Abstract
To assess potential effects of genetically modified (GM) potatoes on the abundance and diversity of rhizobacteria with in vitro antagonistic activity in relation to natural variability among cultivars, two GM potato lines accumulating the carotenoid zeaxanthin in their tubers, the parental cultivar and four additional commercial cultivars were planted at two field sites in Germany. Rhizosphere samples were taken at three developmental stages of the plants. A total of 3,985 bacteria isolated from the rhizosphere were screened for their in vitro antagonistic activity towards Rhizoctonia solani, Verticillium dahliae and Phytophthora infestans using a dual-culture assay. Genotypic characterisation, 16S rRNA gene sequencing and antifungal metabolite analysis was performed to characterize the 595 antagonists obtained. The 16S rRNA gene-based identification of in vitro antagonists revealed strong site-dependent differences in their taxonomic composition. This study showed that the site was the overriding factor determining the proportion and diversity of antagonists from the rhizosphere of potato while the effect of the genetic modification on the proportion of antagonists obtained did not exceed natural variability among the five commercial cultivars tested.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The rhizosphere, which is defined as part of the soil influenced by the plant, has a central role as interface between the soil and the plant (Hiltner 1904; Brimecombe et al. 2001). In the rhizosphere the abundance and metabolic activity of some soil microorganisms is enhanced in response to root exudates and deposits. In turn rhizosphere microorganisms contribute to plant nutrition, health and growth and can induce systemic resistance (Van Loon et al. 1998; Whipps 2001). Cultivation-independent studies (16S rRNA gene fingerprints) have shown that the rhizosphere often harbors a reduced number of dominant bacterial populations whose relative abundance clearly differs from bulk soil (Costa et al. 2006; Marilley et al. 1998; Smalla et al. 2001). Differences in exudate level and composition, together with rhizosphere microsite location, root-mass and structure, but also the nutritional status of the plant are assumed to contribute to individual microbial communities of distinct plant species (Gyamfi et al. 2002; Marschner et al. 2001; Yang and Crowley 2000). Some studies reported on differences in the rhizosphere as well as endophyte bacterial community composition among cultivars of the same plant species (Adams and Kloepper 2002; Dunfield and Germida 2001; Siciliano and Germida 1999). But not only the plant species itself is of vital importance, also the plant developmental stage, soil properties, climatic conditions and agricultural management practice are assumed to have a major influence on the composition of rhizosphere microbial communities (Bossio et al. 1998; Crecchio et al. 2004; Kennedy et al. 2005; Rasche et al. 2006).
Antagonistic microorganisms that reside in the rhizosphere are able to defend the plants against pathogens. Several traits are assumed to play an important role for antagonistic activity: (i) antibiosis, implying the inhibition of a pathogen through the release of antibiotics, toxins or biosurfactants (Fravel 1988), (ii) competitive root colonization (Lugtenberg et al. 2001), and (iii) production of extra-cellular cell wall degrading enzymes like ß-1,3 glucanase, chitinase or cellulase (Chernin and Chet 2002).
Although the application of GM plants in agriculture and forestry is of increasing importance (James 2007) the overall impacts of GM crops on soil quality are, however, still poorly understood. Despite their importance for soil and plant health, the response of soil microbes to the application of GM crops has not been addressed adequately. Unintended side-effects due to altered characteristics of the modified plants cannot be a priori excluded and thus their impact has to be evaluated case-by-case. Genetic modifications of plants often result in phenotypic changes and may also result in alteration of the root exudation which in turn could cause changes in the structural and functional diversity of plant-associated microorganisms. Potential effects of GM crops on beneficial rhizobacteria might be of particular interest as their relative abundance and diversity could affect plant health and ecosystem sustainability. Only few studies attempted to assess potential impacts of GM plants on antagonistic species (Lottmann et al. 1999; Lottmann et al. 2000; Lottmann and Berg 2001). However, these studies compared only the two GM potato lines to a GM control and the parental cultivar.
In the present study two GM potato lines which accumulate the carotenoid zeaxanthin in their tubers (Römer et al. 2002) were investigated under field conditions. Although the two constructs used in this study do not harbor new genes and the modifications are based on antisense respectively co-suppression of an existing pathway (zeaxanthin epoxidase) and should only be active in the potato tuber, it could not be excluded that root exudation might be changed as a result of the genetic modification. Indeed the root length, carbohydrate pattern and pH in the rhizosphere of both GM lines were changed compared to the wild type. Interestingly, also differences between both GM lines were observed (Neumann et al. personal communication). These data clearly indicate that a modification of a single gene can influence the plant phenotype in many ways. The two GM potato lines with increased zeaxanthin level in their tubers were grown together with their parental cultivar and four additional commercial cultivars in a randomised field trial with six replicated plots per cultivar and GM line at two sites in Germany. Rhizosphere samples were taken at three developmental stages. In this study we aimed to assess potential effects of GM potatoes on the abundance and composition of bacteria with in vitro antagonistic activity towards three potato pathogens (Rhizoctonia solani, Verticillium dahliae, Phytophthora infestans) and relate these to natural variability among the potato cultivars investigated. Furthermore, this study provides insights into the phenotypic and genotypic diversity of bacterial isolates with in vitro antagonistic activity.
Materials and methods
Potato cultivars and GM lines
For genetic modifications Solanum tuberosum L. cultivar ‘Baltica’ was used. The construction of the two GM zeaxanthin-accumulating potato lines through co-suppression (SR47/00#18, further referred to as SR47) and antisense (SR48/00#17, further referred to as SR48) was described in detail by Römer et al. (2002). The promoter used in both GM lines were tuber specific and thus no increased zeaxanthin levels in the above ground parts of the plants were expected. This could be confirmed by studying the expression of the zeaxanthin epoxidase in different plant compartments (Wenzel et al. personal communication). The accumulation of zeaxanthin in the tuber reached up to 40 µg/g dry weight (dw) (SR47) and 17 µg/g dw (SR48) compared to 0.2 µg/g dw of the wildtype. In addition to the GM lines and the parental cultivar ‘Baltica’, the potato cultivars ‘Selma’, ‘Désirée’, ‘Ditta’ (all used for food production) and ‘Sibu’ (used for industrial purposes) were grown in the field.
Field design
The two sites used for this study are located in southern Germany. The Roggenstein soil was characterized by 26.1% sand, 44.0% silt, 28.1% clay, Corg: 1.1%, Nt: 0.1% and pH 6.6. The Oberviehhausen soil contained 54.6% sand, 31.3% silt, 14.1% clay, Corg: 1.9%, Nt: 0.2%, and had a pH of 6.5. The climatic conditions in 2005 (Roggenstein) and 2006 (Oberviehhausen) are available from the weather station located nearby both field sites. Plant protection measures were performed according to agricultural practice.
The experimental setup was designed as a randomized field trial: Each plot had a size of 9 × 3 m, resulting in four rows with a total of 40 potatoes. The cultivars and GM lines were grown in six replicated plots, of which four plots were sampled. The data presented in this study are based on the field trials in 2005 (Roggenstein) and 2006 (Oberviehhausen).
Isolation of root-associated bacteria
Sampling of the plants was carried out at three developmental stages, young plants (EC30), flowering plants (EC60) and senescent plants (EC90) according to Hack et al. (1993). The GM lines showed a delayed emergence but at EC60 no obvious differences between the GM lines and commercial cultivars were observed. Therefore, we decided to sample not completely randomized but to select plants in a comparable growth stage at EC30. From each of the plots the roots of five plants were combined as a composite sample and transported to the laboratory. To extract the root-associated bacteria, 10 g of the root material of each plot was transferred into sterile stomacher bags. Thirty ml Milli-Q water was added and samples were homogenized for 60 s in a Stomacher laboratory blender (Seward, West Sussex, UK) at high speed. This step was repeated three times. The resulting suspensions were serially diluted with 0.85% NaCl and plated onto R2A (Merck, Darmstadt, Germany) supplemented with cycloheximide (100 µg/ml) to avoid the growth of fungi. After 5 days of incubation at 28°C colony forming units (CFU) were determined and 100 visually different colonies were picked from each cultivar and GM line, resulting in a total of 700 isolates per sampling time and site. The isolates were purified and stored at −70°C.
Screening for bacteria with in vitro antagonistic potential
The bacterial isolates obtained were screened in a dual-culture in vitro assay for their antagonistic potential towards three major potato pathogens: Rhizoctonia solani Kühn AG3 (Basidiomycete with a chitin-glucan cell wall), Verticillium dahliae Kleb. ELV25 (Ascomycete with a chitin-glucan cell wall) and Phytophthora infestans [Mont.] De Bary strain 20/01 (Oomycete with a cellulose-glucan cell wall). Activity of the isolates against R. solani and V. dahliae was determined on Waksman agar (WA), containing 5 g of bactopeptone (Merck, Darmstadt, Germany), 10 g glucose (Merck), 3 g meat extract (Chemex, München, Germany), 5 g NaCl (Merck) and 20 g bacto-agar (Becton Dickinson, NJ, USA) per l distilled water. Agar disks with mycelia were directly cut out from WA plates with R. solani (grown for 7 days) and placed between the streaks of four isolates. V. dahliae was grown in liquid culture in Czapek Dox medium (Difco, Detroit, MI, USA). 150 µl of the suspension containing hyphal fragments were plated onto the agar and after surface drying the bacteria were streaked on the same plate. Inhibition zones were measured after 5 days of incubation at room temperature as described by Berg et al. (2002). The dual-culture assay against P. infestans was carried out on pea-agar (modified according to Hollomon 1965). For this purpose, 125 g frozen peas were cooked in 500 ml distilled water for 20 min. The broth obtained after filtration was adjusted with distilled water to 1 l after adding 15 g bacto-agar. Agar disks from pea-agar plates with P. infestans grown for 10–12 days were cut out and pre-grown on the pea-agar test plates for 1–2 days before streaking the bacterial isolates. Inhibition zones were determined after 7 days at room temperature. Every bacterial strain was tested twice independently.
Extraction of genomic DNA
Genomic DNA from the in vitro antagonistic strains was extracted using the Qiagen Genomic DNA Extraction Kit (Hilden, Germany) for cell lysis and the MoBio UltraCleanTM 15 DNA Purification Kit (Carlsbad, CA, USA) for DNA-preparation, as described by the manufacturers.
Genotypic characterization using ARDRA and BOX-PCR genomic fingerprinting
Amplified Ribosomal DNA Restriction Analysis (ARDRA) was performed by amplification of the 16S rRNA gene with primers U8-U27 (5’-AGAGTTTGATC(A/C)TGGCTCAG-3’) and 1492–1514 (5’-TACGG(C/T)TACCTTGTTACGACTT-3’) (Weissburg et al. 1991). Amplicons obtained were digested with the enzymes Hin6I and Bsh1236I (Fermentas) for 3 h at 37°C and 300 rpm in a thermomixer (Eppendorf, Thermomixer comfort). Fifteen µl of the resulting fragments were separated in a 4% agarose gel (NuSieve 3:1) in 0.5 × TBE buffer for 4 h, stained with ethidium bromide and visualized using a UV transilluminator. Antagonists displaying similar ARDRA-patterns (cut-off level 85%) were further analyzed using BOX-PCR genomic fingerprinting. BOX-PCR fingerprints were performed using the BOX_A1R primer as described by Rademaker and De Bruijn (1997). Eight µl of the PCR-product were separated in a 1.5% agarose gel in 0.5 × TBE buffer for 4 h, stained with ethidium bromide and photographed under UV.
Identification of bacterial in vitro antagonists
Antagonists with either individual ARDRA patterns or different BOX patterns (cut-off level 85%) were identified by partial sequencing (approx. 700 bp) of the 16S rRNA gene. The sequences obtained were aligned with reference 16S rRNA gene sequences using the nucleotide basic local alignment and search tool (BLASTN) from NCBI databases.
Enzymatic activity, production of siderophores and N-acyl-homoserine-lactone (AHL)-production
Protease activity (casein degradation) was determined from clearing zones in skim milk agar (400 ml sterilised skim milk mixed with 1/10 TSB and 16 g bacto-agar at 55°C) after 5 days of incubation at 28°C. β-glucanase and cellulase activity were tested using chromogenic AZCL and reamazolbrilliant blue R (AZO) substrates, respectively (Megazyme International, Ireland). Formation of blue and white haloes was determined after 5 days at 28°C. Chitinolytic activity was tested in chitin minimal medium as described by Berg et al. (2001). Clearance haloes indicating β-1,4-glucosamine polymer degradation were determined after 7 days of incubation at room temperature. Production of siderophores under Fe3+-limited conditions was analyzed using the plate assay developed by Schwyn and Neilands (1987). Orange haloes, indicating a reduction of iron(III) to iron(II) were measured after 3 days of incubation at room temperature. AHL-production was investigated in a cross-streak assay using the bioluminescent sensor plasmid pSB403 in E. coli to detect 3-oxo-C6-HSL molecules produced by the bacterial antagonists (Winson et al. 1998). The sensor strain Chromobacterium violaceum CV026 (McClean et al. 1997) changes its color to purple in the presence of short chain AHLs. Serratia plymuthica HROC48 (Liu et al. 2007) served as a positive control in both assays.
Statistical analysis
Significance tests for CFU values were performed using ANOVA and Tukey Test. Significance tests for numbers of in vitro antagonists (Chi square exact) were performed with Statistical Product and Service Solutions for Windows v. 15.0. (SPSS Inc., Chicago, IL, USA). Computer-assisted evaluation of both ARDRA and BOX-PCR genomic fingerprints was performed using the GelCompar II program version 4.5. (Applied maths, Kortrijk, Belgium). Cluster analysis was performed using the Dice correlation matrix and the unweighted pair group method using average linkages.
Nucleotide sequence accession numbers
The nucleotide sequences determined in this study were deposited in the GenBank database under accession numbers EU834335-EU834701.
Results
Colony forming units of bacterial isolates from the potato rhizosphere
Significant differences in the CFU counts (p ≤ 0.05) were revealed at EC60 in Roggenstein and at EC30 and EC60 in Oberviehhausen. These differences were detected for the GM lines as well as for some of the commercial cultivars (Table 1).
In vitro antagonistic activity of bacterial isolates towards R. solani AG3, V. dahliae ELV25 and P. infestans
A total of 1,946 isolates originating from the rhizosphere of potatoes grown at the site in Roggenstein and 2,039 isolates from the potato rhizosphere in Oberviehhausen were tested in dual culture assays for their in vitro antagonistic activity. The total numbers of antagonists obtained from the Roggenstein site revealed significant differences both between GM lines and commercial cultivars, as well as between different commercial cultivars for all three pathogens tested (Table 2). The majority of in vitro antagonists towards all three pathogens was obtained from the cultivar ‘Désirée’. For the Oberviehhausen site significant differences in the total numbers of antagonists were observed for R. solani and P. infestans, whereas the numbers of antagonists towards V. dahliae revealed no significant differences between the cultivars and GM lines. Significant differences could be observed both between GM lines and commercial cultivars and between different commercial cultivars (Table 2).
At both sites significantly more isolates showed in vitro antagonistic activity towards P. infestans than towards R. solani and V. dahliae. Moreover, the numbers of antagonists towards P. infestans were significantly higher in Oberviehhausen than in Roggenstein (data not shown).
Genotypic characterization of the antagonists using ARDRA and BOX-PCR
A total of 243 and 339 ARDRA-patterns were generated from rhizosphere isolates of EC60 and EC90 potato plants grown in Roggenstein and Oberviehhausen, respectively. EC30 isolates of both sites were excluded from further identification, because the roots of all cultivars and GM lines were very heterogeneous in size at that sampling time. Furthermore, only the in vitro antagonists displaying either activity towards the Oomycete P. infestans alone or additionally towards one or both of the tested fungi were further analyzed since they accounted for the vast majority of antagonists (243 of 253 in Roggenstein and 352 of 368 in Oberviehhausen, respectively, data not shown). In vitro antagonists displaying identical ARDRA-patterns (cut-off level 85%) were further characterized by BOX-PCR, which offers a higher resolution below the species. Isolates displaying unique ARDRA-patterns or one representative isolate of a group sharing the same BOX-pattern were identified by 16S rRNA gene sequencing.
Identification of bacterial in vitro antagonists from both sampling sites
Based on the characterization by ARDRA and BOX patterns, the 16S rRNA genes of 192 and 295 in vitro antagonistic isolates from Roggenstein and Oberviehhausen, respectively were selected for 16S rRNA gene sequencing (approx. 700 bp). Although both sampling sites revealed the highest proportion of in vitro antagonists towards P. infestans, the composition of antagonistic bacteria revealed strong differences between the sites (Table 3). Most of the 229 isolates identified from the Roggenstein site (66.4%) were affiliated to the class of Gammaproteobacteria (n = 152 isolates), the majority of them showing the highest similarity to the genus Pseudomonas (n = 83 isolates), belonging to 17 different species (see Table S2, supplementary material). Fifty antagonists from Roggenstein (21.8%) were assigned to the High G+C Gram-positives, 40 isolates of which belonged to the genus Streptomyces (11 different species). In contrast to the Roggenstein site, only 69 (21.6%) of the 319 antagonists from the Oberviehhausen site were affiliated to the Gammaproteobacteria, with 36 isolates belonging to the genus Pseudomonas (9 different species). In Oberviehhausen the majority of antagonists from the rhizosphere (46.7%) were assigned to the High G+C Gram-positives (n = 149 isolates) with most sequences displaying the highest similarity to the genus Streptomyces (135 isolates belonging to 43 different species). The second largest group of antagonists from the potato rhizosphere in Oberviehhausen (29.2%) was assigned to the phylum Firmicutes (n = 93 isolates) with 87 isolates belonging to the genus Bacillus (4 different species). In contrast, only 24 antagonists (10.5%) from Roggenstein were assigned to the phylum Firmicutes, with 12 isolates belonging to Bacillus, all of them assigned to the species Bacillus pumilus (see Table S2, supplementary material).
Cell wall degrading enzymatic activity, siderophore- and AHL-production
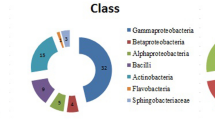
A large proportion of the antagonists displayed glucanase-, cellulase- and protease-activity, while isolates with chitinase-activity were less frequently detected. Strong site-dependent differences were found mainly for glucanase and siderophore production, reflecting the proportion of dominant bacterial genera (Pseudomonas, Streptomyces and Bacillus) in the collection of rhizobacteria with in vitro antagonistic activity from both sites. As revealed by principal component analysis, these three genera separated due to their enzymatic properties (Fig. 1). The proportion of antagonists producing short chain AHLs also exhibited remarkable differences between both sites, with the numbers in the rhizosphere of Roggenstein exceeding those of Oberviehhausen three to four times (data not shown). The majority of antagonists producing AHLs belonged to the genus Pseudomonas. AHL production was detected for antagonists identified as Ps. thivervalensis, Ps. corrugata, Ps. brassicacearum and Ps. stutzeri with both indicator strains while only E. coli pSB403 responded to the AHL molecules produced by antagonists assigned to Ps. chlororaphis, Ps. putida and Ps. jessenii. In Oberviehhausen, only antagonists identified as Ps. brassicacearum produced AHLs in vitro. Antagonists identified as Dickeya chrysanthemi were the only AHL producers which were obtained from both sites. While AHL producing antagonists identified as Pectobacterium carotovorum were found only in the Roggenstein collection, P. atrosepticum was retrieved from the rhizosphere of Oberviehhausen.
Principal components analysis (PCA) ordination biplot of in vitro antagonistic Pseudomonas (circle), Streptomyces (square) and Bacillus (triangle) isolates and their respective enzymatic activity and siderophore production properties (arrows). Antagonists were grouped in the ordination diagram according to Euclidian distances calculated on the basis of their enzymatic and siderophore traits. Arrows pointing in the same direction indicate a positive correlation between enzymatic properties and phylogenetic assignment of the antagonists. Blue symbols: antagonists isolated from the Roggenstein site, black symbols: antagonists isolated from the Oberviehhausen site.
Comparison of BOX-profiles of the same species from different sites
Only for a few species sufficiently high numbers of antagonists were obtained from both sites to allow a comparison of their phenotypic and genotypic diversity which was influenced by sites, cultivars and plots.
Almost the same numbers of antagonists from both sites were identified as Ps. fluorescens (Roggenstein: n = 20; Oberviehhausen: n = 18). The genetic diversity of all Ps. fluorescens antagonists was compared by BOX PCR fingerprints. The results revealed both a high intraspecific diversity for each site as well as strong site-dependent differences between the profiles (Fig. 2). Within one site profiles with more than 80% similarity were found not only for isolates from the same replicate plots, but also for different plots of the same cultivar, yet rarely for different cultivars.
Dendrogram based on BOX-PCR fingerprints of in vitro antagonistic isolates from the potato rhizospheres in Roggenstein (R) and Oberviehhausen (O), identified by 16S rRNA gene-sequencing as Pseudomonas fluorescens. The UPGMA algorithm was applied to the similarity matrix generated with the Dice coefficient. Strain code: site, sampling time (2 = EC60, 3 = EC90), strain number, cultivar/GM line (Bal = ‘Baltica’, SR47 = ‘Baltica’ co-suppresion, SR48 = ‘Baltica’ antisense, Sel = ‘Selma’, Des = ‘Désirée’, Dit = ‘Ditta’, Sib = ‘Sibu’), plot (1 to 4) (original gel see Fig. S1)
A considerable number of antagonists were identified as B. pumilus with 13 and 80 isolates from the potato rhizospheres of Roggenstein and Oberviehhausen, respectively. The comparison of the BOX-profiles revealed clear site-specific differences, with a very stable profile for the majority of isolates from the Oberviehhausen site (68/80), whereas the profiles of the remaining isolates were clearly distinct from these. Some representative isolates displaying the stable BOX-profile are shown in Fig. 3. B. pumilus isolates with identical BOX-patterns were obtained from all four analyzed plots of the cultivars ‘Selma’, ‘Désirée’, ‘Ditta’ and ‘Sibu’, as well as from one plot of the parental cultivar ‘Baltica’ and the GM line SR47, and from two plots of the GM line SR48. Compared to the Oberviehhausen site, isolates obtained from Roggenstein showed completely different profiles, except for one isolate which displayed the same BOX-profile as the majority of isolates from Oberviehhausen (Fig. 3).
Dendrogram based on BOX-PCR fingerprints of in vitro antagonistic isolates from the potato rhizospheres in Roggenstein (R) and Oberviehhausen (O), identified by 16S rRNA gene-sequencing as Bacillus pumilus. The UPGMA algorithm was applied to the similarity matrix generated with the Dice coefficient. Strain code as described in Fig. 2 (original gel see Fig. S2)
Twenty-six and 11 antagonists from the rhizosphere of potato plants grown in Roggenstein and Oberviehhausen, respectively, were assigned to Dickeya chrysanthemi. Eight antagonists obtained exclusively from Roggenstein were identified as Pectobacterium carotovorum. Two antagonists assigned to P. atrosepticum were found in the collection of antagonists from Oberviehhausen. Interestingly, all D. chrysanthemi strains from the Roggenstein site were isolated from all four plots planted with the cultivar ‘Désirée’ (EC60: n = 3; EC90: n = 23), while most D. chrysanthemi isolates from the Oberviehhausen site originated from the cultivar ‘Ditta’ (EC60 n = 2; EC90 n = 7) and only two from ‘Désirée’. All antagonists identified as P. carotovorum originated from one plot of the GM line SR48. For all 26 D. chrysanthemi strains of the cultivar ‘Désirée’ from Roggenstein the same BOX-patterns were obtained (Fig. 4). Seven of nine D. chrysanthemi strains obtained from the cultivar ‘Ditta’ from the Oberviehhausen site displayed the same profile as the strains in Roggenstein and two showed a distinct pattern. The two D. chrysanthemi isolates obtained from ‘Désirée’ in Oberviehhausen shared the same pattern with the strains from the same cultivar in Roggenstein. The eight strains identified as P. carotovorum formed two separate clusters with five and three strains, respectively. The two strains identified as P. atrosepticum formed a tight cluster which was very distant from the other Dickeya and Pectobacterium isolates (data not shown).
Dendrogram based on BOX-PCR fingerprints of in vitro antagonistic isolates from the potato rhizospheres in Roggenstein (R) and Oberviehhausen (O), identified by 16S rRNA gene-sequencing as D. chrysanthemi. The UPGMA algorithm was applied to the similarity matrix generated with the Dice coefficient. Strain code as described in Fig. 2 (original gel see Fig. S3)
Discussion
The rhizosphere was recently described as an important microenvironment for potentially antagonistic bacteria (Berg et al. 2002; Berg et al. 2006) because bacterial populations growing in response to the easily available root exudates are often r-strategists which frequently also produce cell wall degrading extracellular enzymes to improve their competitive behavior in soil (Raaijmakers et al. 2008). Although the present study was mainly aiming to assess potential effects of GM potato lines on the group of beneficial bacteria and to relate these effects to natural variability among cultivars, its experimental design also allowed insights into the diversity of antagonistic rhizobacteria in dependence on the site. However, only for a few species (Ps. fluorescens, B. pumilus and P. chrysanthemi) the number of isolates was large enough to compare the diversity in relation to these factors and to assess their field distribution.
The CFU-counts determined at three developmental plant stages were comparable to those reported by Lottmann et al. (1999) for the rhizosphere of genetically modified, T4-lysozyme producing potato plants, respective control lines and their parental cultivar. In the present study significant differences in the CFU-values were always transient and not stable throughout the whole season.
To assess whether the proportion and composition of plant beneficial bacteria in the rhizosphere of GM potatoes was changed compared to the parental cultivar and the four commercial cultivars, similar numbers of bacterial isolates were retrieved and tested for their in vitro antagonistic activity towards three major potato pathogens. The average proportion of in vitro antagonists towards R. solani and V. dahliae was comparable to results reported by other authors (Berg et al. 2002; Krechel et al. 2002; Lottmann et al. 1999). For the Roggenstein site the total numbers of antagonists obtained revealed significant differences both between GM lines and commercial cultivars, as well as between different commercial cultivars for all three pathogens tested. This was mainly due to the 26 D. chrysanthemi strains with strong antagonistic activity that were recovered from the cultivar ‘Désirée’ at EC90, thus leading to exceptionally higher numbers of antagonists for this cultivar. In Oberviehhausen, significant differences were observed both among commercial cultivars and GM lines and among different commercial cultivars towards R. solani and P. infestans. However, in all cases significant differences found were transient (see Table S1, supplementary material). This finding is consistent with results from other studies which indicated that changes due to the genetic modification of the plants were either transient (Donegan et al. 1995; Dunfield and Germida 2003; Griffiths et al. 2000) or negligible compared to natural factors like field site (Dunfield and Germida 2001) or season (Gyamfi et al. 2002; Heuer et al. 2002). Also altered plant characteristics, independent from the genetic modification were suggested as a possible reason for observed differences (Donegan et al. 1995; Siciliano et al. 1998). Siciliano and Germida (1999) found significant differences in the rhizosphere communities between cultivars of GM Brassica napus cv. Quest to cv. Excel and Brassica rapa cv. Parkland. However, their analysis focussed only on the flowering stage of the plants and the finding could not be generalized for other GM canola varieties (Dunfield and Germida 2001). In the present study the individual soil characteristics and cropping histories of both sites had the strongest influence on the composition of their inhabiting bacterial communities as reflected in the composition of in vitro antagonists. To date several studies showed that the soil type has a strong influence on the composition of microbial communities in the rhizosphere (Costa et al. 2006; Girvan et al. 2003; Latour et al. 1999; Marschner et al. 2001; Sessitsch et al. 2001). A remarkably high proportion of the rhizobacterial isolates from both sites displayed an in vitro antagonistic potential against the Oomycete P. infestans. However, the in vitro assay only evaluates the activity towards the mycelium of the Oomycete and not towards its zoospores which represent the infectious part (Lifshitz et al. 1985). The results of the dual-culture assays towards R. solani and V. dahliae also have to be interpreted with care as in vitro tests do not necessarily correlate with the in situ antagonistic activity (Sari et al. 2006). Although comparable numbers of in vitro antagonists were retrieved from the rhizosphere of potatoes grown at both sites, their taxonomic composition and the proportion of antagonists producing siderophores, glucanase or AHLs showed strong site-dependent differences. One main difference was the proportion of in vitro antagonists affiliated to the Gammaproteobacteria, especially the genus Pseudomonas which was much higher in Roggenstein than in Oberviehhausen (Table 3). Pseudomonas species are supposed to benefit directly from root exudates provided by the plant and are therefore more often found in the rhizosphere than in other microenvironments (Costa et al. 2006; Raaijmakers et al. 2008). Also in the studies by Berg et al. (2002, 2006) the largest proportion of Verticillium dahliae antagonists was affiliated to the genus Pseudomonas. The diversity of isolated Pseudomonas species from the Roggenstein site was quite remarkable and several species such as Ps. thivervalensis, Ps. putida and Ps. corrugata were only obtained from this site. Moreover, this genus accounted for the high number of isolates from the Roggenstein site producing siderophores. A huge diversity of siderophores formed by Pseudomonas species has been reported (e.g. Cornelis and Matthijs 2002; Meyer et al. 2002). In a habitat like soil that is poor in disposable iron, siderophores can be important in rhizosphere colonization (Raaijmakers et al. 1995) and in competing with plant pathogens (De Boer et al. 2003; Lemanceau et al. 1992; Leong 1986). The comparison of BOX-fingerprints generated for Ps. fluorescens isolates revealed both a high genotypic diversity within each site but also between both sites. As reviewed by Picard and Bosco (2008) such variability is important in rhizosphere competence and biocontrol ability. Only few isolates from different plots of the same cultivar or of different cultivars at one field site displayed identical BOX-patterns. The endemism revealed by the use of BOX-PCR was also reported by Cho and Tiedje (2000) who argued that geographic isolation is important in bacterial diversification. Although the same cultivars and GM lines were grown in Oberviehhausen, antagonistic isolates belonged most frequently to the High G+C Gram-positives and Firmicutes. The BOX-patterns of B. pumilus showed that a genetically highly similar population colonized the rhizosphere of the five different potato cultivars and of the two GM lines in Oberviehhausen (Fig. 3). As B. pumilus antagonists were retrieved from the rhizosphere of plants belonging to different cultivars grown in different plots this indicates that this population shows a homogeneous distribution at this site and obviously successfully colonized the rhizosphere of potatoes independent from the cultivar or the genetic modification. Interestingly, one B. pumilus isolate of ‘Ditta’ from the Roggenstein site shared the same BOX-pattern as the B. pumilus antagonists from Oberviehhausen.
The affiliation of the majority of in vitro antagonists in this study is similar to those reported for antagonistic or rhizocompetent bacteria from other studies (Berg et al. 2002, 2006; Juhnke et al. 1987). The presence of antagonists among the dominant cultured bacteria might also mirror their ability to form colonies on plates. The use of members of the genus Pseudomonas for biological control of plant pathogens as well as for plant growth promotion has been reported in several studies (Bloemberg and Lugtenberg 2001; Haas and Défago 2005; Mercado-Blanco and Bakker 2007; O’Sullivan and O’Gara 1992; Weller 1988). The genus Bacillus contains many species that were reported to have in vitro antagonistic activity. B. pumilus was recently described as an antagonist of Gaeumannomyces graminis var. tritici, causing take-all disease of wheat (Sari et al. 2007), but also as a plant growth promoting rhizobacterium inducing systemic resistance against P. infestans on tomato plants (Yan et al. 2002). Kloepper et al. (2004) reviewed several studies reporting on disease reduction by both B. pumilus and B. subtilis species in different hosts. Strains of both species are also available as commercial biocontrol products (Schisler et al. 2004). Also Streptomyces species have been described to reduce plant diseases caused by R. solani (Yuan and Crawford 1995), V. dahliae (Berg et al. 2000) and P. infestans (Malajczuk 1983) and many other plant pathogens.
Furthermore, it is interesting to note that among the in vitro antagonists obtained from both sites several isolates belonged to known potato pathogens, namely the species Dickeya chrysanthemi and Pectobacterium carotovorum as well as S. scabies, S. turgidiscabies and S. acidiscabies. The 26 strains identified as D. chrysanthemi from the potato rhizosphere of Roggenstein were obtained from all four replicates of the cultivar ‘Désirée’ indicating either that infected tubers were planted or a root exudate dependent enrichment in the rhizosphere of ‘Désirée’. Strains identified as P. carotovorum were obtained from one replicate of the GM line SR48. Although both strains are known as opportunistic pathogens, causing tuber soft rot and blackleg in the field but also tuber decay in storage (Pérombelon 2002), no infection symptoms were observed in the respective plant material retrieved from the field. Because all the strains obtained for SR48 originated from the same plot, this might either be the result of a local infection or a consequence of the pooling of the root material. Pathogenic strains of the genera Dickeya and Pectobacterium typically produce a broad range of extracellular enzymes which were described as major pathogenicity factors and are expressed in a cell-density dependent manner (Hélias et al. 2000; Kotoujansky 1987). The production of extracellular enzymes by the isolates might have contributed to their antagonistic activity. To prove whether the in vitro antagonistic Dickeya and Pectobacterium isolates showed pathogenicity in plants, they were tested in a potato slice assay and indeed the strains caused the typical symptoms (data not shown). Interestingly, even though the vast majority of the different Dickeya and Pectobacterium species produced extracellular enzymes and all strains produced AHLs, isolates belonging to P. carotovorum did not antagonize R. solani and V. dahliae in vitro (data not shown). Only D. chrysanthemi showed antagonistic activity towards all three pathogens tested, suggesting additional traits responsible for this antagonism.
Overall, in this study the effects of GM potatoes on rhizobacteria with in vitro antagonistic activity did not exceed natural variability among cultivars. The site was found to be the overriding factor determining the composition and diversity of in vitro antagonists in the rhizosphere of field grown potato plants.
References
Adams PD, Kloepper JW (2002) Effect of host genotype on indigenous bacterial endophytes of cotton (Gossypium hirsutum L.). Plant Soil 240:181–198. doi:10.1023/A:1015840224564
Berg G, Kurze S, Buchner A, Wellington EM, Smalla K (2000) Successful strategy for the selection of new strawberry-associated rhizobacteria antagonistic to Verticillium wilt. Can J Microbiol 46:1128–1137. doi:10.1139/cjm-46-12-1128
Berg G, Fritze A, Roskot N, Smalla K (2001) Evaluation of potential biocontrol rhizobacteria from different host plants of Verticillium dahliae Kleb. J Appl Microbiol 91:963–971. doi:10.1046/j.1365-2672.2001.01462.x
Berg G, Roskot N, Steidle A, Eberl L, Zock A, Smalla K (2002) Plant-dependent genotypic and phenotypic diversity of antagonistic rhizobacteria isolated from different Verticillium host plants. Appl Environ Microbiol 68:3328–3338. doi:10.1128/AEM.68.7.3328-3338.2002
Berg G, Opelt K, Zachow C, Lottmann J, Götz M, Costa R, Smalla K (2006) The rhizosphere effect on bacteria antagonistic towards the pathogenic fungus Verticillium differs depending on plant species and site. FEMS Microbiol Ecol 56:250–261. doi:10.1111/j.1574-6941.2005.00025.x
Bloemberg GV, Lugtenberg BJJ (2001) Molecular basis of plant growth promotion and biocontrol by rhizobacteria. Curr Opin Plant Biol 4:343–350
Bossio DA, Scow KM, Gunapala N, Graham KJ (1998) Determinants of soil microbial communities: effects of agricultural management, season, and soil type on phospholipid fatty acid profiles. Microb Ecol 36:1–12. doi:10.1007/s002489900087
Brimecombe MJ, de Leij FAAM, Lynch JM (2001) The effect of root exudates on rhizosphere microbial populations. In: Pinton R, Varanini R, Nannipieri P (eds) The rhizosphere. Marcel Dekker, New York, pp 95–140
Chernin L, Chet I (2002) Microbial enzymes in biocontrol of plant pathogens and pests. In: Burns R, Dick R (eds) Enzymes in the environment: activity, ecology, and applications. Marcel Dekker, New York, pp 171–225
Cho J-C, Tiedje JM (2000) Biogeography and degree of endemicity of fluorescent Pseudomonas strains in soil. Appl Environ Microbiol 66:5448–5456. doi:10.1128/AEM.66.12.5448-5456.2000
Cornelis P, Matthijs S (2002) Diversity of siderophore-mediated iron uptake systems in fluorescent pseudomonads: not only pyoverdines. Environ Microbiol 4:787–798. doi:10.1046/j.1462-2920.2002.00369.x
Costa R, Falcão Salles J, Berg G, Smalla K (2006) Cultivation-independent analysis of Pseudomonas species in soil and in the rhizosphere of field-grown Verticillium dahliae host plants. Environ Microbiol 8:2136–2149. doi:10.1111/j.1462-2920.2006.01096.x
Crecchio C, Ambrosoli R, Gelsomino A, Minati JL, Ruggiero P (2004) Functional and molecular responses of soil microbial communities under differing soil management practices. Soil Biol Biochem 36:1873–1883. doi:10.1016/j.soilbio.2004.05.008
De Boer M, Bom P, Kindt F, Keurentjes JJB, van der Sluis I, van Loon LC, Bakker PAHM (2003) Control of Fusarium wilt of radish by combining Pseudomonas putida strains that have different disease-suppressive mechanisms. Phytopathology 93:626–632. doi:10.1094/PHYTO.2003.93.5.626
Donegan KK, Palm CJ, Fieland VJ, Porteous LA, Ganio LM, Schaller DL, Bucao LQ, Seidler RJ (1995) Changes in levels, species and DNA fingerprints of soil microorganisms associated with cotton expressing the Bacillus thuringiensis var. kurstaki endotoxin. Appl Soil Ecol 2:111–124. doi:10.1016/0929-1393(94)00043-7
Dunfield KE, Germida JJ (2001) Diversity of bacterial communities in the rhizosphere and root interior of field-grown genetically modified Brassica napus. FEMS Microbiol Ecol 38:1–9. doi:10.1111/j.1574-6941.2001.tb00876.x
Dunfield KE, Germida JJ (2003) Seasonal changes in the rhizosphere microbial communities associated with field-grown genetically modified canola (Brassica napus). Appl Environ Microbiol 69:7310–7318. doi:10.1128/AEM.69.12.7310-7318.2003
Fravel DR (1988) Role of antibiosis in the biocontrol of plant diseases. Annu Rev Phytopathol 26:75–91
Girvan MS, Bullimore J, Pretty JN, Osborn AM, Ball AS (2003) Soil type is the primary determinant of the composition of the total and active bacterial communities in arable soils. Appl Environ Microbiol 69:1800–1809. doi:10.1128/AEM.69.3.1800-1809.2003
Griffiths BS, Geoghegan IE, Robertson WM (2000) Testing genetically engineered potato, producing the lectins GNA and ConA, on non-target soil organisms and processes. J Appl Ecol 37:159–170. doi:10.1046/j.1365-2664.2000.00481.x
Gyamfi S, Pfeifer U, Stierschneider M, Sessitsch A (2002) Effects of transgenic glufosinate-tolerant oilseed rape (Brassica napus) and the associated herbicide application on eubacterial and Pseudomonas communities in the rhizosphere. FEMS Microbiol Ecol 41:181–190. doi:10.1111/j.1574-6941.2002.tb00979.x
Haas D, Défago G (2005) Biological control of soil-borne pathogens by fluorescent Pseudomonads. Nat Rev Microbiol 3:307–319. doi:10.1038/nrmicro1129
Hack H, Gal H, Klemke T, Klose R, Meier U, Stauß R, Witzenberger A (1993) Phänologische Entwicklungsstadien der Kartoffel (Solanum tuberosum L.). Nachrichtenbl Deut Pflanzenschutzd 45:11–19
Hélias V, Andrivon D, Jouan B (2000) Internal colonization pathways of potato plants by Erwinia carotovora ssp. atroseptica. Plant Pathol 49:33–42. doi:10.1046/j.1365-3059.2000.00431.x
Heuer H, Kroppenstedt RM, Lottmann J, Berg G, Smalla K (2002) Effects of T4 lysozyme release from transgenic potato roots on bacterial rhizosphere communities are negligible relative to natural factors. Appl Environ Microbiol 68:1325–1335. doi:10.1128/AEM.68.3.1325-1335.2002
Hiltner L (1904) Über neuere Erfahrungen und Probleme auf dem Gebiete der Bodenbakteriologie unter besonderer Berücksichtigung der Gründüngung und Brache. Arb DLG 98:59–78
Hollomon DW (1965) A medium for the direct isolation of Phytophthora infestans. Plant Pathol 14:34–35. doi:10.1111/j.1365-3059.1965.tb00617.x
James C (2007) Global status of commercialized Biotech/GM crops: 2007. ISAAA Brief No. 37. ISAAA, Ithaca, NY.
Juhnke ME, Mathre DE, Sands DC (1987) Identification and characterization of rhizosphere-competent bacteria of wheat. Appl Environ Microbiol 53:2793–2799
Kennedy NM, Gleeson DE, Connolly J, Clipson NJW (2005) Seasonal and management influences on bacterial community structure in an upland grassland soil. FEMS Microbiol Ecol 53:329–337. doi:10.1016/j.femsec.2005.01.013
Kloepper JW, Ryu C-M, Zhang S (2004) Induced systemic resistance and promotion of plant growth by Bacillus spp. Phytopathology 94:1259–1266. doi:10.1094/PHYTO.2004.94.11.1259
Kotoujansky A (1987) Molecular genetics of pathogenesis by soft-rot Erwinias. Annu Rev Phytopathol 25:405–430. doi:10.1146/annurev.py.25.090187.002201
Krechel A, Faupel A, Hallmann J, Ulrich A, Berg G (2002) Potato-associated bacteria and their antagonistic potential towards plant-pathogenic fungi and the plant-parasitic nematode Meloidogyne incognita (Kofoid & White) Chitwood. Can J Microbiol 48:772–786. doi:10.1139/w02-071
Latour X, Philippot L, Corberand T, Lemanceau P (1999) The establishment of an introduced community of fluorescent pseudomonads in the soil and in the rhizosphere is affected by the soil type. FEMS Microbiol Ecol 30:163–170. doi:10.1111/j.1574-6941.1999.tb00645.x
Lemanceau P, Bakker PAHM, de Kogel WJ, Alabouvette C, Schippers B (1992) Effect of pseudobactin 358 production by Pseudomonas putida WCS358 on suppression of Fusarium wilt of carnations by nonpathogenic Fusarium oxysporum Fo47. Appl Environ Microbiol 58:2978–2982
Leong J (1986) Siderophores: their biochemistry and possible role in the biocontrol of plant pathogens. Annu Rev Phytopathol 24:187–209. doi:10.1146/annurev.py.24.090186.001155
Lifshitz R, Simonson C, Scher FM, Kloepper JW, Rodrick-Semple C, Zaleska I (1985) Effect of rhizobacteria on the severity of Phytophthora root rot of soybean. Can J Plant Pathol 8:102–106
Liu X, Bimerew M, Ma Y, Müller H, Ovadis M, Eberl L, Berg G, Chernin L (2007) Quorum-sensing signaling is required for production of the antibiotic pyrrolnitrin in a rhizospheric biocontrol strain of Serratia plymuthica. FEMS Microbiol Lett 270:299–305. doi:10.1111/j.1574-6968.2007.00681.x
Lottmann J, Berg G (2001) Phenotypic and genotypic characterization of antagonistic bacteria associated with roots of transgenic and non-transgenic potato plants. Microbiol Res 156:75–82. doi:10.1078/0944-5013-00086
Lottmann J, Heuer H, Smalla K, Berg G (1999) Influence of transgenic T4-lysozyme-producing potato plants on potentially beneficial plant-associated bacteria. FEMS Microbiol Ecol 29:365–377. doi:10.1111/j.1574-6941.1999.tb00627.x
Lottmann J, Heuer H, de Vries J, Mahn A, Düring K, Wackernagel W, Smalla K, Berg G (2000) Establishment of introduced antagonistic bacteria in the rhizosphere of transgenic potatoes and their effect on the bacterial community. FEMS Microbiol Ecol 33:41–49. doi:10.1111/j.1574-6941.2000.tb00725.x
Lugtenberg BJJ, Dekkers LC, Bloemberg GV (2001) Molecular determinants of rhizosphere colonization by Pseudomonas. Annu Rev Phytopathol 39:461–490. doi:10.1146/annurev.phyto.39.1.461
Malajczuk N (1983) Microbial antagonism to Phytophthora. In: Erwin DC, Bartnicki-Garcia S, Tsao PH (ed) Phytophthora: its biology, taxonomy, ecology, and pathology. APS Press, pp 197–218
Marilley L, Vogt G, Blanc M, Aragno M (1998) Bacterial diversity in the bulk soil and rhizosphere fractions of Lolium perenne and Trifolium repens as revealed by PCR restriction analysis of 16S rDNA. Plant Soil 198:219–224. doi:10.1023/A:1004309008799
Marschner P, Yang C-H, Lieberei R, Crowley DE (2001) Soil and plant specific effects on bacterial community composition in the rhizosphere. Soil Biol Biochem 33:1437–1445. doi:10.1016/S0038-0717(01) 00052-9
McClean KH, Winson MK, Fish L, Taylor A, Chhabra SR, Camara M, Daykin M, Lamb JH, Swift S, Bycroft BW, Stewart GSAB, Williams P (1997) Quorum sensing and Chromobacterium violaceum: exploitation of violacein production and inhibition for the detection of N-acylhomoserine lactones. Microbiol 143:3703–3711
Mercado-Blanco J, Bakker PAHM (2007) Interactions between plants and beneficial Pseudomonas spp.: exploiting bacterial traits for crop protection. Antonie Van Leeuwenhoek 92:367–389. doi:10.1007/s10482-007-9167-1
Meyer J-M, Geoffroy VA, Baida N, Gardan L, Izard D, Lemanceau P, Achouak W, Palleroni NJ (2002) Siderophore typing, a powerful tool for the identification of fluorescent and nonfluorescent Pseudomonads. Appl Environ Microbiol 68:2745–2753. doi:10.1128/AEM.68.6.2745-2753.2002
O’Sullivan D, O’Gara F (1992) Traits of fluorescent Pseudomonas spp. involved in suppression of plant root pathogens. Microbiol Rev 56:662–676
Pérombelon MCM (2002) Potato diseases caused by soft rot Erwinias: an overview of pathogenesis. Plant Pathol 51:1–12. doi:10.1046/j.0032-0862.2001
Picard C, Bosco M (2008) Genotypic and phenotypic diversity in populations of plant-probiotic Pseudomonas spp. colonizing roots. Naturwissenschaften 95:1–16. doi:10.1007/s00114-007-0286-3
Raaijmakers JM, van der Sluis I, Koster M, Bakker PAHM, Weisbeek PJ, Schippers B (1995) Utilization of heterologous siderophores and rhizosphere competence of fluorescent Pseudomonas spp. Can J Microbiol 41:126–135
Raaijmakers JM, Paulitz TC, Steinberg C, Alabouvette C, Moënne-Loccoz Y (2008) The rhizosphere: a playground and battlefield for soilborne pathogens and beneficial microorganisms. Plant Soil 2008. doi:10.1007/s11104-008-9568-6
Rademaker JLW, de Bruijn FJ (1997) Characterization and classification of microbes by REP-PCR genomic fingerprinting and computer-assisted pattern analysis. In: Caetano-Anollés G, Gresshoff PM (eds) DNA-markers: protocols, applications and overviews. Wiley, New York, N.Y, pp 151–171
Rasche F, Hödl V, Poll C, Kandeler E, Gerzabek MH, van Elsas JD, Sessitsch A (2006) Rhizosphere bacteria affected by transgenic potatoes with antibacterial activities compared with the effects of soil, wild-type potatoes, vegetation stage and pathogen exposure. FEMS Microbiol Ecol 56:219–235. doi:10.1111/j.1574-6941.2005.00027.x
Römer S, Lübeck J, Kauder F, Steiger S, Adomat C, Sandmann G (2002) Genetic engineering of zeaxanthin-rich potato by antisense inactivation and co-suppression of carotenoid epoxidation. Metab Eng 4:263–272. doi:10.1006/mben.2002.0234
Sari E, Etebarian HR, Roustaei A, Aminian H (2006) Biological control of Geaumannomyces graminis on wheat with Bacillus spp. J Plant Pathol 5:307–314. doi:10.3923/ppj.2006.307.314
Sari E, Etebarian HR, Aminian H (2007) The effects of Bacillus pumilus, isolated from wheat rhizosphere, on resistance in wheat seedling roots against the take-all fungus, Geaumannomyces graminis var. tritici. J Phytopathol 155:720–727. doi:10.1111/j.1439-0434.2007.01306.x
Schisler DA, Slininger PJ, Behle RW, Jackson MA (2004) Formulation of Bacillus spp. for biological control of plant diseases. Phytopathology 94:1267–1271. doi:10.1094/PHYTO.2004.94.11.1267
Schwyn B, Neilands JB (1987) Universal chemical assay for the detection and determination of siderophores. Anal Biochem 160:47–56. doi:10.1016/0003-2697(87) 90612-9
Sessitsch A, Weilharter A, Gerzabek MH, Kirchmann H, Kandeler E (2001) Microbial population structures in soil particle size fractions of a long-term fertilizer field experiment. Appl Environ Microbiol 67:4215–4224. doi:10.1128/AEM.67.9.4215-4224.2001
Siciliano SD, Germida JJ (1999) Taxonomic diversity of bacteria associated with the roots of field-grown transgenic Brassica napus cv. Quest, compared to the non-transgenic B. napus cv. Excel and B. rapa cv. Parkland. FEMS Microbiol Ecol 29:263–272. doi:10.1111/j.1574-6941.1999.tb00617.x
Siciliano SD, Theoret CM, de Freitas JR, Hucl PJ, Germida JJ (1998) Differences in the microbial communities associated with the roots of different cultivars of canola and wheat. Can J Microbiol 44:844–851. doi:10.1139/cjm-44-9-844
Smalla K, Wieland G, Buchner A, Zock A, Parzy J, Kaiser S, Roskot N, Heuer H, Berg G (2001) Bulk and rhizosphere bacterial communities studied by denaturing gradient gel electrophoresis: plant-dependent enrichment and seasonal shifts revealed. Appl Environ Microbiol 67:4742–4751. doi:10.1128/AEM.67.10.4742-4751.2001
Van Loon LC, Bakker PAHM, Pieterse CMJ (1998) Systemic resistance induced by rhizosphere bacteria. Annu Rev Phytopathol 36:453–483. doi:10.1146/annurev.phyto.36.1.453
Weissburg WG, Barns SM, Pelletier DA, Lane DJ (1991) 16S ribosomal DNA amplification for phylogenetic study. J Bacteriol 173:697–703
Weller DM (1988) Biological control of soilborne plant pathogens in the rhizosphere with bacteria. Annu Rev Phytopathol 26:379–407. doi:10.1146/annurev.py.26.090188.002115
Whipps JM (2001) Microbial interactions and biocontrol in the rhizosphere. J Exp Bot 52:487–511
Winson KW, Swift S, Fish L, Throup JP, Jorgensen F, Chhabra SR, Bycroft BW, Williams P, Stewart GSAB (1998) Construction and analysis of luxCDABE-based plasmid sensors for investigating N-acyl homoserine lactone-mediated quorum sensing. FEMS Microbiol Lett 163:185–192. doi:10.1111/j.1574-6968.1998.tb13044.x
Yan Z, Reddy MS, Ryu C-M, McInroy JA, Wilson M, Kloepper JW (2002) Induced systemic protection against tomato late blight elicited by plant growth-promoting rhizobacteria. Phytopathology 92:1329–1333. doi:10.1094/PHYTO.002.92.12.1329
Yang CH, Crowley DE (2000) Rhizosphere microbial community structure in relation to root location and plant iron nutritional status. Appl Environ Microbiol 66:345–351. doi:10.1128/AEM.66.1.345-351.2000
Yuan WM, Crawford DL (1995) Characterization of Streptomyces lydicus WYEC108 as a potential biocontrol agent against fungal root and seed rots. Appl Environ Microbiol 61:3119–3128
Acknowledgements
This work was funded by grant 0313277B from the Bundesministerium für Bildung und Forschung. The authors thank U. Zimmerling, A. Büttner and G. Czeplie for valuable assistance in the lab and Dr. F. Niepold for providing the P. infestans isolate. We thank Dr. Holger Heuer and Dr. Siegfried Kropf for discussion in statistical questions. I.-M. Jungkurth is gratefully acknowledged for reading the manuscript. The authors would like to thank J. Dennert and F.X. Maidl (Technical University of Munich) for the perfect management of the experimental plots in Roggenstein and Oberviehhausen. The authors are highly thankful to G. Wenzel (Technical University of Munich) for providing the plant material of the transgenic lines.
Author information
Authors and Affiliations
Corresponding author
Additional information
Responsible Editor: Petra Marschner.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Fig. S1
BOX-PCR fingerprints of in vitro antagonists identified as Pseudomonas fluorescens isolated from the potato rhizospheres in Roggenstein (A) and Oberviehhausen (B), respectively. ST = Standard (1 Kb Plus DNA Ladder). Strain code: cultivar/GM line (Bal = ‘Baltica’; SR47 = ‘Baltica’ co-suppression; SR48 = ‘Baltica’ antisense; Sel = ‘Selma’; Des = ‘Désirée’; Dit = ‘Ditta’; Sib = ‘Sibu’), sampling time (2 = EC60, 3 = EC90), number of replicate plot analysed (1 to 4). * of isolates that were obtained two times from the same plot and showed identical BOX patterns only one representative isolate is shown. (DOC 1,344 kb)
Fig. S2
BOX-PCR profiles of in vitro antagonists identified as Bacillus pumilus isolated from the potato rhizospheres in Roggenstein and Oberviehhausen, respectively. (A) BOX-profiles of a subset of isolates from the potato rhizosphere in Roggenstein. (B) BOX-profiles displayed by the majority of isolates (68/80) identified as Bacillus pumilus isolated from the potato rhizosphere in Oberviehhausen. ST = Standard (1 Kb Plus DNA Ladder). Strain code: cultivar/GM line (Bal = ‘Baltica’; SR47 = ‘Baltica’ co-suppression; SR48 = ‘Baltica’ antisense; Sel = ‘Selma’; Des = ‘Désirée’; Dit = ‘Ditta’; Sib = ‘Sibu’); sampling time (2 = EC60, 3 = EC90), number of replicate plot analysed (1 to 4). * of isolates that were obtained several times from the same plot and showed identical BOX patterns only one representative isolate is shown. (DOC 1,275 kb)
Fig. S3
BOX-profiles of isolates identified as Pectobacterium chrysanthemi (lanes 2–11), P. carotovorum (lanes 12–14) and P. atrosepticum (lane 15) from the potato rhizospheres in Roggenstein (R) and Oberviehhausen (O), respectively. ST = Standard (1 Kb Plus DNA Ladder). Strain code: cultivar/GM line (SR47 = ‘Baltica’ co-suppression; SR48 = ‘Baltica’ antisense; Des = ‘Désirée’; Dit = ‘Ditta’), sampling time (2 = EC60, 3 = EC90), number of replicate plot analysed (1 to 4). * of isolates that were obtained several times from the same plot and showed identical BOX patterns only one representative isolate is shown. (DOC 892 kb)
Table S1
Proportions of in vitro antagonistic isolates in the potato rhizospheres of the sampling site in Roggenstein (A) and Oberviehhausen (B) towards Rhizoctonia solani AG3, Verticillium dahliae ELV25 and Phytophthora infestans as determined by dual-culture assays. (DOC 93 kb)
Table S2
Phylogenetic affiliation of identified in vitro antagonists from both sampling sites determined by partial 16S rRNA gene sequence analysis. Numbers in parenthesis indicate the numbers of isolates that were identified based on the same BOX-fingerprint (>85% similarity). Rog = Roggenstein, Ovh = Oberviehhhausen; EC60 = flowering plants, EC90 = senescent plants; Bal = ‘Baltica’, SR47 = ‘Baltica’ co-suppression, SR48 = ‘Baltica’ antisense, Sel = ‘Selma’, Des = ‘Désirée’, Dit = ‘Ditta’, Sib = ‘Sibu’. Numbers following the cultivar/GM line indicate the plot (1 to 4). (DOC 1,098 kb)
Rights and permissions
About this article
Cite this article
Weinert, N., Meincke, R., Gottwald, C. et al. Effects of genetically modified potatoes with increased zeaxanthin content on the abundance and diversity of rhizobacteria with in vitro antagonistic activity do not exceed natural variability among cultivars. Plant Soil 326, 437–452 (2010). https://doi.org/10.1007/s11104-009-0024-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11104-009-0024-z