Abstract
MRI imaging is crucial for knee joint analysis in osteoarthritis (OA) diagnosis. The segmentation and thickness estimation of knee cartilage are vital steps for OA assessment. Most deep learning algorithms typically produce a single segmentation mask or rely on architectural modifications like Dropout to generate multiple outputs. We propose an alternative approach using Denoising Diffusion Models (DDMs) to yield multiple variants of segmentation outputs for knee cartilage segmentation and thus offer a mechanism to study predictive uncertainty in unseen test data. We further propose to integrate sparsity adaptive losses to supervise the diffusion process to handle intricate knee cartilage structures. We could empirically validate that DDM-based models predict more meaningful uncertainties when compared to Dropout based mechanisms. We have also quantitatively shown that DDM-based multiple segmentation generators are resilient to noise and can generalize to unseen data acquisition setups.
Access provided by Autonomous University of Puebla. Download conference paper PDF
Similar content being viewed by others
1 Introduction
MRI imaging can capture the structural details of the knee joint highlighting fine morphological changes better than any other imaging modality [2]. The clinical diagnostic protocol for Osteoarthritis (OA) is generally carried out by analyzing MRI scans to delineate the knee cartilages, followed by thickness calculation. The delineation of knee cartilages is often subjective, due to their resemblance to tissue features surrounding the cartilages. When building segmentation algorithms for such structures, having a single annotation restricts the learning, leading to closer mimicking of the available annotation. This also affects the predictive power of segmentation on unseen data.
Deep learning-based algorithms usually result in a single output segmentation, which is typically a single or multi-channel softmax output representing voxel-wise classification posterior probability. If the model has Dropout layers, using them at test-time results in random masking of the layer’s inputs, offering an architectural mechanism to obtain variations at the output. We seek to produce an alternative approach for generating multiple segmentation outputs and study it in comparison with the Monte Carlo Dropout technique.
Denoising diffusion models (DDM) [5, 10, 11] are a new generative method that has emerged as high-quality image generators. They use a learned parametrized iterative denoising process which is the reverse of a Markovian diffusion process to yield a ‘sample’, and various inverse problems involving image restoration and synthesis have been demonstrated building upon the DDM sampling framework. Specifically, they offer strong sample diversity and faithful mode coverage of the learned data distribution. Both of these are valuable in generalizing segmentation to unseen data under the aleatoric uncertainty of training annotations.
Related Work. U-Net based architectures such as nnUNet [6, 8] represent standard baselines in automatic knee cartilage segmentation. Going beyond architectural adaptability, the need for precise segmentation of certain localized and sparse structures led to Attention-based transformer models such as TransUNet [3] which encode strong global context by treating the image features as sequences. Towards supporting application-specific requirements such as thickness measurements, PCAM [8], introduces a morphologically constrained module to ensure continuity in the cartilage segmentation.
DDM-based segmentation models [14,15,16] can generate multiple samples which are variants of label maps. This is because the input to DDMs is a noised image, and by changing the additive noise, a slightly different sample is yielded at the output. By supervising the diffusion model to generate outputs close to a specified single annotation, we aim to study the characteristics of the generated multiple outputs with regard to two capabilities: First, handling noisy MRI scans, Second, handling data acquisition variabilities.
For supervising the diffusion process towards segmentation, we have adopted the Diff-UNet [16], and build upon it to address the sparse and intricate characteristics of knee cartilage, which exhibits less inter-tissue variability. Our contributions are:
-
a method of leveraging the stochastic capabilities of Diff-UNet to yield multiple variants of segmentation maps for knee cartilage segmentation, offering a mechanism to study predictive uncertainty in unseen test data.
-
integration of sparsity adaptive losses to supervise the diffusion process, which has shown quantitative improvement in the segmentation of cartilages in the presence of noise, and for scans acquired from a different setup.
2 Methods
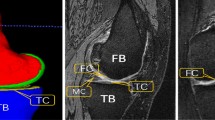
Diffusion UNet. We adopt a new diffusion-based segmentation model Diff-UNet, due to its superior tri-fold capabilities: First, Diff-UNet enables volumetric prediction of the segmentation maps, which is essential to capture the complete structure of the cartilages and enforce consistency across multiple 2D slices, which are inherently sparse in appearance. Second, Diff-UNet enables multi-label prediction of the segmentation maps, which is vital in labeling the different cartilages which share similar tissue appearances. Diff-UNet enables volumetric multi-label prediction of segmentation maps (\(x_{0}\)) of dimension \(N \times W \times H\) by converting it to multi-channel labels (\(X_{0}\)) through one-hot encoding. The iterative noising process generates \(X_{t}\) and \(\hat{X_{0}}\) at each time step, followed by learning to denoise \(X_{t}\) to \(X_{t-1}\), integrating MRI volume \(M \in \mathbb {R}^{1 \times N \times W \times H}\) using bi-phased integration: concatenating M with \(X_{t}\) and employing an additional encoder for multi-scale feature maps. The architectural flow of Diff-UNet is represented in Fig. 1.
Third, in Diff-UNet the losses for supervision are enforced on \(\hat{X_{0}}\) predicted at each time step. This is unlike other diffusion models for segmentation which usually do not enforce constraints directly on \(\hat{X_{0}}\), making the Diff-UNet capable of precise structural mapping.
Loss Integration (Diff-UNet\({_L}\)). The enforcement of losses on the predicted \(\hat{X_{0}}\) enables the incorporation of additional losses, which is necessary to better adapt to the sparse knee cartilage structures. We formulate Diff-UNet\({_L}\) by the addition of boundary enforcement loss (\(L_{BD}\)) [7], focal loss (\(L_{Focal}\)), and Hausdorff distance-based loss (\(L_{HD}\)) [9], along with the existing Diff-UNet losses: MSE loss, Dice loss, and BCE loss (\(L_{Diff-UNet}\)). Surface losses \(L_{BD}\), \(L_{HD}\) are added as both the structures of interest femoral cartilage and tibial cartilage share an adjacent boundary which is difficult for the model to differentiate the dilating boundary. In order to mitigate the challenge posed by the class imbalance problem, we incorporate the \(L_{Focal}\) loss, which is designed to tackle the inherent size variation between the tibial, femoral cartilage structures and the non-cartilage regions within the MRI scan. In Fig. 3 the effect of loss integration is indicated by the differences in the segmentation output.
The top row (a) displays variations in samples from Diff-UNet\({_L}\) and TransUNet. STAPLE was applied to five samples from each model, and a similarity metric (sensitivity) was calculated between the samples and the STAPLE output. The plots show that TransUNet samples exhibit minimal variation, while Diff-UNet\({_L}\) samples have a wider spread with some outliers. The second row provides a clearer visualization of the variations, with TransUNet showing concentrated variation and Diff-UNet\({_L}\) exhibiting meaningful spread. The third row illustrates two consecutive slices with a noticeable abrupt change in GT labels in the right femoral region, where Diff-UNet\({_L}\) displays more variability in that region.
The top block represents the output of our model compared to the Diff-UNet, TansUNet, and our model, TransUNet in the cross-dataset setup. S-O implies a model trained on the SKM dataset and inferred on the OZ dataset. The second block represents the output of our model compared with TransUNet in two noisy setups. The blue boxes depict better performance of our model, The pink boxes depict better performance of TransUNet. (Color figure online)
Multiple Generations and Uncertainty Estimation. The stochastic nature of DDMs enables the generation of multiple segmentation outputs (\(\hat{X^{i}_0}\)) while the deterministic class of models like TransUNet enables stochastic generations can be obtained if the Dropout technique is used. While Dropout based uncertainty stems from the change of configuration of the models, the DDM-based uncertainty highlights the model’s uncertainty about the underlying true data distribution. Based on these differences, we aim to investigate the following questions, First, “What are the differences between the samples which are generated from inherent stochastic models and the Dropout simulated ones?”, Second, “Are the variations within the samples meaningful and resemble the variations which can naturally occur during manual annotator based segmentations?” To address these questions, we have proposed the following experimental formulation. We utilized a group of segmentation samples, denoted as \(\hat{X^{i}_0}\), which were processed through the STAPLE [13] algorithm. This allowed us to generate a consensus-based segmentation mask called \(E_{1}\). This was utilized to measure the similarity between each sample \(\hat{X^{i}_0}\) and \(E_{1}\), referred to as \(STAPLE_{sm}\). A higher degree of similarity between \(\hat{X^{i}_0}\) and \(E_{1}\) indicates reduced variability among the samples produced by the model. These \(STAPLE_{sm}\) was calculated for both diffusion-based model and deterministic segmentation models. We have estimated the Uncertainity from the ensemble of the segmentation samples.
Noise Resilience and Generalisation. The segmentation of sparse cartilage structures in knee MRI becomes more challenging when the acquisition is noisy. To assess the noise resilience capability of the Diff-UNet\({_L}\), we have simulated noisy knee MRI scans by introducing Gaussian noise \( \mathcal {N}(\mu , \sigma ^2)\) and Rician noise \(\text {R}(\mu , \sigma ^2)\). The adaptability of the models in different acquisition setups is very essential for deploying models in practical use cases. To evaluate the generalizability of Diff-UNet\({_L}\), we trained it on one dataset and tested its performance on other datasets (cross-dataset setup). This cross-dataset setup poses higher variation within the set due to the OZ dataset being DESS and the SKM dataset being qDESS.
Thickness Estimation. One of the crucial aspects of assessing OA is estimating the thickness of cartilage. In order to better quantify and visualize the segmentation results in terms of clinically relevant metrics, we have adopted a simple yet efficient thickness estimation from [12]. This method creates a refined 3D model, split the mesh into inner and outer components, and computes thickness using the nearest neighbor method. The thickness maps are visualized through 2D projection.
3 Experimental Setup
Datasets. We have made use of two publicly available datasets knee MRI datasets OAI ZIB [1] (OZ) and SKM-Tea dataset [4] (SKM). OZ includes 507 3D DESS MR data with a sagittal acquisition plane with a voxel spacing of \(0.3645 \times 0.3645 \times 0.7\) mm. SKM has 155 3D knee MRI volumes acquired using a 5-min 3D quantitative double-echo in steady-state (qDESS) sequence. The voxel spacing is \(0.3125 \times 0.3125 \times 0.8\) mm. In order to ensure consistency in OZ data we have adopted the following standardization protocol. We center crop the Region Of Interest (ROI) of the volume with a dimension of \(256 \times 256 \times 120\), perform Non-local means filtering, and Normalise intensity levels across volumes. For the SKM dataset such intra-volume variability doesn’t exists within a volume, so we have applied only an ROI cropping protocol similar to the OZ dataset. We have considered training and test split as given within the datasets.
Metrics. Dice Similarity Coefficient (DSC), Average symmetric surface Distance (ASSD) are adopted for quantitative analysis between predicted and ground truth. The \(STAPLE_{sm}\) is evaluated by calculating the sensitivity between the samples (\(\hat{X^{i}_0}\)) and STAPLE output (\(E_{1}\)). In order to visually highlight the variances of the samples we have considered Mean Subtracted \(STAPLE_{sm}\) while plotting as in Fig. 2.
Implementational Details. We have implemented our methods in the PyTorch framework. We assigned higher weightage to sparsity and boundary constraints in the loss function (\(L_{BD}\), \(L_{Focal}\), \(L_{HD}\)) where \(\lambda _{1}=2\), as compared to \(L_{Diff-Unet}\) where \(\lambda _{2}=1\). For the model, we have adopted similar parameters as used in the Diff-UNet implementation [16]. We have compared with our performance with nnUNet [6] TransUNet [3], nnUNet+PCAM [8], Diff-UNet [16], Diff-UNet\(_{L}\). For the noisy and generalization case we have compared between TransUNet and Diff-UNet\(_{L}\). The STAPLE-based uncertainty estimation utilized 5 samples per volume. For TransUNet, the Dropout probability is 0.3. We have introduced noise within the volumes by adding Gaussian and Rician noise with \( \mathcal {N}(\mu =0, \sigma ^2=0.01)\) and \(\text {R}(\mu =0, \sigma ^2=0.01)\) parameters respectively. For the cross-dataset setup, we have trained the models on OZ dataset and inferred on SKM dataset and vice-versa.
4 Results
The Fig. 3 qualitatively shows the effect of the additional losses integrated with the Diff-UNet. The addition of losses has ensured better consistency within the femoral and tibial cartilage for Diff-UNet\({_L}\), as highlighted in the first row of Fig. 3 with blue boxes. From Table 1 we can infer that the results of our model are comparable to the baselines. The mean error thickness values, comparing GT with respect to Diff-UNet\({_L}\) and TransUNet femoral cartilage is 0.073 mm & 0.061 mm and tibial cartilage is 0.073 mm & 0.058 mm.
Multiple Segmentation and Uncertainty. From the box plots of Mean subtracted \(STAPLE_{sm}\) for TransUNet and Diff-UNet\({_L}\) in Fig. 2(a), it is clearly quantifiable that the variance of Diff-UNet\({_L}\) is much higher TransUNet. The median of the box plots of the Diff-UNet\({_L}\) is higher than that of TransUNet for all the samples. The qualitative visualization of the variations is in Fig. 2(b). In Fig. 2(c), our model effectively detects the uncertain regions in the left femoral regions, which were unmarked by annotators in the first slice but marked in the following slice. This consecutive slice comparison highlights the presence of uncertainty in that specific region. These uncertain regions are well demarcated by our model but missed by TransUNet.
Resilience to Noise. From Table 2, it is clearly evident Diff-UNet\({_L}\) has better performance than TransUNet in both the noise addition setup. Although in both the cases of Femoral and Tibial cartilage, Diff-UNet\({_L}\) has better quantification of results, in the latter case the relative increment is much higher when compared to the former. The appearance of the tibial cartilage in MRI scans is more sparse in nature when compared to the Femoral ones, so they have been more affected by the addition of noise. The qualitative visualisation of the results are in the lower block of the Fig. 3.
Generalisation in Cross-Dataset Setup. From Table 3 it is indicative that Diff-UNet\({_L}\) compared to TransUNet, performs better when the model was trained on OZ dataset & was tested on the SKM dataset and vice versa. Despite the cross-dataset setup, the model has shown incremental performance. The qualitative visualization of the results are in the lower block of the Fig. 3. From the Fig. 4 is observable that the overall structure of the cartilages predicted by the Diff-UNet\({_L}\) is relatively smooth.
5 Discussions
The integration of losses has shown better performance mostly in predicting the cartilages since they are sparse structures in MRI and need additional enforcement. The better quantification of variances and qualification of uncertainty maps from our model are due to DDM’s capability of providing meaningful variations when there is allowable stochasticity. This is further attributed to the model’s capacity to generalize beyond specific annotations and adapt to the intrinsic structures present in the scans, despite being trained on a single annotation. Diff-UNet\({_L}\) outperforms in noisy setups due to DDMs’ unique denoising-based sampling process, enabling better adaptation to noisy conditions during mapping from Gaussian to target distributions. The better generalization of the model is due to the fact that DDMs can better capture the distributional properties of the target without being biased to a certain set of data shown to the model during training.
6 Conclusion
Our proposed DDM-based multiple segmentation generator has shown to have a higher variability within the regions of generations which are natural causes of uncertainty while manual annotation. We have quantitatively and qualitatively verified that diffusion-based models better highlight uncertainty than Droput-based techniques. We have shown that after the addition of Gaussian and Rician noise, our model has better DSC % as compared to TransUNet. Also, in the cross-dataset setup, our method has better performance.
References
Ambellan, F., Tack, A., Ehlke, M., Zachow, S.: Automated segmentation of knee bone and cartilage combining statistical shape knowledge and convolutional neural networks: data from the osteoarthritis initiative. Med. Image Anal. 52, 109–118 (2019)
Braun, H.J., Gold, G.E.: Diagnosis of osteoarthritis: imaging. Bone 51(2), 278–288 (2012)
Chen, J., et al.: TransUNet: transformers make strong encoders for medical image segmentation. arXiv preprint arXiv:2102.04306 (2021)
Desai, A.D., et al.: SKM-TEA: a dataset for accelerated MRI reconstruction with dense image labels for quantitative clinical evaluation. arXiv preprint arXiv:2203.06823 (2022)
Ho, J., Jain, A., Abbeel, P.: Denoising diffusion probabilistic models. In: Advances in neural information processing systems, vol. 33, pp. 6840–6851 (2020)
Isensee, F., Jaeger, P.F., Kohl, S.A., Petersen, J., Maier-Hein, K.H.: nnU-Net: a self-configuring method for deep learning-based biomedical image segmentation. Nat. Methods 18(2), 203–211 (2021)
Kervadec, H., Bouchtiba, J., Desrosiers, C., Granger, E., Dolz, J., Ayed, I.B.: Boundary loss for highly unbalanced segmentation. In: International Conference on Medical Imaging with Deep Learning, pp. 285–296. PMLR (2019)
Liang, D., Liu, J., Wang, K., Luo, G., Wang, W., Li, S.: Position-prior clustering-based self-attention module for knee cartilage segmentation. In: Medical Image Computing and Computer Assisted Intervention-MICCAI 2022: 25th International Conference, Singapore, 18–22 September 2022, Proceedings, Part V, pp. 193–202. Springer, Cham (2022). https://doi.org/10.1007/978-3-031-16443-9_19
Ma, J., et al.: How distance transform maps boost segmentation CNNs: an empirical study. In: Arbel, T., Ayed, I.B., de Bruijne, M., Descoteaux, M., Lombaert, H., Pal, C. (eds.) Medical Imaging with Deep Learning. Proceedings of Machine Learning Research, vol. 121, pp. 479–492. PMLR, 06–08 July 2020. http://proceedings.mlr.press/v121/ma20b.html
Peng, W., Adeli, E., Zhao, Q., Pohl, K.M.: Generating realistic 3D brain MRIs using a conditional diffusion probabilistic model. arXiv preprint arXiv:2212.08034 (2022)
Pinaya, W.H., et al.: Brain imaging generation with latent diffusion models. In: Mukhopadhyay, A., Oksuz, I., Engelhardt, S., Zhu, D., Yuan, Y. (eds.) MICCAI Workshop on Deep Generative Models, vol. 13609, pp. 117–126. Springer, Cham (2022). https://doi.org/10.1007/978-3-031-18576-2_12
Sahu, P., et al.: Reproducible workflow for visualization and analysis of osteoarthritis abnormality progression. In: Proceedings of the International Workshop on Quantitative Musculoskeletal Imaging (QMSKI) (2022)
Warfield, S.K., Zou, K.H., Wells, W.M.: Simultaneous truth and performance level estimation (staple): an algorithm for the validation of image segmentation. IEEE Trans. Med. Imaging 23(7), 903–921 (2004)
Wolleb, J., Sandkühler, R., Bieder, F., Valmaggia, P., Cattin, P.C.: Diffusion models for implicit image segmentation ensembles. In: International Conference on Medical Imaging with Deep Learning, pp. 1336–1348. PMLR (2022)
Wu, J., Fang, H., Zhang, Y., Yang, Y., Xu, Y.: MedSegDiff: medical image segmentation with diffusion probabilistic model. arXiv preprint arXiv:2211.00611 (2022)
Xing, Z., Wan, L., Fu, H., Yang, G., Zhu, L.: Diff-UNet: a diffusion embedded network for volumetric segmentation. arXiv preprint arXiv:2303.10326 (2023)
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2024 The Author(s), under exclusive license to Springer Nature Switzerland AG
About this paper
Cite this paper
Mudiyam, V., Das, A., Ram, K., Sivaprakasam, M. (2024). Diffusion Model Based Knee Cartilage Segmentation in MRI. In: Mukhopadhyay, A., Oksuz, I., Engelhardt, S., Zhu, D., Yuan, Y. (eds) Deep Generative Models. MICCAI 2023. Lecture Notes in Computer Science, vol 14533. Springer, Cham. https://doi.org/10.1007/978-3-031-53767-7_20
Download citation
DOI: https://doi.org/10.1007/978-3-031-53767-7_20
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-031-53766-0
Online ISBN: 978-3-031-53767-7
eBook Packages: Computer ScienceComputer Science (R0)