundantly in usually cream coloured sporodochia, production of oval or ellipsoidal 0–1 septate microconidia on long monophialides in round false heads and the presence of chlamydospores produced single or in chains intercalary in the hyphae and terminally on short lateral branches (Leslie and Summerell 2006; Nelson et al. 1983). Based on the sexual crosses however, Matuo and Snyder (1973) determined that F. solani comprised at least seven heterothallic biological species designated as mating populations (MPs I–VII) of Nectria haematococca. Each biological species represented one of the host-specific forms and included F. solani ff. sp. cucurbitae (MP I), batatas (MP II), mori (MP III), xanthoxyli (MP IV), cucurbitae race 2 (MP V), pisi (MP VI) and robiniae (MP VII) (Aoki et al. 2014). To date, at least seven additional specialized forms have been described based on their specific pathogenicity on a particular host plant and without known sexual morphs (Aoki et al. 2014; Bueno et al. 2014; Chung et al. 2011). However, morphological and biological species concepts applied to F. solani faced many limitations (Cai et al. 2011; Taylor et al. 2000) and recognizing species boundaries within the FSSC remained a challenge.
Studies conducted over the past two decades employing genealogical concordance phylogenetic species recognition has significantly improved the taxonomic resolution in multiple cryptic species and has become a widely applied method for species delimitation in fungal groups that reproduce both sexually and/or asexually (Aoki et al. 2014; Schroers et al. 2016; Taylor et al. 2000). These studies revealed that the morphospecies F. solani comprise at least 60 phylogenetically distinct species distributed among three biogeographically structured clades (Nalim et al. 2011; O’Donnell 2000). The largest and the most diverse group belongs to clade 3 that shows a cosmopolitan distribution and accommodates all seven biological species (Matuo and Snyder 1973), supporting their genealogical exclusivity (Aoki et al. 2014). It is important to note, however, that several different generic names, mostly referring to sexual stages, have been applied to F. solani in line with previous nomenclature rules that dictated that the teleomorphic name of a fungus has preference over its anamorphic name. In accordendance with changes in the International Code of Nomenclature for algae, fungi and plants (ICN; McNeil et al. 2012) that resulted in abandoning dual nomenclature of fungi, allowing both teleomorphic or anamorphic name to be applied to each taxon, Geiser et al. (2013) proposed to preserve the long standing use of the name Fusarium over all competing teleomorphic names. Although Neocosmospora is a teleomorph name that was found deeply nested within the FSSC clade 3 and represents correct teleomorhic name for this group of fungi, the treatment of Geiser et al. (2013) has received broad and robust support from plant pathologists and other users worldwide. Recently, Lombard et al. (2015), later followed by Sandoval-Denis et al. (2017), recombined all species of the FSSC into Neocosmospora excluding many agriculturally and medically relevant species from the genus Fusarium. We, however, consider that the name Fusarium is well established in mycological literature and therefore support the unitary use of the name Fusarium in the consensus proposal by Geiser et al. (2013).
In the present study, morphological characters and phylogenetic relationships of strains collected from compost and from a vascular tissue of wilting branch of hibiscus (Hibiscus sp.) are evaluated based on DNA sequences of 6 gene loci. The main objectives of the study were to: (i) determine systematic and phylogenetic relationships of collected strains to one another and to related species from the FSSC, and (ii) determine whether various legumes are capable to symptomatically or asymptomatically host the fungi. Our results indicate that the examined strains represent two novel, distinct species, in the complex which we formally describe here.
Materials and methods
Fungal cultures
Strains used in this study were isolated from yard waste compost and a vascular tissue of diseased hibiscus (Hibiscus sp.) branch showing severe wilt symptoms and dieback of the branches. One of the strains was collected in 2014 from yard waste compost commercially produced in Hannover (Lower Saxony, Germany), and three additional strains were isolated in 2015 from infected hibiscus grown in Witzenhausen (Hessen, Germany). Isolations were performed following the methods of Baćanović (2015) and each Fusarium isolate was examined and morphologically identified as F. solani according to Leslie and Summerell (2006). All cultures obtained in this study are maintained at the Internal Culture Collection of Ecological Plant Protection Department at University of Kassel. Type specimens of the newly proposed taxa were deposited in the reference collection of the Westerdijk Fungal Biodiversity Institute—KNAW, Utrecht, the Netherlands (CBS numbers) with replicates maintained in the culture collection at the Leibniz Institute DSMZ-German Collection of Microorganisms and Cell Cultures, Braunschweig, Germany (DSM numbers).
DNA extraction
Genomic DNA was isolated from cultures actively growing on half strength potato dextrose agar (PDA) (19.5 g PDA l−1 and 10 g agar l−1, Sigma Aldrich, Steinheim, Germany) using the UltraClean Microbial DNA isolation kit (Mo Bio Laboratories, Carlsbad, California) according to the instructions of the manufacturer. All DNA extracts were diluted 200–500 times in milli-Q water and stored at 4 °C before use.
PCR amplification and sequencing
Portions of the following six gene fragments were chosen for multilocus sequence typing: the internal transcribed spacer (ITS) region and domains D1/D2/D3 of the nuclear large-subunit (LSU) rRNA, the translation-elongation factor 1 alpha (tef1), β-tubulin (tub2), and RNA polymerase II the largest (rpb1) and the second largest subunit (rpb2).
The PCR amplification and sequencing of the tef1 and rpb2 loci was conducted using the primer pairs, EF1 and EF2 (O’Donnell et al. 1998), and RPB2-5F2 and fRPB2-7cR (Sung et al. 2007; Liu et al. 1999), respectively. The ITS, LSU, tub2, and rpb1 genes were amplified using primer pairs ITS1 and ITS5 (White et al. 1990), LROR and LR5 (Rehner and Samuels 1994; Vilgalys and Hester 1990), T1 and Bt-2b (O’Donnell and Cigelnik 1997; Glass and Donaldson 1995), and RPB1-Ac and RPB1-Cr (Matheny et al. 2002; Stiller and Hall 1997), for each locus respectively. The PCR’s were performed in a 2720 Thermal Cycler (Applied Biosystems, Foster City, California) in a total volume of 12.5 μl.
For ITS and LSU the PCR mixture contained 1 µl of diluted genomic DNA, 7.69 µl of milli-Q water, 1.25 µl of PCR buffer, 0.63 µl of 50 mM MgCl2, 0.63 µl of 99.9% DMSO, 0.7 µl of 1 mM dNTP’s, 0.25 µl of each primer (10 pMol per µl stock) and 0.1 Unit Taq DNA polymerase E (Genaxxon Bioscience, Biberach, Germany). For tub2 the PCR reaction contained 1 µl of diluted genomic DNA, 8.9 µl of milli-Q water, 1.25 µl of PCR buffer, 0.25 µl of 50 mM MgCl2, 0.5 µl of 1 mM dNTP’s, 0.25 µl of each primer (10 pMol per µl stock) and 0.1 Unit Taq DNA polymerase E (Genaxxon Bioscience, Biberach, Germany). The rpb1 PCR amplification mixture contained 2 µl of diluted genomic DNA, 6.06 µl of milli-Q water, 1.25 µl of PCR buffer, 1.26 µl of 50 mM MgCl2, 0.63 µl of 99.9% DMSO, 0.7 µl of 1 mM dNTP’s, 1 µl mM of each primer (10 pMol per µl stock) and 0.1 Unit Taq DNA polymerase E (Genaxxon Bioscience, Biberach, Germany).
Conditions for amplification for all loci were an initial denaturation step of 5 min at 95 °C, followed by 40 cycles of denaturation, annealing and elongation and a final elongation step of 7 min at 72 °C. For the ITS, LSU and tub2 amplification the 40 cycles consisted of 30 s at 95 °C, 30 s at 52 °C and 1 min at 72 °C. For rpb1, the amplification consisted of 5 cycles of 30 s at 95 °C, 30 s at 56 °C and 1 min at 72 °C, 5 cycles of 30 s at 95 °C, 30 s at 54 °C and 1 min at 72 °C, and 30 cycles of 30 s at 95 °C, 30 s at 55 °C and 1 min at 72 °C.
The PCR products were analysed by electrophoresis on a 1% (w/v) agarose gel containing 0.1 μg ethidium bromide ml−1 in 1 × TAE buffer (0.4 M Tris, 0.05 M NaAc, 0.01 M EDTA, pH 7.85) and visualized under UV light. Hyperladder I (Bioline, Luckenwalde, Germany) was applied as size standard following the recommendations of the manufacturer. The sequence products were purified through Sephadex G-50 Superfine columns (Sigma Aldrich, St. Louis, MO) in a 96-well MultiScreen HV plate (Millipore, Billerica, MA), and sequenced in both directions using the aforementioned primer pairs. The reactions were performed with an ABI Prism 3730xl DNA Analyser (Applied Biosystems) using the BigDye Terminator v3.1 Cycle Sequencing Ready Reaction Kit (Life Technologies Europe BV, Applied Biosystems, Bleiswijk, The Netherlands).
Phylogenetic analyses
Obtained raw sequences were assembled and edited in MEGA v. 6.06 software (Tamura et al. 2013). Sequences of the individual loci were aligned using MAFFT (Katoh and Standley 2013) on the web server of the European Bioinformatics Institute (EMBL EBI) (http://www.ebi.ac.uk/Tools/msa/mafft/) (Li et al. 2015), and the alignments were checked and manually corrected if necessary in MEGA v. 6.06. To establish the identity of the isolates to the species level, different phylogenetic analyses were conducted. First, a single alignments were constructed for the ITS, tef1 and rpb2 gene sequences. The analysis included 43 sequences for ITS, 46 sequences for tef1 and 45 sequences for rpb2 loci. Second, a multi-locus sequence analyses was conducted using combined datasets of ITS, tef1 and rpb2 gene sequences. Phylogenetic inference was based on two independent algorithms: RaxML (ML) and Bayesian analyses (BI) which were run on the CIPRES Science Gateway portal (Miller et al. 2010) using RaxML v. 8.2.9 and MrBayes v. 3.2.6, respectively. Evolutionary models were calculated using MrModelTest v. 2.3 (Nylander 2004) selecting the best-fit model for each data partition according to the Akaike criterion. For ML analyses, the default parameters were used and BS was carried out using the rapid bootstrapping algorithm (BS) with the automatic halt option. Bayesian analyses included two parallel runs of 5,000,000 generations, with the stop rule option and a sampling frequency set to every 1000 generations. The 50% majority rule consensus trees and posterior probability (PP) values were calculated after discarding the first 25% of the samples as burn-in. The resulting trees were plotted using FigTree v. 1.4.2 and MEGA v. 6.06. The individual gene datasets were assessed for incongruence before being concatenated by checking their individual phylogenies for conflicts between clades with significant ML and BI support. All the Fusarium strains used in the phylogenetic analyses including detailed information for strains from this study and assigned GenBank accession numbers are listed in Table 1.
Morphological characterization and growth rates
Colony morphology, pigmentation and odour were determined from cultures grown on potato dextrose agar (39 g PDA l−1, Sigma Aldrich, Steinheim, Germany), malt extract agar (MEA; 15 g malt extract l−1, 1.5 g peptone from soybean l−1 and 8 g agar l−1, Sigma Aldrich, Steinheim, Germany), synthetic low nutrient agar (SNA; Nirenberg 1976) and carnation leaf agar (CLA; Leslie and Summerell 2006). Cultures were prepared by placing 5 mm agar plugs in the centre of the PDA, MEA, SNA (+filter paper) and CLA plate and incubated for 10 days at 23 °C in darkness under constant blacklight blue (BLB) fluorescent light (F40, range 315–400 nm with the peak at 365 nm). Colony colours and odour were determinate primarily from cultures grown on PDA and MEA, and are rated according to a standardized colour atlas of (Kornerup and Wanscher 1978). Conidia and conidiophores were documented either from colonies grown on SNA and CLA, and/or from slide cultures prepared by transferring a small amount of mycelia on the edges of SNA agar blocks (ca. 1 cm2) and overlaying the blocks with a No. 1 cover glass. After 10 days of incubation (described above), microscopic characters were examined in mounts of lactic acid with cotton blue under Zeiss Axiosskop2 plus microscope equipped with a Jenoptik ProgRes® digital camera (JENOPTIK, Germany). Minimum of 30 measurements (unless indicated differently) were made per structure using CapturePro 2.8 (JENOPTIK, Germany) software. The reported measurements include the minimum and maximum values in brackets, and the mean values plus and minus the standard deviation.
Average radial growth rates were determined at various temperatures from cultures grown on PDA, MEA and SNA following the methods of Aoki et al. (2003) with some modifications. Briefly, 5 mm diameter plugs were taken from the actively growing edge of a 15 days old PDA colony and placed mycelium side down 1 cm from the edge of a fresh PDA, MEA and SNA plate. Cultures were incubated in 9 cm plastic Petri dishes at five temperature regimes (15–35 °C) at 5 °C intervals in darkness. Colony radius was measured daily for 4 days in four directions around the colony. Average mycelial growth rates were calculated from three replicate plates for each respective temperature and expressed as diametric growth per 24 h.
Aggressiveness test on legumes
To determine the host range and plant response to inoculations, two representing strains (CBS 142480 and CBS 142481) were evaluated for aggressiveness on 19 plant cultivars and accession belonging to 7 legume genera in a greenhouse bioassay (Table 2). Prior to planting seeds of Crotalaria, Glycine, Pisum and Vicia spp. were surface sterilized with 70% ethanol for 5 min, whereas the seed of the remaining plant cultivars and accessions were treated with 97% sulphuric acid for 4 min. Seeds were then thoroughly washed under running tap water and germinated for 48 h on wet filter paper in a glass Petri dishes (8 × 1.5 cm) at room temperature. Two germinated seeds were transplanted into 300 ml pots filled with autoclaved sand and inoculated with 2 × 104 spores g−1 substrate. Five replicate pots arranged in completely randomized design were used per treatment.
The inoculum and targeted spore concentration was prepared from cultures incubated in malt extract broth (MEB) as described in Šišić et al. (2017). The pots were inoculated 24 h after transplanting and kept in the greenhouse at 19/16 °C day/night temperature regime and 16 h photoperiod (provided with 400 W high-pressure sodium lamps). Pots were watered daily with tap water and fertilized with complex N:P:K fertilizer Wuxal Super (8:8:6 + microelements). A total of 120 mg of N l−1 of substrate was divided into six equal portions and added weekly over the course of the experiment.
After 59 days of growing, plants were removed from pots, and the roots were separated from the above ground biomass. Above ground plant parts of each pot were weighted and dried at 105 °C until constant weight was attained. Roots were washed under running tap water, and root rot severity was assessed using a visual 0-8 score scale based on external root tissue discoloration levels as described in Šišić et al. (2018a). Briefly, disease severity rating (DSR) 0 = no symptoms, 1 = streaks at the transition zone, epicotyl or hypocotyl, 2 = brown lesion cover up to 50% of root perimeter, 3 = brown-black lesion cover 51 to 99% of root perimeter, 4 = black lesion cover 100% of stem perimeter, 5 = black lesion spread up to 30–49% of the tap root, 6 = black lesions spread up to 50 to 70% of the tap root, 7 = black lesions spread > 70% of the tap root, 8 = dead plant.
Fulfilment of Koch’s postulates
To confirm that infection was the result of the inoculated strains, the fungi were re-isolated from the root pieces using the protocol described in Šišić et al. (2017). Briefly, three to six randomly selected roots from each treatment were surface-sterilized for 10 s in 1% NaOCl, thoroughly washed in distilled water and placed on filter paper under a laminar flow hood for 1 h to dry. Subsequently, the roots were cut into approximately 1 cm long fragments, placed in Petri dishes containing half strength PDA medium and incubated under alternating cycles of 12 h BLB fluorescent light and 12 h darkness. After 10–15 days of incubation, fungal colonies developing from the root pieces were sub-cultured separately in Petri dishes containing PDA and SNA agar, incubated as described previously, and identified based on cultural characteristics and microscopic examination of conidiogenous cells (Leslie and Summerell 2006). To additionally verify species identity, representative F. solani isolates were randomly selected from different inoculation treatments and a portion of the tef1 gene region was amplified and sequenced as described previously. These sequences were then compared with the tef1 gene sequences of the inoculated reference strains using the MEGA V.6.06.
Evaluation of the host range
Cultural methods in combination with disease severity data were used to determine the host range of the inoculated strains on tested plants. The response of the tested legume cultivars and accessions to inoculations was determined according to criteria described in Šišić et al. (2018b) previously adopted from Kolander et al. (2012). The accessions were considered symptomatic hosts if the inoculated isolate was re-isolated from surface sterilized roots, the average disease severity rating was higher than 2 and significantly greater than in the un-inoculated control plants. Accessions were also considered symptomatic if DSR < 2 but there was a significant reduction in mean plant biomass of inoculated plants compared to the corresponding control. Some of the control plants showed moderate symptoms on the roots (mean DSR > 2) caused by factors other than inoculation with Fusarium conidia, and in this case the host was considered susceptible if the final disease severity level of inoculated treatments was significantly higher than that of the corresponding control plants. The accessions were considered asymptomatic hosts if disease rating was less than or equal to 2, there was no significant reduction in biomass, and the fungus was re-isolated from the root parts following surface sterilization.
Data analysis
All statistical analyses were done using R statistical software (version 3.3.1; R Core Team 2013). To determine whether biomass of inoculated treatments differed significantly from biomass of the corresponding non-inoculated controls, data were analysed with one-way analysis of variance (ANOVA) and Dunnett’s posthoc test (Bretz et al. 2011). The normal and homogeneous distribution of residuals was examined using Shapiro–Wilks-W-Test and Levene’s test, respectively. When necessary data were log10 transformed before analysis. Differences in root rot severity ratings of inoculated and non-inoculated treatment within each plant species were compared using the non-parametric Pairwise Wilcoxon Rank Sum Tests (Conover 1999).
Results
Phylogenetic analysis
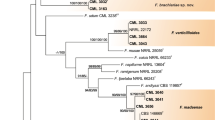
BLASTn searches against the GenBank database, using ITS, tef1 and rpb2 sequences of the isolates under study showed high similarities with members affiliated to the FSSC, but with undescribed species. Searches using ITS sequences resulted in Blast matches with maximum identity scores with 99% between F. solani FSSC44 (strain YIMPH300045) and the CBS 142481 (compost strain), and 99% between Fusarium sp. FSSC44 (strain GJS 09-1459) and the CBS 142480 (hibiscus strain). The rbp2 sequences for both strains CBS 142481 and CBS 142480 matched with Fusarium sp. FSSC44 (strain GJS 09-1459) with 99% similarity. The tef1 sequences matched with Fusarium sp. FSSC44 (strain GJS 09-1459) with 99% for both strains. Due to the limited sequence data available in GenBank for F. solani complex fungi, the tub2, LSU and rpb1 gene regions, these loci could not meaningfully be included in the phylogenetic inference. Thus, multi-locus sequence analysis was primarily based on the ITS, rpb2 and tef1 sequences. The analysis was conducted in order to position the isolates in the F. solani complex. The analysis included 43 ITS sequences and 45 sequences of rpb2 and 46 tef1 spanning all known species and genotypes of the FSSC and outgroup taxon F. staphyleae (NRRL 22316). Each gene was analysed separately prior to multi locus analysis. In gene trees of ITS, tef1 and rpb2 separately, strain CBS 42481 and CBS 142480 were found as members of monophyletic clades supported by high bootstrap values (Figs. 1, 2, 3). The ITS phylogeny was not able to clearly distinguish five of the most important lineages/haplotypes, FSSC29, FSSC28a, FSSC27a, while it could distinguish clearly between the CBS 42481 and CBS 142480 (FSSC44) and were found monophyletic clades supported by high bootstrap value (BS = 99%) (Fig. 1). The other two markers; rpb2 and tef1 were the only loci clearly identifying all the environmental and clinically significant lineages/haplotypes and species including the new clades; the CBS 42481 and CBS 142480 (FSSC44) with bootstrap value (BS = 99–100%) (Figs. 1, 2, 3). The final analysis included 2064 bp characters from three loci (ITS, rpb2 and tef1) of 46 strains including the outgroup taxa F. staphyleae, in order to establish the phylogenetic position of CBS 142481 and CBS 142480 (FSSC44) that formed distinct, new lineages in the individual phylogenies. The new fully supported lineages were phylogenetically and morphologically different from their sister clades, which includes F. keratoplasticum, F. falciforme and the others species (Fig. 4). Multi-locus sequence typing using three conserved loci commonly used for phylogenetic analysis revealed one was closely related to the previously known haplotype, but undescribed and unnamed, FSSC 44 clade (CBS 142480), while the other isolate was not similar to any previously described FSSC isolate (CBS 142481). This group of isolates includes CBS 142481 (compost strain) in one clade, and CBS 142480, KU90.15 and KU90.1.15 (hibiscus strains) in different clade (FSSC44). Both species formed highly supported clades closely related but distinct from other clades, representing a previously undescribed phylogenetically-supported species. These two clades are described below as two new species, Fusarium stercicola and Fusarium witzenhausenense.
Phylogenetic tree generated by Bayesian inference (BI) and Maximum Likelihood (ML) of the Fusarium solani species complex inferred from ITS rDNA dataset. Bootstrap support values from Bayesian posterior probabilities and Maximum Likelihood (RaxML) above 0.8 and 80%, respectively, are indicated at the nodes. Examined strains and names of newly proposed taxa are indicated in red. Ex-type strains are indicated with superscript “T”. The tree is rooted to NRRL 22316, F. staphyleae
Phylogenetic tree generated by Bayesian inference (BI) and Maximum Likelihood (ML) of the Fusarium solani species complex inferred from tef1 dataset. Bootstrap support values from Bayesian posterior probabilities and Maximum Likelihood (RaxML) above 0.8 and 80%, respectively, are indicated at the nodes. Examined strains and names of newly proposed taxa are indicated in red. Ex-type strains are indicated with superscript “T”. The tree was rooted to NRRL 22316, F. staphyleae
Phylogenetic tree generated by Bayesian inference (BI) and Maximum Likelihood (ML) of the Fusarium solani complex inferred from rbp2 dataset. Bootstrap support values from Bayesian posterior probabilities and Maximum Likelihood (RaxML) above 0.8 and 80%, respectively, are indicated at the nodes. Examined strains and names of newly proposed taxa are indicated in red. Ex-type strains are indicated with superscript “T”. The tree was rooted to NRRL 22316, F. staphyleae
Phylogenetic tree generated by Bayesian inference (BI) and maximum Likelihood (ML) of the Fusarium solani complex inferred from the combined ITS rDNA, tef1 and rbp2 datasets. Bootstrap support values from Bayesian posterior probabilities and Maximum Likelihood (RaxML) above 0.8 and 80%, respectively, are indicated at the nodes. Examined strains and names of newly proposed taxa are indicated in red. Ex-type strains are indicated with superscript “T”. The tree was rooted to NRRL 22316, F. staphyleae
Taxonomy
Fusarium stercicola A. Šišić, A. M. S. Al-Hatmi, J. Baćanović-Šišić, S. A. Ahmed & M. R. Finckh sp. nov.—MycoBank MB823581; Fig. 5.
Morphological description of Fusarium stercicola CBS 142481 (DSM 106211). a Growth on PDA; b growth on MEA; c Sporodochia after 10 days appearing as orange slimy masses on pieces of carnation leaf placed on CLA; d, e multiseptate macroconidia; f short Monophialides with false head; g, h erect conidiophores producing 0–1 septate conidia; i Monophialides and microconidia; j Chlamydospores. Scale bar = 10 µm
Etymology Latin, stercus = compost, and lat., incola = inhabitant, referring to the substrate from which the ex-type strain of this fungus was collected.
Holotype dried specimen in herbarium CBS H-23352; living ex-type strain CBS 142481 = DSM 106211, isolated from composted yard waste plant debris, Hannover, Germany.
Colonies on PDA after 10 days (d) incubated at 23 °C under constant BLB fluorescent light showing net average radial growth rates of 34 mm. Colony surface white (Kornerup and Wanscher; K&W 4A1) to yellowish white (K&W 4A2), aerial mycelium white, cottony, forming abundantly through the colony. Reverse pigmentation yellow (K&W 3A6) at the center fading to yellowish-white (K&W 3A2) at the margin. Colony margin entire to undulate (Fig. 5a). Odour strong moldy. On MEA after 10 d at 23 °C and constant BLB fluorescent light colonies showing net average radial growth rates of 39 mm. Colony surface white (K&W 4A1) at the center fading to yellowish white (K&W 4A2) at the margin, radially striated, aerial mycelium white, cottony and more densely formed at the colony center becoming flat towards the margin. Reverse pigmentation overall pale orange (K&W 5A3) with irregular grey-reddish (K&W 8C4) pigmented areas in the agar. Colony margin entire to undulate (Fig. 5b). Odour moldy.
Colonies after 10 d at 23 °C under constant BLB fluorescent light showing net radial mycelial growth of 28 mm on SNA. Aerial mycelium white and more densely formed over the filter paper. On CLA, aerial mycelium scant, produced mainly on and around carnation leaf pieces and nearly invisible elsewhere. Pale orange sporodochia produced slowly and sparsely on pieces of carnation leaves (Fig. 5c). Scant production of erect conidiophores, tapering uniformly from base to tip from the agar surface and aerial mycelium (66.1–) 75.8–121.0 (–165.2) µm long and (3.0–) 3.5–4.9 (–6.2) µm wide at base. Single phialids comprise terminal (22.6–) 36.5–62.3 (–84.2) µm of the conidiophores. Tip of the phialid with periclinical thickening, collarete at most slightly flared (Fig. 5d–i).
Conidia often held in clear, colourless drops of liquid, 0, 1, 2, 3 (− 4, 5) septate: zero-septate ellipsoidal (6.4–) 8.2–12.4 (− 15.4) × (2.6–) 3.1–3.9 (− 4.7) µm; one-septate (12.5–) 14.8–19.8 (− 2.2) × (3.7–) 4.1–5.1 (− 5.5) µm; two septate (19.0–) 20.5–23.5 (− 24.8) µm; three septate slightly curved, apical cell rounded and undeveloped foot cell (23.1–) 26.1–31.0 (− 34.0) × (4.5–) 4.9–5.8 (− 6.3) µm; 4, 5-septate (n = 11) slightly curved with a slightly hooked apical cell and barely notched basal cell (36.3–) 37.3–44.0 (− 46.7) × (4.4–) 4.5–6.6 (− 7.8) (Fig. 5d–i).
Chlamydospores formed abundantly after 10 days, unicellular, ovoid to ellipsoidal, single or in chains, terminal on hyphae or intercalary, pale brown in colour, consisting of enlarged, thick-walled vegetative cells: unicellular (6.1–) 7.0–9.7 (− 11.5) × (4.4–) 5.2–7.2 (− 8.5) µm; multicellular (10.1–) 11.4–14.8 (− 17.1) in diam (Fig. 5j).
Radial mycelial growth on PDA, MEA and SNA after 4 days in constant dark at five different temperatures: colonies showing optimal growth at 25 °C on all culture media of 8.6, 10.0, and 10.1 mm/d on PDA, MEA and SNA, respectively (Fig. 7).
Fusarium witzenhausenense A. Šišić, A. M. S. Al-Hatmi, J. Baćanović-Šišić, S. A. Ahmed & M. R. Finckh sp. nov.—MycoBank MB 823582; Fig. 6.
Morphological description of Fusarium witzenhausenense CBS 142480 (DSM 106212). a Growth on PDA; b growth on MEA; c white sporodochia appearing after 10 days on pieces of carnation leaf placed on CLA; d–i Conidiophores producing 0–3 septate conidia; j small phialidic peg with conidia; k, l macro and micro conidia. Bars: d, e = 100 µm; f, g, h, i = 50 µm; j, k, l = 10 µm
Etymology Refers to Witzenhausen, the city in central Germany, where the ex-type strain of the fungus was collected.
Holotype dried specimen in herbarium CBS H-23351; living ex-type strain CBS 142480 = DSM 106212, isolated from vascular tissue of infected hibiscus (Hibiscus sp.) showing severe wilt symptoms and dieback of the branches, Witzenhausen, Germany.
Colonies on PDA after 10 d incubated at 23 °C under constant BLB fluorescent light showing net average radial growth rates of 33 mm. Colony surface white (K&W 4A1) in the middle around initial inoculum and yellowish-white (K&W 4A2) elsewhere. Aerial mycelium white, cottony and more densely formed at the colony centre becoming flat towards margin. Reverse pigmentation often absent. Colony margin entire to undulate (Fig. 6a). Odour mouldy. On MEA after 10 d at 23 °C and constant BLB fluorescent light colonies showing net average radial growth rates of 41 mm. Colony surface brownish-grey (K&W 8D3) at the centre and yellowish white (K&W 4A2) at the margin, with radial V shaped stripes composed of dense white flat mycelium stretching from colony centre to colony edge. Aerial mycelium scarce, colony fine with abundant production of cream slimy sporodochia at the margins. Reverse pigmentation reddish-grey (K&W 8B2) in the centre fading to yellowish white (K&W 4A2) at the margin with irregularly distributed brownish-grey pigmented (K&W 8D2) areas. Colony margin entire to undulate (Fig. 6b). Odour sweet to mouldy.
Colonies after 10 d at 23 °C under constant BLB fluorescent light showing average net radial mycelial growth of 33 mm on SNA. Aerial mycelium white to cottony and somewhat more densely formed over the filter paper. Conidiophores formed more densely on and around fitter paper than more distant from filter paper. On CLA little or no aerial mycelium. White collared sporodochia abundantly formed on pieces of carnation leaves, and often seen as flower like structures (Fig. 6c). Abundant production of erect, mononematous conidiophores from the agar surface and aerial mycelium. Mononematous conidiophores, unbranched or occasionally branched once or twice (120.6–) 190.6–323.4 (− 377.7) µm long and (8.4–) 11.0–16.2 (− 17.7) µm wide at base, terminating into subcilindrical (10.5–) 21.6–47.6 (− 57.2) µm long and (3.0–) 3.1–3.9 (− 4.4) wide phialide. Tip of the phialide with inconspicuous periclinical thickening, collarette not flared (Fig. 6d–i).
Conidia abundantly formed on SNA and CLA and often held in a clear, colourless drops of liquid 0, 1 and 3 septate: zero-septate ellipsoidal or with a rounded apex and truncate base, (6.2–) 7.1–10.5 (− 13.4) × (3.1–) 3.5–4.5 (− 5.5) µm; one-septate ellipsoidal, (13.5–) 16.2–21.0 (–22.5) × (3.8–) 4.8–5.6 (− 6.1) µm; three septate slightly curved or arcuate with a rounded apical cell and poorly developed basal cell (25.4–) 28.0–36.0 (− 41.3) × (4.8–) 5.3–6.3 (− 6.9) µm (Fig. 6d–l). Chlamydospores absent.
Radial mycelial growth on PDA, MEA and SNA after 4 days in constant dark at five different temperatures: colonies showing optimal growth at 25 °C on all culture media of 9.2, 11.1, and 11.2 mm/d on PDA, MEA and SNA, respectively (Fig. 7).
Host range
Responses of tested legumes to inoculation with F. stercicola and F. witzenhausenense varied greatly (Table 2). Both fungi were re-isolated from surface sterilized roots of 18 out of the 19 accessions and plant cultivars tested. Among the 18 successfully colonized legumes by F. stercicola, 6 were symptomatic and 12 were asymptomatic hosts. Symptomatic hosts included one Pisum sativum cv. Santana, and one accession (acc.) of Lathyrus dymenum acc. 1660, Medicago orbicularis acc. 479, Trifolium subterraneum acc. 1065, and Vicia ervilia acc. 1531. Among these, the highest mean disease severity rating (DSR) was observed on P. sativum cv. Santana and V. ervilia acc. 1531 of 4.1 and 4.3, respectively. Among 12 asymptomatic hosts, L. sativus acc. 1668 developed DSR > 2, however due to contamination in the un-inoculated control by other pathogenic fungal species (data not shown) which resulted in a non-significant difference between inoculated and corresponding un-inoculated control treatment, the accession was classified as asymptomatic host. As assessed by plating surface sterilized root segments, Crotalaria spectabilis was the only plant species found not to host tested F. stercicola isolate CBS 142481 (Table 2). Compared to the corresponding controls, inoculation with F. stercicola in general had neutral or somewhat positive biomass effects on the tested legumes with, significant biomass increase only in T. subterraneum cv. Campeda (Fig. 8).
Effect of Fusarium stercicola (strain CBS 142481 = DSM 106211) and F. witzenhausenense (strain CBS 142480 = DSM 106212) inoculation on dry plant biomass of 19 leguminous cultivars (cv.) and accessions (acc.) evaluated in greenhouse bioassay. Effects on the dry plant biomass of inoculated plants are given relative to the un-inoculated control treatment performance which was set at 100% (green line). Error bars show mean ± standard deviation of inoculated treatments. Asterisks above bars indicate significant difference between inoculated treatment and the corresponding un-inoculated control according to Dunnett’s t test at P < 0.05. (Color figure online)
Similar results were also observed for F. witzenhausenense. Nine accessions were found to be symptomatic and nine asymptomatic hosts, and one Glycine max cv. Amandine was classified as non-host for the fungus (Table 2). As for F. stercicola, the highest mean disease severity rating was observed on P. sativum cv. Santana and V. ervilia acc. 1531, with DSR of 4.5 and 6.3, respectively. In contrast to F. stercicola, however, there was general trend of biomass decrease following inoculation with F. witzenhausenense isolate CBS 142480, with a significant reduction compared to corresponding control observed only in P. sativum cv. Santana (Fig. 8).
Discussion
In the current study we have identified and described two additional lineages of the FSSC, represented by German isolates CBS 142481 (DSM 106211) and CBS 142480 (DSM 106212) collected from commercially produced yard waste compost and a vascular tissue of wilting branch of hibiscus, respectively. Multi-locus sequence typing using three conserved loci commonly used for phylogenetic analysis of the genus Fusarium revealed that CBS 142480 was closely related to the previously known haplotype FSSC44 clade, but unnamed and undescribed yet, while the other isolate CBS 142481 was not similar to any previously described lineages/haplotypes or species within the FSSC complex. Both species received strong monophyletic bootstrap support in the individual analysis of each gene (Figs. 1, 2, 3) and combined loci (Fig. 4). The current recommendation for to assess species level identification within the genus Fusarium, using tef1 and rpb2 sequences, compared with curated reference sequences in the following databases as FUSARIUM-ID (http://isolate.fusariumdb.org; Geiser et al. 2004) and Fusarium MLST (http://www.westerdijkinstitute.nl/Fusarium/; O’Donnell et al. 2015; 2016). In our study, these two loci have shown high resolving power and allowed for a distinguishing of the CBS 142480 and CBS 142481 as distinct clades. At the same time, ITS sequences were found not to possess sufficient rate of inter-specific variability. Although, ITS rDNA as the recommended barcode for fungi has many advantages, in particular the large number of reference sequences available in GenBank (Schoch et al. 2012), for most species complexes, however, this gene is insufficiently variable to discriminate molecular siblings (Balajee et al. 2009; Summerbell et al. 2005). Thus, closely related species may have identical ITS sequences and/or several ITS types may be found within a group of species, which can lead to incorrect phylogenetic inferences.
The morphological and molecular phylogenetic data independently showed that these strains represent distinct populations of the FSSC, both nested within Clade 3 (Nalim et al. 2011; O’Donnell 2000). Although, species within the F. solani complex share many morphological features (Aoki et al. 2014), we have found several unique morphological characters in colony morphology and conidia that can be used to distinguish newly described taxa. While both species have similar growth dynamics on PDA, MEA and SNA, F. stercicola and F. witzenhausenense are distinguished by the phenotypic differences in colony characters and production of pigments on PDA and MEA. F. stercicola produces abundant white mycelium through the colony on PDA and MEA, yellow to yellowish white pigments on PDA, and pale orange pigments with distinct grey-reddish patches on MEA. In contrast, F. witzenhausenense does not produce pigments on PDA, but produces abundant cream slimy sporodochia at the margins of the MEA colony, and reddish-grey to yellowish white pigments with irregularly distributed brownish-grey pigmented patches in the agar. F. stercicola can also be differentiated from F. witzenhausenense by the color of sporodochia produced on CLA, which are orange in F. stercicola and white in F. witzenhausenense. The former species also produced chlamydospores and 4- to 5-septate conidia which were not be observed in F. witzenhausenense. Furthermore, both species can be differentiated from the generic type, Fusisporium (Fusarium) solani s. str., by the presence of distinguishable pigments on artificial media, their conidiophore sizes, that are much longer in F. witzenhausenense in particular, and by the size, and shape of the basal and apical cells of macroconidia (Schroers et al. 2016).
In addition to morphological and molecular phylogenetic data, the newly described taxa also have different ecological affinities. F. stercicola was collected from commercialized yard waste compost suggesting high competitive ability of the species and its saprophic nature. In addition, F. stercicola abundantly produced chlamydospore on artificial media, the structures generally known to be highly persistent in the environment for considerable periods of time (Isaac 1992). Several different Fusarium species were, therefore, reported to survive the active thermophilic composting phase, the trait attributed mainly to their ability to produce heat resistant chlamydospores (Bollen et al. 1989; Termorshuizen et al. 2005). Thus, the compost colonization by F. stercicola may have occurred at any composting phase or post maturation compost handling. On the one hand, these results suggest considerable ecological plasticity of the species, and on the other, having in mind wide spread usage of this commercialized compost, it is likely that the fungus is distributed over wider areas of Germany and can be found in variety of habitats under diverse pedo-climatic conditions. In contrast, isolates of F. witzenhausenense were recovered from infected vascular tissue of hibiscus indicating significant pathogenic potential of the species and its association with vascular wilts in hibiscus. Although we did not confirm pathogenicity of the recovered isolates on the same host plant, previous studies that have established F. solani sensu lato as one of the major causal agent of root rots and wilts of hibiscus (Hassan et al. 2014).
In addition, greenhouse aggressiveness assays performed in this study showed that isolates of F. stercicola and F. witzenhausenense are able to symptomatically and asymptomatically infect roots of a various legume hosts. These results suggest that both species are not host specific, and are ecologically fit to sustain stable populations in variety of habitats. Differences in the host range between the species and general trend across the study of positive and negative biomass response of the tested legumes to inoculations with F. stercicola and F. witzenhausenense, respectively, also suggest that the newly described taxa may differ in their lifestyle and colonizing strategies in the environment.
We conclude that the strains examined here represent two novel species within the already phylogenetically diverse FSSC. Molecular phylogenetic data presented combined with the greenhouse bioassay and species specific cultural characters should assist disease specialists in more precisely determining host range, distribution and ecology of these fungi. Furthermore, as plants of the family Fabaceae are very distant to the originally sourced material from which the new taxa were recovered, taken together our results suggest that F. stercicola and F. witzenhausenense are able to colonize various ecological niches and point to plant debris and soil as probable and common environmental sources of both species.
Data availability
Sequences of all strains used in this study were submitted to GenBank database. Representative fungal strains were deposited in the culture collection of the Westerdijk Fungal Biodiversity Institute—KNAW, Utrecht, the Netherlands with replicates maintained in the culture collection at the Leibniz Institute DSMZ-German Collection of Microorganisms and Cell Cultures, Braunschweig, Germany. All fungal and plant materials are maintained at University of Kassel and are available for research purposes.
References
Al-Hatmi AMS, Hagen F, Menken SBJ et al (2016) Global molecular epidemiology and genetic diversity of Fusarium, a significant emerging group of human opportunists from 1958 to 2015. Emerg Microbes Infect 5:e124. https://doi.org/10.1038/emi.2016.126
Aoki T, O’Donnell K, Homma Y et al (2003) Sudden-death syndrome of soybean is caused by two morphologically and phylogenetically distinct species within the Fusarium solani species complex—F. virguliforme in North America and F. tucumaniae in South America. Mycologia 95:660–684
Aoki T, O’Donnell K, Geiser DM (2014) Systematics of key phytopathogenic Fusarium species: current status and future challenges. J Gen Plant Pathol 80:189–201. https://doi.org/10.1007/s10327-014-0509-3
Appel O, Wollenweber HW (1910) Grundlagen einer Monographie der Gattung Fusarium (Link). Arbeit Biol f Land u -Forstwt 8:207
Baćanović J (2015) Pathogens occurring in the winter pea—maize—winter wheat rotation, their host specificity and the potential of compost in suppressing foot and root disease of peas. Dissertation, University of Kassel, Witzenhausen. http://nbn-resolving.de/urn:nbn:de:hebis:34-2015091749047
Balajee SA, Borman AM, Brandt ME et al (2009) Sequence-based identification of Aspergillus, Fusarium, and Mucorales species in the clinical mycology laboratory: where are we and where should we go from here? J Clin Microbiol 47:877–884. https://doi.org/10.1128/JCM.01685-08
Bollen GJ, Volker D, Wijnen AP (1989) Inactivation of soil-borne plant pathogens during small-scale composting of crop residues. Eur J Plant Pathol 95:19–30
Bretz F, Hothorn T, Westfall PH (2011) Multiple comparisons using R. CRC Press, Boca Raton
Bueno CJ, Fischer IH, Rosa DD et al (2014) Fusarium solani f. sp. passiflorae: a new forma specialis causing collar rot in yellow passion fruit. Plant Pathol 63(2):382–389. https://doi.org/10.1111/ppa.12098
Cai L, Giraud T, Zhang N et al (2011) The evolution of species concepts and species recognition criteria in plant pathogenic fungi. Fungal Divers 50:121–133. https://doi.org/10.1007/s13225-011-0127-8
Chung WC, Chen LW, Huang JH et al (2011) A new ‘forma specialis’ of Fusarium solani causing leaf yellowing of Phalaenopsis. Plant Pathol 60:244–252. https://doi.org/10.1111/j.1365-3059.2010.02376.x
Conover WJ (1999) Practical nonparametric statistics. Wiley, New York
Geiser DM, Jiménez-Gasco MMD, Kang S et al (2004) FUSARIUM-ID v. 1.0: a DNA sequence database for identifying Fusarium. Eur J Plant Pathol 110:473–479
Geiser DM, Aoki T, Bacon CW et al (2013) One fungus, one name: defining the genus Fusarium in a scientifically robust way that preserves longstanding use. Phytopathology 103:400–408
Glass NL, Donaldson GC (1995) Development of primer sets designed for use with the PCR to amplify conserved genes from filamentous ascomycetes. Appl Environ Microbiol 61:1323–1330
Hassan N, Shimizu M, Hyakumachi M (2014) Occurrence of root rot and vascular wilt diseases in Roselle (Hibiscus sabdariffa L.) in Upper Egypt. Mycobiology 42:66. https://doi.org/10.5941/myco.2014.42.1.66
Isaac S (1992) Fungal plant interaction. Chapman & Hall, London
Katoh K, Standley DM (2013) MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol 30:772–780
Kolander TM, Bienapfl JC, Kurle JE et al (2012) Symptomatic and asymptomatic host range of Fusarium virguliforme, the causal agent of soybean sudden death syndrome. Plant Dis 96:1148–1153
Kolattukudy PE, Gamble DL (1995) Nectria haematococca: pathogenesis and host specificity in plant diseases. In: Kohmoto K, Singh US, Singh RP (eds) Pathogenesis and host specificity in plant pathogenic fungi and nematodes. Elsevier, Oxford, pp 83–102
Kornerup A, Wanscher JH (1978) Methuen handbook of colour. Eyre Methuen, London
Leslie JF, Summerell BA (2006) The Fusarium laboratory manual. Blackwell publishing, Ames
Li W, Cowley A, Uludag M et al (2015) The EMBL-EBI bioinformatics web and programmatic tools framework. Nucleic Acids Res 43(W1):W580–W584. https://doi.org/10.1093/nar/gkv279
Liu YJ, Whelen S, Hall BD (1999) Phylogenetic relationships among ascomycetes: evidence from an RNA polymerse II subunit. Mol Biol Evol 16:1799–1808
Lombard L, van der Merwe NA, Groenewald JZ et al (2015) Generic concepts in Nectriaceae. Stud Mycol 80:189–245. https://doi.org/10.1016/j.simyco.2014.12.002
Matheny PB, Liu YJ, Ammirati JF et al (2002) Using RPB1 sequences to improve phylogenetic inference among mushrooms (Inocybe, Agaricales). Am J Bot 89:688–698
Matuo T, Snyder WC (1973) Use of morphology and mating populations in the identification of formae speciales in Fusarium solani. Phytopathology 63:562–565
McNeil J, Barrie FR, Buck WR, et al. (2012) International Code of Nomenclature for algae, fungi and plants (Melbourne Code). In: Eighteenth International Botanical Congress Melbourne, Australia: A. R. G. Gantner Verlag KG. Regnum Vegetabile 154. http://herbario.udistrital.edu.co/herbario/images/stories/international%20code%20of%20nomenclature.pdf
Miller MA, Pfeiffer W and Schwartz T (2010) Creating the CIPRES Science Gateway for inference of large phylogenetic trees. In: Gateway computing environments workshop (GCE), 2010, IEEE, pp 1–8
Nalim FA, Samuels GJ, Wijesundera RL et al (2011) New species from the Fusarium solani species complex derived from perithecia and soil in the Old World tropics. Mycologia 103:1302–1330. https://doi.org/10.3852/10-307
Nelson PE, Tousson TA, Marasas WFO (1983) Fusarium species: an illustrated manual for identification. Pennsylvania State University Press, University Park
Nirenberg HI (1976) Untersuchungen über die morphologische und biologische Differenzierung in der Fusarium Sektion Liseola. Mitteilungen der Biologischen Bundesanstalt für Land- und Forstwirtschaft (Berlin-1. Dahlem) 169:1–17. https://doi.org/10.1002/jpln.19771400220
Nylander JAA (2004) MrModeltest v25. Program distributed by the author. Evolutionary Biology Centre, Uppsala University, Uppsala, p 2
O’Donnell K (2000) Molecular phylogeny of the Nectria haematococca-Fusarium solani species complex. Mycologia 92:919. https://doi.org/10.2307/3761588
O’Donnell K, Cigelnik E (1997) Two divergent intragenomic rDNA ITS2 types within a monophyletic lineage of the fungus Fusarium are nonorthologous. Mol Phylogenet Evol 7:103–116
O’Donnell K, Cigelnik E, Nirenberg HI (1998) Molecular systematics and phylogeography of the Gibberella fujikuroi species complex. Mycologia 90:465. https://doi.org/10.2307/3761407
O’Donnell K, Sutton DA, Fothergill A et al (2008) Molecular phylogenetic diversity, multilocus haplotype nomenclature, and in vitro antifungal resistance within the Fusarium solani species complex. J Clin Microbiol 46:2477–2490. https://doi.org/10.1128/JCM.02371-07
O’Donnell K, Ward TJ, Robert V et al (2015) DNA sequence-based identification of Fusarium: current status and future directions. Phytoparasitica 43:583–595. https://doi.org/10.1007/s12600-015-0484-z
O’Donnell K, Sutton DA, Wiederhold N et al (2016) Veterinary Fusarioses within the United States. J Clin Microbiol 54:2813–2819. https://doi.org/10.1128/JCM.01607-16
R Core Team (2013) A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. http://www.R-project.org/
Rehner SA, Samuels GJ (1994) Taxonomy and phylogeny of Gliocladium analysed from nuclear large subunit ribosomal DNA sequences. Mycol Res 98:625–634
Sandoval-Denis M, Guarnaccia V, Polizzi G et al (2017) Symptomatic Citrus trees reveal a new pathogenic lineage in Fusarium and two new Neocosmospora species. Persoonia 2018:1–25. https://doi.org/10.3767/persoonia.2018.40.01
Schoch CL, Seifert KA, Huhndorf S et al (2012) Nuclear ribosomal internal transcribed spacer (ITS) region as a universal DNA barcode marker for Fungi. Proc Natl Acad Sci USA 109:6241–6246. https://doi.org/10.1073/pnas.1117018109
Schroers HJ, Samuels GJ, Zhang N et al (2016) Epitypification of Fusisporium (Fusarium) solani and its assignment to a common phylogenetic species in the Fusarium solani species complex. Mycologia 108:806–819. https://doi.org/10.3852/15-255
Šišić A, Baćanović J, Finckh MR (2017) Endophytic Fusarium equiseti stimulates plant growth and reduces root rot disease of pea (Pisum sativum L.) caused by Fusarium avenaceum and Peyronellaea pinodella. Eur J Plant Pathol 148:271–282. https://doi.org/10.1007/s10658-016-1086-4
Šišić A, Baćanović-Šišić J, Karlovsky P et al (2018a) Roots of symptom-free leguminous cover crop and living mulch species harbor diverse Fusarium communities that show highly variable aggressiveness on pea (Pisum sativum). PLOS ONE 13:e0191969. https://doi.org/10.1371/journal.pone.0191969
Šišić A, Baćanović-Šišić J, Al-Hatmi AMS et al (2018b) The ‘forma specialis’ issue in Fusarium: A case study in Fusarium solani f. sp. pisi. Scientific Reports. https://doi.org/10.1038/s41598-018-19779-z
Snyder WC, Hansen HN (1941) The species concept in Fusarium with reference to section Martiella. Am J Bot 28:738. https://doi.org/10.2307/2436658
Stiller JW, Hall BD (1997) The origin of red algae: implications for plastid evolution. Proc Natl Acad Sci 94(9):4520–4525
Summerbell RC, Lévesque CA, Seifert KA et al (2005) Microcoding: the second step in DNA barcoding. Philos Trans R Soc B 360:1897–1903. https://doi.org/10.1098/rstb.2005.1721
Sung GH, Sung JM, Hywel-Jones NL et al (2007) A multi-gene phylogeny of Clavicipitaceae (Ascomycota, Fungi): identification of localized incongruence using a combinational bootstrap approach. Mol Phylogenet Evol 44:1204–1223. https://doi.org/10.1016/j.ympev.2007.03.011
Tamura K, Stecher G, Peterson D et al (2013) MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 30:2725–2729. https://doi.org/10.1093/molbev/mst197
Taylor JW, Jacobson DJ, Kroken S et al (2000) Phylogenetic species recognition and species concepts in fungi. Fungal Genet Biol 31:21–32. https://doi.org/10.1006/fgbi.2000.1228
Termorshuizen AJ, van Rijn E, Blok WJ (2005) Phytosanitary risk assessment of composts. Compost Sci Util 13:108–115. https://doi.org/10.1080/1065657X.2005.10702226
Vilgalys R, Hester M (1990) Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. J Bacteriol 172:4238–4246
von Martius CFP (1842) Die Kartoffelepidemie der letzen Jahre, oder die Stockfäule und Raude der Kartoffeln, geschildert und ihren ursächlichen Verhältnissen erörtet. Denkschrift Munchen Akad, Wiss, p 70
White TJ, Bruns T, Taylor J (1990) Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA, Gelfand DH, Sninsky JJ et al (eds) PCR protocols: a guide to methods and applications. Academic Press, San Diego, pp 315–322
Wollenweber HW, Reinking OA (1935) Die Fusarien, ihre Beschreibung, Schadwirkung, und Bekaämpfung. Paul Parey, Berlin
Zhang N, O’Donnell K, Sutton DA et al (2006) Members of the Fusarium solani species complex that cause infections in both humans and plants are common in the environment. J Clin Microbiol 44:2186–2190. https://doi.org/10.1128/JCM.00120-06
Acknowledgements
This research was funded by the Zentralen Forschungsförderung (ZFF) provided by the University of Kassel (Grant Number 1930).
Conflicts of interest
The authors declare that they have no conflict of interest.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Šišić, A., Al-Hatmi, A.M.S., Baćanović-Šišić, J. et al. Two new species of the Fusarium solani species complex isolated from compost and hibiscus (Hibiscus sp.). Antonie van Leeuwenhoek 111, 1785–1805 (2018). https://doi.org/10.1007/s10482-018-1068-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10482-018-1068-y