Abstract
Bacteria and fungi are ubiquitous in the near-surface atmosphere where they may impact on the surrounding environment and human health, especially in coastal megacities. However, the diversity, composition, and seasonal variations of these airborne microbes remain limited. This study investigated the airborne microbes of the near-surface atmosphere in coastal megacity Qingdao over one year. It was found that the sample in summer displayed the highest bacterial and fungal diversity, while sample in winter exhibited the lowest bacterial and fungal diversity. Proteobacteria was the dominating bacteria, and Dothideomycetes was the most dominating fungi in the near-surface atmosphere, which took up 53–76 and 49–78% relative abundance, respectively. However, the bacterial diversity and community composition had significant seasonal variations. These data suggest that a complex set of environmental factors, including landscaping ratio, solar radiation temperature, and marine microorganisms, can affect the composition of airborne microbes in the near-surface atmosphere in coastal megacity. The analysis of the pathogenic microorganisms or opportunistic pathogenic microorganisms existed in the near-surface atmosphere revealed that the relative abundance of pathogenic microorganisms in autumn was the highest. The main pathogenic microorganisms or opportunistic pathogenic microorganisms were Acinetobacter baumannii (accounting for up to 9.93% relative abundance), Staphylococcus epidermidis (accounting for up to 11.26% relative abundance), Mycobacterium smegmatis (accounting for up to 3.68% relative abundance), Xanthomonas oryzae pv. oryzae (accounting for up to 5.36% relative abundance), which may be related to certain human or plant diseases in specific environments or at certain seasons. Therefore, the investigation of airborne microbial communities of near-surface atmosphere in coastal megacities is very important to both the understanding of airborne microbes and public health.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
1 Introduction
The atmosphere is considered to be an extreme environment for microorganisms because of its chemical and physical characteristics, such as high solar radiation, low moisture and nutrients, as well as its large dispersing capability. However, it has long been known that microorganisms are present in the atmosphere, primarily associated with aerosols. These so-called bioaerosols contain bacteria as well as fungi, viruses, algae, protists, and plant materials such as pollen. Accumulating evidence indicates that bacteria and fungi often represent a major proportion of total aerosol microorganisms in the atmosphere (Jaenicke 2005) with thousands to millions of cells per cubic meter of air (Lighthart 2000). A subset of these airborne bacteria and fungi may be plant, animal, and human pathogens or serve as triggers for allergies and allergenic asthma in humans (Bowers et al. 2013). There is also increasing evidence that microbial cells may have important influences on atmospheric conditions (Sesartic et al. 2012). However, most studies related to atmospheric aerosols nowadays focused on their physical and chemical characteristics. There were only a few studies focused on their microbiology characteristics such as the temporal and spatial variability in airborne bacteria and fungi diversity caused by location or seasonal conditions (Fahlgren et al. 2010). To better understand the influences of bacteria and fungi in the aerosol on the surrounding environment, it is important to study the diversity and community composition of bacteria and fungi.
With recent development in molecular biology technologies, researchers can gain unprecedented insight into the diversity and composition of airborne bacteria and fungi. Research from Franzetti indicated that there were large seasonal variations in the airborne bacteria communities in Milan, Italy (Franzetti et al. 2011). In addition, Bowers investigated the spatial variability in airborne bacterial communities across land-use types and it was found that aerosols collected at urban and small towns did harbor distinct airborne bacterial communities (Bowers et al. 2011). Interestingly, Camilla investigated the diversity of airborne bacteria in Kalmar, Sweden (Camilla et al. 2010), and it was found that the dominance of bacteria belonging to the genera Sphingomonas sp. and Pseudomonas sp., while Bowers found that the most dominant bacteria in the inland cities of Denver and Greeley (USA) was Actinobacteria (Bowers et al. 2013). By contrast with the information on the airborne bacteria, little is known on the diversity and composition of airborne fungi, although Bowers found that fungi made up approximately 21% relative abundance of bioaerosols by high-throughput sequencing. The researches mentioned above achieved some useful information; however, our understanding of microbial dynamics in the atmosphere is still limited. There were no researches on seasonal variations of airborne microbial communities in coastal megacity and how such variability impacts on human life. Worldwide, more than half of the total population lives within a range of 100 km from the coast. Therefore, the investigation on airborne microbial communities of near-surface atmosphere in coastal megacities is very important to both the understanding of airborne microbes and public health.
To the best of our knowledge, the study described here represents the most comprehensive temporal analysis of microbial communities in near-surface atmosphere in a coastal megacity to date. The coastal megacity Qingdao, one of the important cities in the east coast of China, is chosen as the target city. Similar to other typical coastal megacities in the world, Qingdao is one of the economic centers. It has nine million permanent residents and, each year, it attracts thirty million visitors from all over the world. The study of microbial communities in near-surface atmosphere in Qingdao can provide valuable references for both the study on airborne microbial communities in other coastal megacities and the prevention of infectious diseases.
2 Methods
2.1 Sampling sites
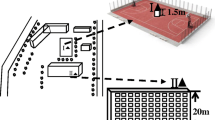
A total of four air samples were collected for four seasons (one air sample for each season) from the intersection of Shandong road and Fushun road located in Shibei District of Qingdao in Shan Dong (N 36°06′7.00″, E120°23′35.80″), China, which is about 4.5 km from the downtown. Shandong road is the most congested street in the city, and Fushun road is also a main road with 70 vehicles passing through every minute. The sampled intersection in this study was surrounding by Qingdao technological university, Disease control center, and a small business center. The environmental factors are detected hourly during sampling. TASI-620 digital temperature and humidity meter was used to detect the temperature and humidity, and TASI-641 anemometer was used to record the wind speed. The average values and fluctuating ranges are listed and shown in Table 1.
2.2 Sampling methods
In a typical sampling, high-volume (100 L min−1) dichotomous aerosol samplers (KC-6120 Andersen sampler) were used to isolate bioaerosols from the atmosphere from 6 am to 6 pm every day for 2 days and combined the first day air sample and the second day air sample into one sample as a representative sample for each season. The sampler was mounted on a platform which is 1.5 m above the ground level and 5 m away from the dairy breakfast stalls and trash can to minimize the collection of heavy dust particles in the food-manufacturing processes to maximize the collection of fine particles distributed in the air. No human activity was near the sampling area while samples were collected. Filter membranes were replaced every day placed in sterile bags, sealed with tape, and refrigerated (−20 °C) for later analysis. All microbial analyses were conducted on 45-mm fiberglass aerosol collection filters, and the airborne particles were collected using douching and ultrasonic wave to 5 mL sterile water to wash incoming air, and aerosols were collected into a sterile 50-mL tube every 3 min with a 2-min transfer-refill cycle for a total of 30 min. The aerosol wash water was centrifuged at 12000 rpm for 10 min, and the sediment was preserved for extraction of environmental DNA.
2.3 DNA extraction and purification
DNA was extracted by using the Fast DNA Spin kit for Soil (OMEGA, China) according to the manufacturer’s instructions. The crude DNA was purified by using Wizard Plus SV Minipreps DNA Purification System (Promega Corporation, USA) and quantified by ethidium bromide UV detection on an agarose gel.
2.4 Amplification of 16S rRNA gene
16S rDNA genes were amplified with the bacterial-specific 16S rDNA primers 1500R (5′-AGAAAGGAGGTGATCCAGCC-3′) and 27F (5′-AGAGTTTGATCCTGGCTC AG-3′) (Wang et al. 2010). 18S rDNA genes were amplified with the fungal-specific 16S rDNA primers EF4f(5′- 5′-GGA AGG GGT GTA TTT ATT Ag–3′)and EF3r(5′-TCC TCT AAA TGA CCA GTTTG-3′)(Smit et al. 1999). Gene was amplified through an initial denaturation at 94 °C for 5 min, followed by 32 cycles of 94 °C for 45 s, 55 °C for 30 s, and 72 °C for 1 min, and completed with an extension period of 10 min at 72 °C. The products with expected size about 1500 bp were pooled and purified with the EZNA TM Gel Extraction Kit (OMEGA, China) according to the manufacturer’s instructions. The purified 16S rDNA gene and 18S rDNA fragments from each sample were ligated into pGM-T vector (MBI Fermentas, USA). The ligation was transformed into E. coli DH5α competent cells and cultured on Luria–Bertani (LB) agar plates containing ampicillin (50 μg/ml), X-gal (20 mg/ml), and IPTG (100 mg/ml). A total of 160 white recombinant colonies were randomly selected from each of the two libraries. The insert fragment was amplified by using the vector-specific M13/PUC primers, M13/PUC F: 5′-GTAAAACGACGGCCAGT-3′, M13/PUC R: 5′- CAGGAAACAGCTATGAC -3′.
2.5 Amplified ribosomal DNA restriction analysis (ARDRA) and sequencing
In order to characterize the diversity of 16S rDNA gene and 18S rDNA within the clone libraries, amplified ribosomal DNA restriction analysis (ARDRA) was performed. The unpurified PCR products were digested with the restriction endonucleases HhaI and RsaI (Promega Corporation, USA). The digested fragments were separated by gel electrophoresis in 3.0% agarose gel with ethidium bromide staining and observed with UV illumination. Fragment patterns were sorted into operational taxonomic units (OTUs) according to the numbers and sizes of fragments, and representative clones from different OTUs were selected for nucleotide sequencing.
2.6 Sequence analyses
Sequences were analyzed for orientation and detection of non-16S rDNA gene sequences using Orientation Checker (Ashelford et al. 2006). The presence of chimeras was assessed by MALLARD (Ashelford et al. 2006). A sequence identified at the 99.9% threshold was discarded as a chimera. The remaining sequences were aligned using CLUSTALW (Thompson et al. 1994). On the basis of the alignment, a distance matrix was constructed using the DNADIST program from PHYLIP ver. 3.66 (http://evolution.genetics.washington.edu/phylip.html) with the default parameters. The resulting matrices were run in Mothur (Schloss et al. 2009) to generate diversity indexes and clustering analyses. The operational taxonomic units (OTUs) were defined with ≥97% identity for clustering analyses.
2.7 Calculation of traditional diversity indices
In order to estimate the genotypic diversity based on ARDRA, the Shannon–Weaver index (H′), Simpson index (D) (Pavao-Zuckerman and Coleman 2007; Chowdhury et al. 2009), Margalef index (M), Evenness index (E), and coverage index (C) were calculated. The formulae for the indices are shown in Table 2.
3 Results
3.1 General characteristics of the bacterial and fungal clone libraries
To assess bacterial diversity, 645 clones were selected from four bacterial libraries which were derived from near-surface atmosphere in urban street at four seasons. Characteristics of clone libraries of bacteria obtain from the near-surface atmosphere in urban street at four seasons are listed in Table 3. Similarity of those sequences ranged from 80 to 100%. Nonredundant analyses identified 286 unique sequences which were assigned into 101 OTUs. Interestingly, 50.5% of these sequences had less than 97% similarity with known sequences, which indicated that they may be potential novel species. The coverage of clone libraries ranged from 76.5 to 96.5%, suggesting that the selected sequences can reasonably represent bacterial communities of individual samples. In order to compare the genotypic diversity, the community richness (Margalef index), community diversity (Shannon–Weaver index, Simpson index), and equitability (Evenness index) were calculated (Table 3). The greater Margalef index, Shannon–Weaver index, and Simpson index means the higher richness and diversity, and the higher Evenness index express the higher species evenness. Table 3 exhibits that the samples which were sampled in spring and summer were observed with significantly higher bacterial diversity than those in autumn and winter, and the samples in summer displayed the highest diversity, while relatively low bacterial diversity was found in winter.
To assess the fungal diversity, 348 clones were selected from four fungal libraries. Characteristics of all the four clone libraries of fungi obtained from the near-surface atmosphere in urban street at four seasons are listed in Table 4. Similarity of these sequences ranged from 87 to 100%. Nonredundant analyses identified 103 unique sequences, which were assigned into 23OTUs. Among these sequences, 56.5% of them had less than 97% similarity with known sequences. The coverage of clone libraries ranged from 86 to 93%, suggesting that the selected sequences can reasonably represent fungal communities of individual samples. In addition, the samples which were sampled in summer also displayed the highest diversity while the samples in winter showed low fungal diversity. The Shannon–Weaver index, Simpson index, and Margalef index from bacteria clone libraries were all higher than those from the fungi clone libraries, which indicated that bacterial diversity from the near-surface atmosphere in urban street was higher than that of fungal clone libraries.
3.2 Phylogeny of bacterial community from the near-surface atmosphere
The analyses of phylogenetic compositions revealed that eight phyla existed in the four clone libraries, and the clone libraries were mainly dominated by 5–7 phyla (Table 5). The clone library of the samples in autumn consisted of only five phyla (Actinomycete, Firmicutes, Cyanophyta, Deinococcus–Thermus, and Proteobacteria). The clone library of the samples in spring was proved to be the most diverse, containing seven major phyla (Actinomycete, Chloroflexi, Firmicutes, Cyanophyta, Bacteroidetes, Deinococcus–Thermus, and Proteobacteria). The most dominant phylum among all libraries was Proteobacteria. In particular, clone libraries of the samples in autumn were shown to be highly dominated by Proteobacteria (76.24%). Actinomycete, Firmicutes, Cyanophyta, and Proteobacteria were detected in all seasons while Planctomycetes emerged only in autumn. These eight phyla existed in the near-surface atmosphere in urban street consisted of 60 genera, and the relative abundance of which is shown in Table 5.
The similarity of bacterial diversity and community composition (at genus level) of the air samples collected in different seasons was analyzed by paired-samples t test. The principle of this computational approach is described as follows. First, the paired-samples statistics for bacterial community composition of the near-surface atmosphere in different seasons were conducted (Table 6). Bacterial clone libraries were constructed using the DNA sample, and 645 clones were selected from four bacterial libraries which were assigned into 60 genera; therefore, N is 60 in Table 6. The relative abundances of certain species in two different seasons are subtracted, and the absolute value of the result is selected. The absolute mean differences of relative abundances of all the species in two different seasons are calculated. The mean differences equal zero if the diversity and community composition of the bacteria in the two different seasons are identical; however, the mean differences are bigger if the differences of diversity and community composition of the bacteria in the different seasons are greater. Then, the paired-samples t test is used to detect the differences (Table 7). In this study, the null hypothesis is that the mean difference is zero, and this means that we suppose the diversity and community composition of the bacteria in two different seasons are identical. If the Sig is no more than 0.05 in the model, we can reject the null hypothesis at 0.05 level. From Tables 9 and 10, it can be seen that the mean differences of relative abundances of all the species in arbitrary two seasons were >2, and Sig (2-tailed) <0.05, and those data indicated that there are significant differences for the bacterial diversity and community composition of the air samples collected in different seasons (Tables 9, 8, 10).
57 OTUs belong to phylum Proteobacteria, and Proteobacteria was dominant in all clone libraries. In the Proteobacteria, there are α-Proteobacteria, β-Proteobacteria, γ-Proteobacteria, and ε-Proteobacteria classes, which include 21, 15, 18, and 3 genus, respectively (Table 3). However, the community composition of Proteobacteria (PB1-57) varied greatly. In spring, β-Proteobacteria was the most dominant with 29.92% relative abundance, and in β-Proteobacteria PB34 and PB24 which were identified as Massilia timonae and Janthinobacterium lividum, respectively, were the main genus. In summer, α-Proteobacteria was the most dominant with Sphingomonas echinoides (OUT PB 18) and Massilia alkalitolerans (OUT PB 25) making up 31.91% of relative abundance. In autumn, γ-Proteobacteria was the most dominant and Acinetobacter haemolyticus (OUT PB38) made up 25.17% of relative abundance. However, there was no dominant class in Proteobacteria in winter, and α-Proteobacteria, β-Proteobacteria, γ-Proteobacteria, and ε-Proteobacteria almost had the same bacteria numbers. In spite of the significant seasonal variations in Proteobacteria, Massilia aurea and Massilia timonae (PB26-29 and PB33-35) were found in the four seasons.
Table 9 shows that all of the other 43 OTUs distributed in seven phyla, and they were phylum Actinomycete, phylum Firmicutes, phylum Cyanophyta, phylum Deinococcus–Thermus, phylum Chloroflexi, phylum Bacteroidetes, and phylum Planctomycetes. The community composition of these nine phyla presented great seasonal variations.
Fourteen OTUS distributed in 14 genera, which all belong to phylum Actinomycete (Table 4). Genera in Actinomycete all had low relative abundance and mainly existed in the clone library of the samples in spring. For example, although nine genera distribute in the clone library of the samples in spring, except Corynebacterium glucuronolyticum (OTU AC5, 4.19%) and Okibacterium fritillariae (OTU AC13, 3.59%), each of them shared no more than 2% relative abundance. In contrast to the clone library of the samples in spring, genera in Actinomycete were less both in the types and in relative abundance than the other three clone libraries. A total of eight genera were detected in those three libraries. Furthermore, except Mycobacterium smegmatis (OTU AC12) and Brachybacterium faecium (OTU AC4) making up 3.68 and 2.65% in clone library of the samples in summer and autumn clone libraries, respectively, the other six genera took no more than 1.5% relative abundance in all the three clone libraries.
Sixteen OTUs belong to phylum Firmicutes, and Firmicutes was stably present in all clone libraries of the samples in four seasons except summer (Table 4). There were two classes, Bacillaceae and Clostridia, in phylum Firmicutes in the four clone libraries. There were ten genera in Bacillaceae, and the community varied greatly in different seasons. In spring, Carnobacterium (OTU BA 1) was found to be one of the predominant taxa in the clone library of the samples in spring (7.78%). In autumn, Staphylococcus epidermidis corresponding to OTU BA8 was one of the predominant taxa with 11.26% relative abundance. In winter, Planomicrobium koreense and Carnobacterium, the representative sequences of which were OTU BA 7 and OTU BA 3, were detected with high relative abundance (17.06 and 7.04%, respectively). There were six genera in Clostridia, and the genera in Clostridia were mainly present in clone library of the samples in spring and they were not detected in autumn. In addition, except Gemmiger formicilis (OTU CL6), the other genera have relatively low abundances in all clone libraries.
Four OTUs belong to phylum Cyanophyta, and Cyanophyta was mainly distributed in clone library of the samples in spring. OTU CY1, the representative sequence of which was identical to Acaryochloris marina, took up 8.98% relative abundance in the spring clone libraries and was one of the predominant taxa. However, all of the other three OTUs in phyla Cyanophyta have relatively low abundances in all clone libraries. Interestingly, the representative sequences of all of the four OTUs showed no more than 92% identity to the closest known species (Acaryochloris marina, Cyanothece sp. ATCC 51142, Microcoleus sp. PCC 7113, Oscillatoria nigro-viridis, respectively). It indicated that species in Cyanophyta in the near-surface atmosphere in urban street were distinctly different from those existed in other environment. Three OTUs belong to phylum Deinococcus–Thermus, and Deinococcus–Thermus was predominant taxa in clone library of the samples in summer. Although only one OTU DE1, which was identical as Deinococcus grandis, was detected in summer clone library, it took up 31.9% relative abundance. This has important significance in that the genera in Deinococcus–Thermus have high resistance to solar radiation.
Two OTUs belong to phylum Chloroflexi, and Chloroflexi was mainly distributed in clone library of the samples in winter. Anaerolinea thermophila (OTU AN1) and Caldilinea aerophila (OTU AN2) accounted for 8.45 and 3.52% relative abundances, respectively, in clone library of the samples in winter. However, genera in Chloroflexi were rarely detected in other seasons. Interestingly, the representative sequences of OTU AN1 and OTU AN2 all showed no more than 85% identity to the closest known species. Considering the length of the sequence in amplified ribosomal DNA restriction analysis (ARDRA) method used in this study is about 1500 bp, which can lead the classification of microorganisms more definite. It indicated that genera in Chloroflexi in the near-surface atmosphere may be new members.
Three OTUs belong to phylum Bacteroidetes, and only one OUT belongs to phylum Planctomycetes. Those four OTUs all had low relative abundance in all seasonal clone libraries.
3.3 Phylogeny of fungal community from the near-surface atmosphere
The analyses of phylogenetic compositions of fungal communities revealed that phylum Ascomycota, Basidiomycota, Phycomycetes, and Chytridiomycetes existed in the four clone libraries. The clone library of the samples in summer showed the most diversification (Table 10). Three phyla were detected in summer clone library, and the clone libraries of the samples in spring, autumn, and winter consisted of two phyla. Four phyla existed in the near-surface atmosphere in urban street consisted of 23 genera. The similarity of fungal diversity and community composition (at genus level) of the air samples collected in different seasons was also analyzed by paired-samples t test. Fungal clone libraries were constructed using the same DNA sample as clone libraries, and 348 clones were selected from four fungal libraries which were assigned into 23 genera; therefore, N is 23 in Table 11 which is different from that in Table 6. From Tables 11 and 12, it can be seen that the mean differences of relative abundances of all the species in arbitrary two seasons were >5, and Sig (2-tailed) <0.04, and those data indicated that there are significant differences for the fungal diversity and community composition of the air samples collected in different seasons.
Pyrenophora were stable and dominant in all clone libraries of the samples in four seasons. The community compositions of fungi revealed an obvious seasonal variation. There were eight genera in the clone library of the samples in spring. Pyrenophora and Grifola were the most dominant genera. OTU F3 in Pyrenophora and OTU F19 in Grifola which were identified as Pyrenophora phaeocomes and Grifola sordulenta were the main species (which took up 47.5 and 30% relative abundance, respectively). There were 11 genera in the clone library of the samples in summer. Besides Pyrenophora (45.54% relative abundance), Catinella was also a dominant genus and it had 24.75% relative abundance. There were seven genera in the clone library of the samples in autumn. Pyrenophora was the most dominant genus (76.54%), and Xylaria was another dominant genus (12.35% relative abundance). There were nine genera in the clone library of the samples in winter. Similar to the compositions of fungal community in the clone library of the samples in autumn, in winter, Pyrenophora was the dominant genus (50.88% relative abundance) and Xylaria was another dominant genus (14.04% relative abundance).
Interestingly, except Catinella olivacea, Pyrenophora phaeocomes, Xylaria hypoxylon, and Grifola sordulenta, all the other species in the clone libraries of the samples in all seasons detected in this study took up no more than 10% relative abundance.
4 Discussion
4.1 Bacterial diversity and community in the megacity atmosphere
Bacterial diversity and community are influenced by many environmental and meteorological factors. The temporal and spatial variability of bacterial communities has also been investigated in different studies. Even though some researchers insisted that airborne microbial communities can be highly variable over short time scales (Fierer et al. 2008), most authors recognized that differences in community structures were larger between seasons than within a season (Bertolini et al. 2013; Bowers et al. 2011; Franzetti et al. 2011). Therefore, this study focuses on the seasonal variations in bacterial and fungal diversity and community composition of the near-surface atmosphere.
This study reveals that 36, 26, 20, and 23 genera were detected in bacteria library of the samples in spring, summer, autumn, and winter, respectively. Among these genera, there were six genera distributed in all clone libraries and they were Paracoccus, Rubellimicrobium, Sphingomonas, Massilia, Acinetobacter, and Pseudomonas. In addition, Proteobacteria accounted for 26 genera and Planctomycetes accounted for only one genus. Regardless of atmospheric conditions, Proteobacteria was the dominant bacteria of all of the air samples collected in four seasons, which agrees well with previous reports on microbial diversity in atmosphere (Bowers et al. 2012). The bacterial diversity and community composition of the air samples collected in different seasons had significant seasonal variations (Tables 5, 6; Fig. 1).
In spring, β-Proteobacteria was the dominant bacteria which make up 30% relative abundance, while all other classes make up less than 15% relative abundance (Fig. 1) individually. The most dominant genus in β-Proteobacteria was Massilia (Table 2), which includes five species, M. alkalitolerans, M. aurea, M. brevitalea, M. plicata and M. timonae. This is different with other study on bacterial community composition of the near-surface atmosphere (Yamaguchi et al. 2014; Bowers et al. 2009; Franzetti et al. 2011). And the possible reasons for the highest relative abundance of Massilia in airborne bacteria were also speculated. The source tracking analysis of airborne bacteria indicates that leaf and soil sources dominated in the spring and early summer months (Triky-Dotan et al. 2010). The surface of leaf and rhizosphere was detected to have a large proportion of the Massilia, especially in rhizosphere (up to 85% relative abundance) at early stages of succession (Ferrari et al. 2005), which may be due to the in vitro attributes of Massilia related to plant growth promotion, including IAA production (Kuffner et al. 2010), siderophore production (Hrynkiewicz et al. 2010), and in vitro antagonism toward Phytophthora infestans (Weinert et al. 2010). As it is well known, Qingdao is a beautiful city with a relatively high landscaping ratio (40%) and surrounded by large areas of agricultural land which may result in the higher relative abundance of Massilia in airborne bacteria.
In summer, Deinococcus–Thermus (32%) was the most dominant class followed by α-Proteobacteria (29%) and β-Proteobacteria(20%). Deinococcus possesses highly proficient DNA double-strand break (DSB) repair mechanisms, such as homologous recombination (HR) (Daly and Minton 1995), extended synthesis-dependent strand annealing (ESDSA) (Zahradka et al. 2006; Slade et al. 2009) and single-strand annealing (SSA) (De La Tour et al. 2011). All of these repair mechanisms enable Deinococcus to accurately reassemble its genome from hundreds of DNA fragments produced by irradiation. In addition, Deinococcus evolved to have very efficient nonenzymatic and enzymatic antioxidant defenses which specifically protect proteins against oxidative damages (Rémi et al. 2015; Ghosal et al. 2005; Daly 2009). Therefore, Deinococcus is one of the most radioresistant organisms on earth which can survive under extreme doses of radiation (1000-fold higher than most vertebrates) (Munteanu et al. 2015). Besides extreme irradiation environment, Deinococcus spp. can survive other extreme environments such as desiccation, hyperthermy, and hypothermy environments. Therefore, although the atmosphere is an extreme environment with intense solar radiation which may lead to lethal effects for most microbes, Deinococcus–Thermus can survive and become the most dominant class in the atmosphere of near surface in summer. In addition to Deinococcus, Sphingomonas echinoides in α-Proteobacteria also has a high relative abundance (22.09%), which agrees well with the results of Franzetti’s report (Franzetti et al. 2011). The species Sphingomonas echinoides has been detected in soil, water, and various extreme environments. They can grow and multiply easily in soil and rhizosphere. In addition, Sphingomonas echinoides can adapt to the changing environment very well and it can also survive in hyperthermy, hypothermy, and ultraviolet ray irradiation, especially in nutrient-deficient conditions. In conclusion, the main bacteria existed in the near-surface atmosphere exhibit extreme environment-resistant abilities. Therefore, hyperthermy and solar radiation may play important roles on the microbial diversity and community composition of the near-surface atmosphere in summer.
In autumn, γ-Proteobacteria (40%) was the most dominant class followed by α-Proteobacteria (31% relative abundance). There were three genera in class γ-Proteobacteria in the clone library of samples in autumn, and Acinetobacter was in the ascendant (37.08% relative abundance). This was similar to the bacterial community composition of the near-surface atmosphere in coastal city Osaka, Japan (Yamaguchi et al. 2014). There were four species affiliated with Acinetobacter. They were A. baumannii, A. haemolyticus, A. lwoffii, and A. oleivorans. Acinetobacter spp. is an aerobic mesophilic bacterium, and it was often detected in water and soil. Acinetobacter spp. can multiply rapidly in atmosphere with more particulates, especially with high relative humidity. Eight species in α-Proteobacteria were detected in autumn, of which Sphingomonas kaistensis (8.61%) and Brevundimonas subvibrioides (9.26%) were dominant. Beside species in γ-Proteobacteria and α-Proteobacteria, Staphylococcus epidermidis in Bacillaceae was also dominant in autumn. Staphylococcus epidermidis has a wide distribution in environment, especially in skin, mucous membrane, hair, and feather of human beings and animals. From what was discussed above, it can be seen that bacteria that multiply rapidly in comfortable conditions or distribute widely, such as Acinetobacter spp. and Staphylococcus epidermidis, were dominant in autumn.
Different from the other seasons, in winter, Firmicutes has higher relative abundance, while Chloroflexi, α-Proteobacteria, β-Proteobacteria, γ-Proteobacteria, and ε-Proteobacteria shared approximately equal relative abundances (10–15%), which was similar to findings reported previously (Bowers et al. 2009; Franzetti et al. 2011). On the species level, Planomicrobium koreense was dominant in winter. Planomicrobium was able to survive hypothermy and high salinity, and they were detected in coastal soil sediment, glacier, and seafood frequently (Yoon et al. 2001; Dai et al. 2005; Zhang et al. 2009; Luo et al. 2014). Arcobacter nitrofigilis was also dominant in winter. Arcobacter nitrofigilis is of phylogenetic interest because of its lifestyle as a symbiotic organism in a marine environment in contrast to many other Arcobacter species which are associated with warm-blooded animals. In conclusion, the dominant bacteria existed in the near-surface atmosphere in winter were normally existed in marine environment. Therefore, marine bacteria may play important roles on the microbial diversity and community composition of the near-surface atmosphere in winter. Qingdao is a coastal city with low environmental temperature in winter, which may lead to the high relative abundance of Planomicrobium koreense and Arcobacter nitrofigilis in atmosphere.
This study also analyzed the pathogenic bacteria or opportunistic pathogenic bacteria existed in the near-surface atmosphere. Thirteen species, the 16S rDNA sequences of which show identity to the pathogenic bacteria or opportunistic pathogenic bacteria, were detected in the atmosphere (Table 13). From Table 11, it can be seen that the relative abundance of pathogenic bacteria or opportunistic pathogenic bacteria in autumn (24.49%) was higher than that in other three seasons (7–9%). The reason may be that the environmental conditions were comfortable in autumn and pathogenic bacteria or opportunistic pathogenic bacteria with wide distributions and strong reproduction abilities multiplied rapidly in the ambient environment which may be possible sources of those pathogens and it leads to the high relative abundances of pathogen in the near-surface atmosphere.
Although 13 pathogenic bacteria or opportunistic pathogenic bacteria were detected in the near-surface atmosphere, the main pathogenic bacteria or opportunistic pathogenic bacteria were Acinetobacter baumannii, Staphylococcus epidermidis, Xanthomonas oryzae pv. Oryzae and Mycobacterium smegmatis. Acinetobacter baumannii is abundant in normal flora of skin and mucous membranes in humans and responsible for opportunistic infections such as upper respiratory tract infections, pneumonia, meningitis, septicemia, and urinary tract infections, particularly in intensive care units (ICUs) (Shahcheraghi et al. 2011). Furthermore, A. baumannii is resistant to a wide range of antibiotics. Staphylococcus epidermidis is a commensal bacterium of human skin, nasal, and oral mucosa. However, in recent decades, it has emerged as a common cause of hospital-acquired infections, being responsible for 40–90% of infections associated with indwelling devices in seriously ill or immunocompromised patients (Rogers et al. 2009; von Eiff et al. 2002). The gram-negative plant pathogenic Xanthomonas oryzae pv. oryzae (Xoo) is the causal agent of bacterial blight disease on rice (NiÑo Liu et al. 2006). Bacterial blight is the most serious bacterial disease of rice in tropical Asian countries where high-yielding rice cultivars are often highly susceptible. It has the potential to reduce rice yields by as much as 50% (Mew 1987). Mycobacterium smegmatis is generally considered a nonpathogenic microorganism; however, in some very rare cases, it may cause disease (Reyrat and Kahn 2001). Beside those four pathogenic bacteria or opportunistic pathogenic bacteria, all of the other nine species in Table 5 can also cause opportunistic infections in human populations.
Currently, it is difficult to establish linkages between pathogenic bacteria or opportunistic pathogenic bacteria and outbreaks of infectious diseases. This long-term temporal analysis of bacterial diversity, especially pathogenic bacteria or opportunistic pathogenic bacteria, in the near-surface atmosphere of coastal megacity provides valuable references for the prevention of infection diseases and is important to human and environmental health.
4.2 Fungal diversity and community in the megacity atmosphere
Fungi took up low relative abundances of bioaerosol (Bowers et al. 2013), but they likely impact those people suffering from allergies and asthma. Fungi in the near-surface atmosphere have not attracted much attention. Therefore, this study investigated fungal diversities. It was found that there were 20 fungal genera in the near-surface atmosphere of coastal megacity. These 20 genera belonged to nine classes. Although Dothideomycetes was the most dominant class in the near-surface atmosphere in all seasons (48.75–78.21%), fungal diversity and community composition had some temporal variations (Tables 11, 12; Fig. 2). In spring, there were five classes in the near-surface atmosphere. Dothideomycetes and Homobasidiomycetes were dominant classes which took up 49 and 35% relative abundances, respectively. In summer, there were seven classes and fungal diversity was the highest. However, the species distribution was poor. For example, Dothideomycetes took up 78% relative abundance while all other classes took up no more than 6% relative abundances. In autumn, Dothideomycetes took up 78% relative abundance, followed by Sordariomycetes (16%) while all of the other classes took up no more than 3% relative abundances. In winter, there were four classes and fungal diversity was the lowest. Beside Dothideomycetes (60% relative abundance), Sordariomycetes was another dominant class in fungi.
On the genus level, Pyrenophora belonging to Dothideomycetes was the most dominated genus, and it took up more than 47% relative abundance in each season. Grifola belonging to Homobasidiomycetes (30% relative abundance) and Catinella belonging to Dothideomycetes (24.5% relative abundance) were also dominated in spring and summer, respectively. This was different with the findings from other researchers. For example, Chang investigated the airborne fungi in buildings and outdoor environments with culture-dependent methods, and it was found that Cladosporium, Penicillium, Aspergillus, and nonsporulating fungi were the most common fungi outdoors in each season in the USA from 1996 to 1998(Chang et al. 2001), while Shelton studied the airborne microorganisms in open-air swine houses with culture-dependent methods and found that Cladosporium was the predominant genus(Shelton et al. 2002). Many researches reveal that Pyrenophora and Xylaria were more often detected on the leaves which indicated that fungi on plant leaves may be one of the main sources of airborne microbes in the near-surface atmosphere in coastal megacity Qingdao. Among those fungi in the near-surface atmosphere, some species are liable to cause different plant diseases. Xylaria hypoxylon can cause plant diseases and lead to wither, and several species in Pyrenophora are important pathogens of cereals (Ellwood et al. 2012). However, this study did not detect pathogenic fungi or opportunistic pathogenic fungi to human being in the near-surface atmosphere.
From all the research above, we can find that fungal diversity is lower compared with bacterial diversity. The possible reasons were also speculated. On the one hand, the researches on the airborne fungi are precious little, which leads to a limited database. In this study, nine representative sequences of OUT (total 23 OUT) showed no more than 85% identity to the closest known species, which indicated that they may be potential novel species. In classifications, placing them in the closest known species may reduce fungal diversity to some degrees. On the other hand, the finite number of clones in clone library may be another reason lead to the low fungal diversity.
5 Conclusion
This study investigated the microbial communities in near-surface atmosphere in a coastal megacity. It was found that Proteobacteria was the dominant bacteria and Dothideomycetes was the most dominant fungi in the near-surface atmosphere in four seasons in the coastal megacity Qingdao. However, bacterial diversity and community composition of the air samples collected in different seasons had significant seasonal variations. The main influence factors of microbial communities in near-surface atmosphere in the coastal megacity may be landscaping ratio, solar radiation, and temperature. In addition, marine microorganisms had some impacts on microbial communities in atmosphere. The analysis of the pathogenic microorganisms or opportunistic pathogenic microorganisms existed in the near-surface atmosphere revealed that the relative abundance of pathogenic microorganisms or opportunistic pathogenic microorganisms in autumn was the highest. The main pathogenic microorganisms or opportunistic pathogenic microorganisms were Acinetobacter baumannii, Staphylococcus epidermidis, ycobacterium smegmatis, spores of Ascomycotina and Pyrenophora, which may be related to human diseases in specific environments or at certain seasons. Therefore, city disease control departments may need to pay more attentions to these pathogenic microorganisms or opportunistic pathogenic microorganisms. It is worth noting that this study may miss a proportion of the bacterial or fungal diversity in the atmosphere, because of the collection efficiencies of microbial samplers used in this study and the finiteness of clone library, which means that there are more pathogenic or opportunistic pathogenic bacteria and fungi in near-surface atmosphere in a coastal megacity than expected.
References
Ashelford, K. E., Chuzhanova, N. A., Fry, J. C., Jones, A. J., & Weightman, A. J. (2006). New screening software shows that most recent large 16S rRNA gene clone libraries contain chimeras. Applied and Environmental Microbiology, 72, 5734–5741.
Bertolini, V., Gandolfi, I., & Ambrosini, R. (2013). Temporal variability and effect of environmental variables on airborne bacterial communities in an urban area of Northern Italy. Applied Microbiology and Biotechnology, 97(14), 6561–6570.
Bowers, R. M., Clements, N., Emerson, J. B., Wiedinmyer, C., Hannigan, M. P., & Fierer, N. (2013). Seasonal variability in bacterial and fungal diversity of the near -surface atmosphere. Environmental Science and Technology, 47(21), 12097–12106.
Bowers, R. M., Lauber, C. L., Wiedinmyer, C., Hamady, M., Hallar, A. G., Fall, R., et al. (2009). Characterization of airborne microbial communities at a high-elevation site and their potential to act as atmospheric ice nuclei. Applied and Environmental Microbiology, 75, 5121–5130.
Bowers, R. M., McCubbin, I. B., Hallar, A. G., & Fierer, N. (2012). Seasonal variability in airborne bacterial communities at a high-elevation site. Atmospheric Environment, 50, 41–49.
Bowers, R. M., Sullivan, A. P., Costello, E. K., Collett, J. L., Jr., Knight, R., & Fierer, N. (2011). Sources of bacteria in outdoor air across cities in the midwestern United States. Applied and Environmental Microbiology, 77, 6350–6356.
Camilla, F., Åke, H., Douglas, N., & Ulla, L. Z. (2010). Annual variations in the diversity, viability, and origin of airborne bacteria. Applied and Environmental Microbiology, 76(9), 3015–3025.
Chang, C. W., Chung, H., & Huang, C. F. (2001). Exposure of workers to airborne microorganisms in open-air swine houses [J]. Applied and Environmental Microbiology, 67(1), 155–161.
Chowdhury, S. K. R., Sangle, G. V., & Xie, X. (2009). Effects of extensively oxidized low-density lipoprotein on mitochondrial function and reactive oxygen species in porcine aortic endothelial cells. American Journal of Physiology-Endocrinology and Metabolism, 298, 89–98.
Dai, X., Wang, Y. N., & Wang, B. J. (2005). Planomicrobium chinense sp. nov., isolated from coastal sediment, and transfer of Planococcus psychrophilus and Planococcus alkanoclasticus to Planomicrobium as Planomicrobium psychrophilum comb. nov. and Planomicrobium alkanoclasticum comb. nov. International Journal of Systematic and Evolutionary Microbiology, 55(2), 699–702.
Daly, M. J. (2009). A new perspective on radiation resistance based on Deinococcus radiodurans. Nature Reviews Microbiology, 7, 237–245.
Daly, M. J., & Minton, K. W. (1995). Interchromosomal recombination in the extremely radioresistant bacterium Deinococcus radiodurans. Journal of Bacteriology, 177, 5495–5505.
De La Tour, C. B., Boisnard, S., Norais, C., Toueille, M., & Bentchikou, E. (2011). The deinococcal DdrB protein is involved in an early step of DNA double strand break repair and in plasmid transformation through its single-strand annealing activity. DNA Repair (Amst), 10, 1223–1231.
Ellwood, S. R., Syme, R. A., & Moffat, C. S. (2012). Evolution of three Pyrenophora cereal pathogens: Recent divergence, speciation and evolution of non-coding DNA. Fungal Genetics and Biology, 49, 825–829.
Fahlgren, C., Hagström, Å., Nilsson, D., & Zweife, U. L. (2010). Annual variations in the diversity, viability, and origin of airborne bacteria. Applied and Environmental Microbiology, 9(76), 3015–3025.
Ferrari, B. C., Binnerup, S. J., & Gillings, M. (2005). Microcolony cultivation on a soil substrate membrane system selects for previously uncultured soil bacteria. Applied and Environmental Microbiology, 71, 8714–8720.
Fierer, N., Liu, Z., Rodriguez-Hernandez, M., Knight, R., Henn, M., & Hernandez, M. T. (2008). Short-term temporal variability in airborne bacterial and fungal populations. Applied and Environmental Microbiology, 74(1), 200–207.
Franzetti, A., Gandolfi, I., Gaspari, E., Ambrosini, R., & Bestetti, G. (2011). Seasonal variability of bacteria in fine and coarse urban air particulate matter. Applied Microbiology and Biotechnology, 90, 745–753.
Ghosal, D., Omelchenko, M. V., Gaidamakova, E. K., Matrosova, V. Y., & Vasilenko, A. (2005). How radiation kills cells: Survival of Deinococcus radiodurans and Shewanella oneidensis under oxidative stress. FEMS Microbiology Reviews, 29, 361–375.
Hrynkiewicz, K., Baum, C., & Leinweber, P. (2010). Density, metabolic activity, and identity of cultivable rhizosphere bacteria on Salix viminalis in disturbed arable and landfill soils. Journal of Plant Nutrition and Soil Science, 173, 747–756.
Jaenicke, R. (2005). Abundance of cellular material and proteins in the atmosphere. Science, 308(5718), 73.
Kuffner, M., De Maria, S., Puschenreiter, M., Fallmann, K., & Wieshammer, G. (2010). Culturable bacteria from Zn- and Cd- accumulating Salix Caprea with differential effects on plant growth and heave metal availability. Journal of Applied Microbiology, 108, 1471–1484.
Lighthart, B. (2000). Mini-review of the concentration variations found in the alfresco atmospheric bacterial populations. Aerobiologia, 16(1), 7–16.
Luo, X., Zhang, J., & Li, D. (2014). Planomicrobium soli sp. nov., isolated from soil. International Journal of Systematic and Evolutionary Microbiology, 64(8), 2700–2705.
Mew, T. W. (1987). Current status and future prospects of research on bacterial blight of rice. Annual review of Phytopathology, 25, 359–382.
Munteanu, A., Uivarosi, V., & Andries, A. (2015). Recent progress in understanding the molecular mechanisms of radioresistance in Deinococcus bacteria. Extremophiles, 19(4), 707–719.
NiÑo Liu, D. O., Ronald, P. C., & Bogdanove, A. J. (2006). Xanthomonas oryzae pathovars: Model pathogens of a model crop. Molecular Plant Pathology, 7(5), 303–324.
Pavao-Zuckerman, M. A., & Coleman, D. C. (2007). Urbanization alters the functional composition, but not taxonomic diversity, of the soil nematode community. Applied Soil Ecology, 35, 329–339.
Rémi, D., Takefumi, O., & Geneviève, C. (2015). Identification of new genes contributing to the extreme radioresistance of deinococcus radiodurans using a Tn5-based transposon mutant library. PLoS ONE. doi:10.1371/journal.pone.0124358.
Reyrat, J. M., & Kahn, D. (2001). Mycobacterium smegmatis: An absurd model for tuberculosis? Trends in Microbiology, 9(10), 472–473.
Rogers, K. L., Fey, P. D., & Rupp, M. E. (2009). Coagulase-negative staphylococcal infections. Infectious Disease Clinics of North America, 23, 73–98.
Schloss, P. D., Westcott, S. L., & Ryabin, T. (2009). Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Applied and Environmental Microbiology, 75, 7537–7541.
Sesartic, A., Lohmann, U., & Storelvmo, T. (2012). Bacteria in the ECHAM5-HAM global climate model. Atmospheric Chemistry and Physics, 12(18), 8645–8661.
Shahcheraghi, F., Abbasalipour, M., Feizabadi, M., Ebrahimipour, G., & Akbari, N. (2011). Isolation and genetic characterization of metallo-β-lactamase and carbapenamase producing strains of Acinetobacter baumannii from patients at Tehran hospitals. Iranian Journal of Microbiology, 3, 68–74.
Shelton, B. G., Kirkland, K. H., & Flanders, W. D. (2002). Profiles of airborne fungi in building and outdoor environments in the Unites Stated [J]. Applied and Environmental Microbiology, 68(4), 1743–1753.
Slade, D., Lindner, A. B., Paul, G., & Radman, M. (2009). Recombination and replication in DNA repair of heavily irradiated Deinococcus radiodurans. Cell, 136, 1044–1055.
Smit, E., Leeflang, P., & Glandorf, B. (1999). Analysis of fungal diversity in the wheat rhizosphere by sequencing of cloned PCR-amplified genes encoding 18S rRNA and temperature gradient gel electrophoresis[J]. Applied and Environmental Microbiology, 65(6), 2614–2621.
Thompson, J. D., Higgins, D. G., & Gibson, T. J. (1994). CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Research, 22, 4673–4680.
Triky-Dotan, S., Ofek, M., Austerweil, M., Steiner, B., & Minz, D. (2010). Microbial aspects of accelerated degradation of metam sodium in soil. Phytopathology, 100, 367–375.
von Eiff, C., Peters, G., & Heilmann, C. (2002). Pathogenesis of infections due to coagulase-negative staphylococci. The Lancet Infectious Diseases, 2, 677–685.
Wang, G. H., Dong, J. D., & Li, X. (2010). The bacterial diversity in surface sediment from South China sea. Acta Oceanology, 29(40), 98–105.
Weinert, N., Meincke, R., Gottwald, C., Radi, V., & Dong, X. (2010). Effects of genetically modified potatoes with increased zeaxanthin content on the abundance and diversity of rhizobacteria with in vitro antagonistic activity do not exceed natural variability among cultivars. Plant and Soil, 326, 437–452.
Yamaguchi, N., Park, J., & Kodama, M. (2014). Changes in the airborne bacterial community in outdoor environments following Asian dust events. Microbes and Environments, 29(1), 82–88.
Yoon, J. H., Kang, S. S., & Lee, K. C. (2001). Planomicrobium koreense gen. nov., sp. nov., a bacterium isolated from the Korean traditional fermented seafood jeotgal, and transfer of Planococcus okeanokoites (Nakagawa et al. 1996) and Planococcus mcmeekinii (Junge et al. 1998) to the genus Planomicrobium. International Journal of Systematic and Evolutionary Microbiology, 51(4), 1511–1520.
Zahradka, K., Slade, D., Bailone, A., Sommer, S., & Averbeck, D. (2006). Reassembly of shattered chromosomes in Deinococcus radiodurans. Nature, 443, 569–573.
Zhang, D. C., Liu, H. C., & Xin, Y. H. (2009). Planomicrobium glaciei sp. nov., a psychrotolerant bacterium isolated from a glacier. International Journal of Systematic and Evolutionary Microbiology, 59(6), 1387–1390.
Acknowledgements
This work was supported by the Natural Science Foundation of Shandong Province (2015ZRB01546),the National Natural Science Foundation of China (No. 31570541, 31170509), and science and technology plan projects for universities in Shandong province(J14LD05), basic research project of Qingdao (15-9-1-64-jch).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Xu, Al., Song, Zw., Lang, Xl. et al. Seasonal variability in bacterial and fungal diversity and community composition of the near-surface atmosphere in coastal megacity. Aerobiologia 33, 555–575 (2017). https://doi.org/10.1007/s10453-017-9489-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10453-017-9489-z