Abstract
N-acetyl-l-glutamate kinase (NAGK) catalyzes the second step of l-arginine biosynthesis and is inhibited by l-arginine in Corynebacterium crenatum. To ascertain the basis for the arginine sensitivity of CcNAGK, residue E19 which located at the entrance of the Arginine-ring was subjected to site-saturated mutagenesis and we successfully illustrated the inhibition-resistant mechanism. Typically, the E19Y mutant displayed the greatest deregulation of l-arginine feedback inhibition. An equally important strategy is to improve the catalytic activity and thermostability of CcNAGK. For further strain improvement, we used site-directed mutagenesis to identify mutations that improve CcNAGK. Results identified variants I74V, F91H and K234T display higher specific activity and thermostability. The l-arginine yield and productivity of the recombinant strain C. crenatum SYPA-EH3 (which possesses a combination of all four mutant sites, E19Y/I74V/F91H/K234T) reached 61.2 and 0.638 g/L/h, respectively, after 96 h in 5 L bioreactor fermentation, an increase of approximately 41.8% compared with the initial strain.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
l-Arginine is an industrially important, semi-essential amino acid with applications in the health food supplement, pharmaceutical and cosmetics industries [1, 16]. At the industrial scale, arginine can be produced by microbial fermentation [21]. It has been almost six decades since arginine production has been explored and studied using microorganisms, such as Bacillus subtilis [11] and Corynebacterium glutamicum [16]. In particular, C. glutamicum has advantages as an l-arginine producer because it carries a strong flux towards l-glutamate formation [5, 20]. Present efforts to further improve the industrial productivity of these strains have taken advantage of significant progress in the development of molecular techniques [7, 25], including using rational metabolic engineering to induce specifically targeted modifications that have been used to enhance product yield [15, 27]. For example, site-directed mutagenesis of CcNAGK was used to generate feedback inhibition-resistant N-acetyl-l-glutamate kinase, which increased l-arginine production to 45.6 g/L in Corynebacterium crenatum [25]. Controlling the transcription levels of argGH in fed-batch fermentation of H-7-GH redistributed the l-arginine metabolic flux and produced 389.9 mM l-arginine with productivity of 5.42 mM h−1 [28]. Arginine operon overexpression, NADPH optimization and increased glucose consumption in fed-batch fermentation resulted in a final strain that produced 87.3 g/L l-arginine with a yield of up to 0.431 g l-arginine/g glucose [13]. Furthermore, systems metabolic engineering has been used to achieve an l-arginine yield of 92.5 g/L [16].
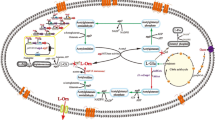
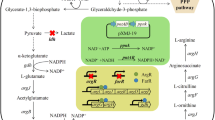
There are three routes for l-arginine biosynthesis from l-glutamate in prokaryotes: the linear pathway, the cycle pathway and a new pathway [4, 21]. In Corynebacteria, l-arginine is synthesized via the recycling pathway in which the acetyl group is recycled from N-acetyl-l-ornithine to l-glutamate and it is regarded as more evolved and economical than others pathway [21]. The cycle pathway involves eight enzymatic reactions in which glutamate is used as a precursor and in which feedback inhibition control is mediated by N-acetyl-l-glutamate kinase (NAGK, encoded by argB). NAGK is a critical rate-limiting enzyme that catalyzes the second reaction in the cycle pathway [9, 17, 26]. Thus, to construct a strain with a high l-arginine yield, l-arginine feedback inhibition of NAGK should be relieved, and NAGK should be overexpressed to obtain higher enzyme activity. NAGKs that are inhibited by arginine are highly similar ring-like hexamers formed by linking three homodimers through the interlacing of an N-terminal mobile kinked α-helix where l-arginine binds [17]. Previous studies have demonstrated that preventing feedback control of NAGK by mutating argB enhances arginine production, as reported by Ikeda et al. [10] and Schendzielorz et al. [18]. Previous work from our group has shown that residues E19, H26, H268 and R209 in CcNAGK have important roles in l-arginine feedback inhibition [24]. Furthermore, mutation studies of CgNAGK and CcNAGK indicated that E19 may be an essential amino acid influencing the affinity of l-arginine, reported by Huang et al. [8] and Zhao et al. [28]. However, there is no report on the mechanism of interaction between l-arginine and the Arginine-ring in NAGK.
In our previous work, a mutant strain C. crenatum SYPA5-5 could accumulate 43.2 g/L l-arginine after cultivation for 96 h in batch fermentation mode under optimal culture conditions and repression of the l-arginine biosynthesis operon by the regulator (encoded by argR) was removed in this mutant strain [23]. Feedback inhibition-resistant NAGK successfully increased the tolerance of C. crenatum to l-arginine in our previous work. Docking analysis revealed that residue E19 is located at the entrance of the Arginine-ring, suggesting that it may have a more important role in feedback inhibition than other sites. In this study, we subjected residue E19 to site-saturated mutagenesis to determine the basis for the arginine sensitivity of CcNAGK and observed greater relief of inhibition in the resulting strain, simultaneously, this could provide a useful reference for removing the feedback inhibition of other enzymes. Hence, we constructed and assessed mutants NAGKE19X (X represents 19 different residues).
In addition, the crystal structures of the NAGKs of different species have shed light on the mechanism of catalysis [14], and we have improved the catalytic efficiency and thermal stability of CcNAGK based on the results of docking analysis and sequence alignment. To investigate the effect on l-arginine yield of improving CcNAGK specific activity and thermostability on the basis of remove feedback inhibition, the multi-mutated NAGKEH3 (including mutations E19Y/I74V/F91H/K234T) which exhibits both deregulated feedback inhibition and enhanced catalytic activity and thermostability was generated. Finally, the recombinant plasmid pDXW10-argBEH3 was introduced into C. crenatum SYPA5-5 to strengthen the l-arginine biosynthesis flux and the fermentation characteristics of the recombinant SYPA-argBEH3 were investigated.
Materials and methods
Strains, plasmids, primers and chemicals
The argB gene (GenBank accession number: AY509864) encoding NAGK from C. crenatum SYPA5-5 was used as template. E. coli JM109 and E.coli BL21 (DE3) were used as hosts for gene cloning and expression, respectively. C. crenatum SYPA5-5 was used as an l-arginine producer. The expression plasmid pET-28a was used for expression and mutagenesis studies. All of the strains, plasmids and primers used in this work are listed in Table 1. The shuttle vector pDXW10 was used for gene expression in C. crenatum SYPA5-5. The restriction enzymes, Prime STAR® HS DNA Polymerase and T4 DNA ligase were purchased from TaKaRa BioCo. (Dalian, China). All other chemicals of high grade were obtained from commercial sources.
Structure simulation and molecular docking
The amino acid sequences of CcNAGK were submitted to SWISS-MODEL Workspace (http://swissmodel.expasy.org/) to obtain a relatively accurate 3D structure, and the predicted residues in the catalytic cavity were obtained using the I-TASSER server (http://zhanglab.ccmb.med.umich.edu/I-TASSER/). The rationality of the model was examined by Profile-3D scoring and a Ramachandran plot. Molecular docking was performed using AutoDock 4.2 software (Scripps Institute, California, USA). The energy interaction value was calculated for each ligand-receptor complex, and the best docking position was chosen through the docking analysis. To select the target residues for mutagenesis, a sequence alignment of CcNAGK with five N-acetyl-l-glutamate kinases with high thermal stability and catalytic activity from different organisms was analyzed using Clustal W2 (http://www.ebi.ac.uk/clustalw/). The structural changes in the l-arginine binding sites were analyzed using PyMOL software [26].
Site-directed mutagenesis of CcNAGK and the construction of recombinant strains
Site-saturated mutagenesis of argB was carried out using overlapping PCR with the amplified argB from C. crenatum SPYA5-5 used as a template. To construct recombinant expression vectors of pET28a-argBH3 and pET28a-argBEH3, the multi-mutated argBH3 and argBE19Y were generated using overlapping PCR. Mutations were introduced using the primers listed in Table 1. Successful introduction of desired mutations was confirmed by DNA sequencing, and then the desired sequences were inserted into multiple cloning sites of pET-28a and transformed to E. coli BL21 (DE3) cells for expression. Subsequently, the multi-mutated argBH3 and argBEH3 were digested and inserted into multiple cloning sites of pDXW10. The recombinant plasmids were transformed into C. crenatum using the electroporation method described by Tauch et al. [22].
Expression and purification of proteins
Recombinant E. coli BL21 cells were cultured at 37 °C to OD = 1.0 (approximately 3 h) and were induced with 1 mM IPTG, after which time the culture was incubated for 10 h at 16 °C. The C. crenatum cells were inoculated into fermentation medium and cultivated for 18 h at 30 °C on 150 rpm, and then collected by centrifugation. The cell paste was suspended in Tris–HCl buffer (pH 8.0) and disrupted on ice by sonication to obtain the intracellular proteins. Recombinant NAGK proteins were purified using a His-Trap HP affinity column as described by Xu et al. [26]. The purified protein was used for activity assays and protein purity was determined by SDS-PAGE analysis (12% acrylamide).
NAGK activity assay
NAGK activity was measured at 37 °C and pH 8.0 (Tris–HCl buffer) for 1 h using the colorimetric assay method of Haas and Leisinger [6]. The standard enzymatic reaction mixture contained the following components, in a total volume of 3 ml aqueous solution: 600 mM Tris–HCl (pH 8.0), 20 mM NAG, 20 mM MgCl2, 20 mM ATP sodium, 150 mM NH2OH·HCl, and enzyme preparation. The reactions were initiated by the addition of enzyme and stopped by the addition of 1 ml N-hydrochloric acid (containing 5% FeCl3·6H2O, 4% trichloroacetic acid). The absorbance of the resulting solution (clarified by centrifugation) was determined at A540. One unit of NAGK was defined as the amount of enzyme that generated 1 µmol NAG hydroxamate per minute. Protein concentration was determined with the Bradford method [2] with bovine serum albumin used as a standard. The specific activity of NAGK was defined as the activity unit per mg protein (U/mg). All assays were performed in triplicate.
Measurement of thermal stabilities
Because the optimal fermentation temperature for l-arginine production is 30 °C in C. crenatum, we determined the thermal stabilities of wild-type and mutant NAGKs at 30, 40 and 50 °C. Thermostability was measured by incubating purified enzyme in Tris–HCl buffer (pH 8.0) for different times at 30, 40 and 50 °C. After incubation, residual activity was measured at 37 °C and pH 8.0. Activity of the enzyme without incubation was defined as 100%. All assays were performed in triplicate.
Determination of kinetic parameters
Apparent K m values were determined from Lineweaver–Burk double-reciprocal plots prepared by varying the concentration of a substrate at a fixed high concentration of the other substrate. To estimate the catalytic ability of wild-type and mutant NAGKs, the catalytic constant k cat value was measured via V max/Et.
l-Arginine feedback inhibition experiments
To investigate the effect of l-arginine concentration on NAGK activity, we added 0–50 mmol/L of l-arginine to the enzymatic reaction mixture and then the activity of the mutated NAGK was determined. The feedback inhibition curve was constructed by changing the l-arginine concentration of. The \(I_{0.5}^{\text{R}}\) is defined as the concentration of l-arginine that causes 50% inhibition. Activity of the enzyme without l-arginine was defined as 100%.
Growth medium and conditions for l-arginine production
Corynebacterium crenatum SYPA5-5 and corresponding recombinant strains were first grown in LBG containing 10 g/L peptone, 10 g/L NaCl, 5 g/L yeast extract and 5 g/L glucose. Following they were inoculated to the seed culture medium and cultured at 30 °C for 24 h in a shake flask. The seed culture medium consisted of 50 g/L glucose, 20 g/L yeast extract, 20 g/L (NH4)2SO4, 1.5 g/L KH2PO4, 0.5 g/L MgSO4·7H2O and 1 g/L CaCO3. The shake flask culture was then transferred into 5 L bioreactor (BIOTECH-5BG, Baoxing Co., China) containing 3 L fermentation medium A, which consisted of 150 g/L glucose, 12 g/L yeast extraction, 20 g/L (NH4)2SO4, 1.5 g/L KH2PO4, 1 g/L KCl, 0.5 g/L MgSO4·7H2O, 20 mg/L FeSO4·7H2O and 20 mg/L MnSO4·H2O, pH 7.0. Glucose was sterilized separately. The aeration rate was controlled at 2.5 vvm for all experiments. Studies in the bioreactor were carried out at an agitation rate of 600 rpm and pH was maintained at 7.0 by flowing aqueous ammonia.
Assays of cell concentration, glucose and l-arginine
Cell concentration was first monitored at 562 nm. Glucose concentration in the media assayed was using a glucose analyzer (Biosensor SBA-50, Shandong, China). The l-arginine concentration was detected by HPLC (DIONEX, UltiMate 3000, USA) equipped with a venusil AA column (Agela Technologies, China). All assays were performed in triplicate cultures.
Results and discussion
Homology modeling and residue functional analysis of CcNAGK
A tertiary structure of monomeric CcNAGK was built based on the crystal structures of the arginine-sensitive NAGK from Mycobacterium tuberculosis, (PDB ID: 2ap9, chain F) using the SWISS-MODEL Workspace (Fig. S1a). The quality of the model was assessed using Ramachandran statistics and Profile-3. Our results showed that 90.8% of amino acids are located in the optimum region of the Ramachandran plot, with 4.1% in the allowed region and 5.1% in the disallowed region (Fig. S1b), and the Profile-3D graph (Fig. S1c) indicated that 90.31% of the residues had an averaged 3D score ≥0.2. Thus, the structure evaluation analysis confirmed that the NAGK model can represent the CgNAGK structure. A previous study to determine the structural basis of feedback control by l-arginine and molecular docking showed that the Arginine-ring was composed of residues E19, H26, R209 and H268. In particular, residue E19 is located at the entrance of the Arginine-ring and was hypothesized to be a key site (shown in Fig. 1b) that might have a greater role in feedback inhibition than the other sites (H26, R209, H268). Consequently, E19 was subjected to site-saturated mutagenesis to ascertain the basis for the arginine sensitivity and to relieve the inhibition to a greater degree. Subsequently, the E19X (with X representing 19 different types of residues) mutants of NAGK were constructed. Furthermore, residues K35, G71, I74, F91, R92, N191, D195 and K252 are located at the substrate binding pocket by analysis of the I-TASSER server and molecular docking, as shown in Fig. 1a. These residues could have an important influence on catalytic activity. Furthermore, residues located in some structures, including turns, important loops and hydrophobic cores, may play important roles in favoring conformational stability. For the first time, it was found that the residue K234, located at the corner of α-helix and β-sheet, might have an important role in conferring properties of CcNAGK. The sequence alignment of CcNAGK with five N-acetyl glutamate kinases from different organisms was shown in Fig. 2. In these residues, the K35, G71, R92, N191, D195 and K252 are highly conserved in enzymes from different bacteria. Thus, absolutely conserved residues were not taken into account in this study. Accordingly, the three residues I74, F91 and K234, which might have an important influence on the catalytic activity and thermal stability of CcNAGK were subjected to site-directed mutagenesis, and the variants of NAGK were termed I74V, F91L, F91H, K234D and K234T. The CcNAGK and variants were expressed and purified, 12% SDS-PAGE analysis showed that none of the point mutations affected protein solubility or purification (shown in Fig. S2).
The tertiary docking structure of the monomeric CcNAGK and substrate, feedback inhibitor l-arginine. Molecular docking was performed using AutoDock software. An overall view of the docking structure is shown on the left side. The magnified structures on the right side are substrate-docking and Arginine-docking sites of CcNAGK. a Substrate-docking site. Spheres and sticks represent bound NAG and ATP, and residues of substrate binding sites are shown in sticks. b Arginine-docking site. Mesh representation of the bound arginine and residues of arginine-binding sites are shown in sticks
Multiple sequence alignment of NAGK homologies. The sequences used were NAGK homologies from Corynebacterium crenatum (CcNAGK), Mycobacterium tuberculosis (M.t-2ap9), Chlamydomonas reinhardtii (C.r-4usj), Thermotoga maritime (T.m-2bty), Arabidopsis thaliana (A.t-2rd5) and Escherichia coli (E.c-1gs5). The NAGK homologies were aligned by using Clustal W2 programs. The mutation sites (E19, I74, F91 and K234) were indicated by blue triangle below (the residues from 120 to 179 and from 240 to 317 were omitted) (color figure online)
l-Arginine feedback inhibition and elaboration of the inhibition-resistant mechanism
The specific activities and \(I_{0.5}^{\text{R}}\) values were determined for wild-type CcNAGK and the mutant E19X enzymes, as shown in Table 2. The specific activities of the mutants with the 19th residue replaced by other residues were similar to that of the native CcNAGK (9.11 U/mg), except for the variants E19W and E19P. The specific activity of NAGKE19W was 4.28 U/mg, approximately half that of CcNAGK. Docking analysis of CcNAGK with l-arginine showed E19 to be located in the outer edge of the protein (as shown in Fig. 1a), while the side chain of Trp in the 19th position was an indolyl group that was annular and bulkier in the E19W mutant. The bulkier side chain in W19 could affect protein conformation while inhibiting protein folding and stability, thereby markedly decreasing specific activity. As previously reported, an increase in chain length results in a sudden decrease in activity, and a bulky group is likely to perturb the local protein conformation [3, 19]. Furthermore, E19P was nearly inactivated, which might be caused by proline restraining the α-helix formation and thus preventing proper folding of NAGK. In fact, proline residues are known to destabilize and distort α-helical strands by their inability to form hydrogen bonds and by their steric hindrance [12].
The \(I_{0.5}^{\text{R}}\) of mutant E19D was similar to that of CcNAGK (0.39 mM), while the \(I_{0.5}^{\text{R}}\) values of other mutants with the 19th residue replaced (by Gln, Asn, Ser, Thr, Cys, Met, Ile, Leu, Val, Ala or Gly) increased approximately 40-fold (Table 2). These results indicate that all of these mutants mitigate l-arginine-mediated inhibition, with the exception of E19D. As is well known, Glu and Asp are acidic amino acids, while the others are not charged. Taking wild-type CcNAGK and variants NAGKE19Q, NAGKE19D and NAGKE19N as examples, structural analysis showed that the structures of Gln and Asn were similar to those of Glu and Asp, respectively. However, the only difference was that the carboxyl groups which provides negatively charged of Glu and Asp were substituted with the amide groups of Gln and Asn (shown in Fig. 3a–d). These results indicate that the combination of l-arginine (guanidyl) and the Arginine-ring depends on whether the 19th residue has a negative charge.
Arginine-binding site of CcNAGK and the feedback inhibition-resistance mechanism was delineated by structural analysis in wild-type and mutant CcNAGKs. Mesh representation of the l-arginine and residues of arginine-binding sites (E19, H26, R209 and H268) were shown in sticks. a The wild-type CcNAGK in E19, b the mutant E19Q, c the mutant E19D, d the mutant E19N,(e the mutant E19R, f the mutant E19Y
Furthermore, the \(I_{0.5}^{\text{R}}\) value of E19R, E19H and E19K exhibited a 60-fold increase in efficiency, suggesting greater relief of feedback inhibition in these mutants. In fact, Arg, His and Lys are basic amino acids and are positively charged, and take E19R for example as shown in Fig. 3e. In this regard, it was found that for feedback inhibition-resistant NAGKs, positively basic amino acid mutants had a greater effect than mutant residues with no charge. The result might be caused by Arg, His and Lys are positively charged residues that are mutually exclusive with the guanidine of l-arginine.
Excitingly, the other three variants, E19Y, E19W and E19F, showed the \(I_{0.5}^{\text{R}}\) values increased approximately 380-fold, which indicated greatly reduced feedback inhibition. Structural analysis of these mutants showed that these side chains of the 19th residues are aromatic-rings, as shown in Fig. 3f for E19Y. The reason for greatly reduced feedback inhibition might be that these residues did not introduce a negative charge, and their bulkier aromatic ring side chain is located at the entrance of the Arginine-ring, preventing l-arginine from interacting with the 19th site while also restricting interaction of arginine and other sites containing H26, R209 and H268. Namely, the bulkier aromatic ring side chain could directly prevent l-arginine access into the Arginine-ring. Consequently, the feedback inhibition of l-arginine was the most deregulated in the E19Y, E19W and E19F mutants.
Taken together, these results indicate that residue E19, which is located at the entrance of the Arginine-ring, controls l-arginine access into the Arginine-ring. The negative charge in the 19th residue is vitally important to the activity of l-arginine, and the alleviation of feedback inhibition is related to the negative charge and side chain structure of the 19th residue. In fact, in consideration of enzyme activity and the effect of deregulation of feedback inhibition, the E19Y mutant is the best choice for further study.
Improved catalytic activity and thermostability of CcNAGK by site-directed mutagenesis
Enzyme activities and kinetic parameters of wild-type and mutant CcNAGK (I74V, F91L, F91H, K234D and K234T) were determined, and the data are shown in Table 3. The specific activities of I74V (12.58 U/mg) and F91H (13.86 U/mg) were 38% and 52% higher than that of wild-type CcNAGK (9.11 U/mg), respectively, while the specific activities of F91L, K234D and K234T were similar to that of CcNAGK. The mutant I74V revealed a 32% decrease in K m(NAG) and a 64% increase in k cat(NAG)/K m(NAG), and F91H showed not only decreases in K m(NAG) and K m(ATP) but also increases in k cat(NAG) and k cat(NAG), thus increasing the specific activity 1.52-fold. These results indicate that substitution of I74 by valine enhances substrate (NAG) affinity, and substitution of F91 by histidine enhances not only substrates (NAG and ATP) affinity but also greatly improves catalytic constants. Furthermore, the thermostability of the enzyme was monitored after incubation in Tris–HCl buffer (pH 8.0) for different durations at 30, 40 and 50 °C. As shown in Fig. 4, the I74V, F91H and K234T mutants exhibited higher thermal stability than wild-type, whereas F91L and K234D were similar to wild-type. Because the variants I74V, F91H and K234T displayed higher specific activity or thermostability than wild-type, they were selected for further study.
Effect of temperature on enzyme stability. Enzymes were incubated in Tris–HCl buffer (pH 8.0) for different durations at 30, 40, and 50 °C, and the residual activities were determined following incubation. Enzyme activity without incubation was defined as 100% (all assays were performed in triplicate, and standard deviations of the biological replicates are represented by error bars)
Construction of CcNAGK multi-mutants
To further improve catalytic activity and thermostability, the multi-mutant H3 (including I74V/F91H/K234T) was constructed and the mutant enzyme were termed as NAGKH3. To obtain an NAGK with improved properties, including not only superior catalytic efficiency and thermalstability but also freedom from l-arginine feedback inhibition, the NAGKEH3 mutant (including E19Y/I74V/F91H/K234T) was generated. The multi-mutants NAGKH3 and NAGKEH3 were abundantly expressed in the soluble form and purified as previously described (Fig. S2). Subsequently, specific activities and \(I_{0.5}^{\text{R}}\) were determined. The specific activities of these mutants were superior to that of wild-type, displaying an approximate increase of 73%. In addition, the \(I_{0.5}^{\text{R}}\) of the NAGKEH3 was raised evidently about 382-fold (data shown in Table 3). Determination of enzymatic properties also showed that the multi-mutants exhibited superior thermal stability (as shown in Fig. 4), which is important for the long fermentation period of l-arginine. Consequently, NAGKEH3 could be considered a feedback inhibition-resistant mutant with a high \(I_{0.5}^{\text{R}}\) value and superior catalytic activity and thermal stability.
Construction of the recombinant C. crenatum SYPA-E19Y and SYPA-EH3
Modulating the gene expression and metabolism of C. crenatum has proven to be an efficient tool for enhancing amino acid production. After inserting the mutant gene argBE19Y into the shuttle vector pDXW10 and constructing the recombinant plasmid pDXW10-argBE19Y, we sought to determine whether the E19Y mutant was effective in relieving l-arginine feedback inhibition and increasing l-arginine biosynthesis in C. crenatum SYPA5-5. The recombinant plasmids were subsequently transformed into C. crenatum and the engineered strain SYPA-E19Y was constructed. Concurrently, to investigate the influence of improving CcNAGK specific activity and thermostability on l-arginine yield, genetic engineering was used to construct the recombinant strain C. crenatumSYPA-EH3, which expresses a NAGK mutant with a combination of the mutant sites E19Y/I74V/F91H/K234T that results in superior catalytic activity and thermal stability and resistance to l-arginine feedback inhibition. To examine the expression level of the mutant NAGKs, the expressions levels of argBE19Y and argBEH3 in C. crenatum were tested using SDS-PAGE. Figure 5 shows that the target protein bands exhibited a molecular weight of about 33 kDa, indicating that the mutant genes had been overexpressed in C. crenatum SYPA5-5, carried by a multi-copy plasmid pDXW10. Additionally, enzyme activities in recombinants were evaluated by comparison with those of wild-type SYPA5-5. The results showed that the total activities of NAGK in SYPA-E19Y and SYPA-EH3 displayed 9.35 and 15.52-fold higher than that of wild-type SYPA5-5, respectively (shown in Table 4).
Improving l-arginine production by improving catalytic activity and thermostability in feedback inhibition-resistant recombinant C. crenatum SYPA-EH3
l-arginine production of SYPA-E19Y reached 55.3 g/L after 96 h in a 5 L bioreactor fermentation, approximately 27.9% higher than that of the initial strain C. crenatum SYPA5-5 (Fig. 6b). We tracked intracellular l-arginine concentrations during the period of fermentation and observed that intracellular l-arginine in C. crenatum SYPA5-5 was at its highest concentration (14.8 mM) at 48 h and was 7.6 mM at the end of fermentation. This result revealed that intracellular CcNAGK activity was feedback-inhibited by l-arginine during the fermentation period. Conversely, the \(I_{0.5}^{\text{R}}\) value of the E19Y mutant was 148.5 mM and increased 382-fold. The l-arginine inhibition curves (Fig. S3) showed constant specific activity of E19Y compared to the wild-type, remaining approximately 99.2% activity in the presence of 20 mM l-arginine (the maximum intracellular l-arginine concentration was 17.8 mM in the recombinant strain, see Supplementary Fig. S4). These results indicate that CcNAGK feedback inhibition in SYPA-E19Y was successfully deregulated. However, the productivity of l-arginine showed a downward trend in the late period of fermentation (72–96 h) compared to the early period (Fig. 7). These results illustrated the hypothesis that l-arginine yield could be markedly increased by preventing l-arginine feedback inhibition of CcNAGK. Metabolic flux through the l-arginine production pathway could be enhanced by mutating the rate-limiting enzyme. Furthermore, l-arginine productivity was markedly decreased in the late period of fermentation might be caused by CcNAGK has a relatively low catalytic activity and thermostability. Consequently, we concluded that while removing l-arginine feedback inhibition of NAGK is one vital method for improving l-arginine production, an equally important mechanism is improving NAGK catalytic activity and thermostability.
Comparison of l-arginine production between C. crenatum SYPA5-5, recombinant SYPA-E19Y and SYPA-EH3. a Cell concentration and glucose concentration. b l-Arginine concentration and l-arginine productivity. C. crenatum SYPA5-5 (open and filled squares); recombinant SYPA-E19Y (open and filled circles) and recombinant SYPA-EH3 (open and filled triangles) (all values the mean of at least three independent experiments, and error bars represent the standard deviations of the biological replicates)
Comparison of l-Arginine productivity and CcNAGK enzyme activity in C. crenatum SYPA5-5, recombinant SYPA-E19Y and SYPA-EH3. a Comparison of l-arginine productivity. b Comparison of CcNAGK enzyme activity. C. crenatum SYPA5-5 (squares), recombinant SYPA-E19Y (circles) and recombinant SYPA-EH3 (triangles) (all assays were performed in triplicate cultures, standard deviations of the biological replicates were represented by error bars)
To further increase l-arginine yield and productivity, the constructed strain C. crenatum SYPA-EH3 was used as a basis to develop a new type of l-arginine producer by genetic engineering. Subsequently, the C. crenatum SYPA5-5, SYPA-E19Y and SYPA-EH3 strains were cultivated for 96 h in the fermentation medium, and their corresponding characteristics were compared. The fermentation curves, including cell concentration, glucose consumption, l-arginine yield and productivity are presented in Figs. 6 and 7. The growth rates of the three strains were similar throughout the entire fermentation period (shown in Fig. 6a), indicating that overexpression of the mutant argB gene did not affect cell growth in C. crenatum. It was also observed that the glucose consumption remained fairly consistent at the beginning of fermentation but accelerated obviously after 36 h, compared with the initial strain as shown in Fig. 6a. This increase results from the removal of feedback inhibition in the metabolic synthesis pathway of l-arginine and the superior l-arginine synthesis of recombinants compared to the initial strain. l-arginine yield and productivity were similar between the strains before 36 h. Then, the recombinants showed a slight advantage in l-arginine production between 36 and 48 h. At the end of fermentation, l-arginine yield increased noticeably to 55.3 and 61.2 g/L in the recombinants SYPA-E19Y and SYPA-EH3, respectively, values that were approximately 27.9 and 41.8% higher than that of SYPA5-5, respectively (Fig. 6b). This result indicates that l-arginine yield can be markedly increased by preventing l-arginine feedback inhibition and improving the catalytic efficiency and thermostability of CcNAGK. We also tracked CcNAGK activity during the fermentation period and found the trends of changing enzyme activity to be similar in all tested strains before 48 h; however, this trend differed after 48 h. In particular, enzyme activity sharply declined in the initial strain and in the recombinant SYPA-E19Y after 72 h, whereas activity only slightly decreased in SYPA-EH3 (Fig. 7a). Importantly, l-arginine productivity sharply declined in both the initial strain and in the recombinant SYPA-E19Y after 72 h, whereas this downward productivity trend was relieved in SYPA-EH3 (Fig. 7b). This result might explain the greater thermostability of the mutant NAGK compared with the relatively low thermostability of wild-type CcNAGK. It was obvious that l-arginine productivity declined gradually in the recombinant SYPA-EH3; therefore l-arginine yield could achieve 56 g/L after 84 h. For the recombinant C. crenatum SYPA-EH3, the fermentation time to achieve the same yield achieved by SYPA-E19Y was shortened to 12 h, and the maximum yield reached 61.2 g/L after 96 h.
Conclusions
In C. crenatum, NAGK is inhibited by l-arginine. In this study, we selected residue E19, located at the entrance of the Arginine-ring, for site-saturated mutagenesis to ascertain the basis for the l-arginine sensitivity of CcNAGK and to deregulate the feedback inhibition. This method could provide a useful reference for removing the feedback inhibition of other enzymes. Ultimately, we obtained the E19Y mutant with the greatest ability to deregulate feedback inhibition. The l-arginine yield of recombinant SYPA-E19Y reached 55 g/L after 96 h in 5 L bioreactor fermentation, approximately 27.9% higher than the yield of the initial strain. After removing the feedback inhibition, the multi-mutated NAGKEH3 was generated to improve the catalytic activity and thermal stability of CcNAGK and to further increase l-arginine yield. NAGKEH3 displayed superior catalytic efficiency and thermostability and deregulated feedback inhibition by l-arginine. Finally, the l-arginine yield of recombinant SYPA-EH3 reached 61.2 g/L, 41.8% greater than that attained in the initial strain. These results illustrate the hypothesis that l-arginine yield may be markedly increased by preventing l-arginine feedback inhibition of CcNAGK. l-arginine productivity could be enhanced by improving the catalytic efficiency and thermostability of CcNAGK in the late period of fermentation, resulting in a further increase in l-arginine yield. These results could shift the paradigm for future strain development for l-arginine fermentation.
References
Alvares TS, Meirelles CM, Bhambhani YN, Paschoalin VM, Gomes PS (2011) l-Arginine as a potential ergogenic aid in healthy subjects. Sports Med 41:233–248. doi:10.2165/11538590-000000000-00000
Bradford MM (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72:248–254
Chaudhary S, Vats ID, Chopra M, Biswas P, Pasha S (2009) Effect of varying chain length between P(1) and P(1′) position of tripeptidomimics on activity of angiotensin-converting enzyme inhibitors. Bioorg Med Chem Lett 19:4364–4366. doi:10.1016/j.bmcl.2009.05.079
Cunin R, Glansdorff N, Pierard A, Stalon V (1986) Biosynthesis and metabolism of arginine in bacteria. Microbiol Rev 50:314–352
Duperray F, Jezequel D, Ghazi A, Letellier L, Shechter E (1992) Excretion of glutamate from Corynebacterium glutamicum triggered by amine surfactants. Biochim Biophys Acta 1103:250–258
Haas D, Kurer V, Leisinger T (1972) N-acetylglutamate synthetase of Pseudomonas aeruginosa. An assay in vitro and feedback inhibition by arginine. Eur J Biochem 31:290–295
Holatko J, Elisakova V, Prouza M, Sobotka M, Nesvera J, Patek M (2009) Metabolic engineering of the l-valine biosynthesis pathway in Corynebacterium glutamicum using promoter activity modulation. J Biotechnol 139:203–210. doi:10.1016/j.jbiotec.2008.12.005
Huang Y, Li C, Zhang H, Liang S, Han S, Lin Y, Yang X, Zheng S (2016) Monomeric Corynebacterium glutamicum N-acetyl glutamate kinase maintains sensitivity to l-arginine but has a lower intrinsic catalytic activity. Appl Microbiol Biotechnol 100:1789–1798. doi:10.1007/s00253-015-7065-4
Huang Y, Zhang H, Tian H, Li C, Han S, Lin Y, Zheng S (2015) Mutational analysis to identify the residues essential for the inhibition of N-acetyl glutamate kinase of Corynebacterium glutamicum. Appl Microbiol Biotechnol 99:7527–7537. doi:10.1007/s00253-015-6469-5
Ikeda M, Mitsuhashi S, Tanaka K, Hayashi M (2009) Reengineering of a Corynebacterium glutamicum l-arginine and l-citrulline producer. Appl Environ Microbiol 75:1635–1641. doi:10.1128/AEM.02027-08
Kisumi M, Kato J, Sugiura M, Chibata I (1971) Production of l-arginine by arginine hydroxamate-resistant mutants of Bacillus subtilis. Appl Microbiol 22:987–991
MacArthur MW, Thornton JM (1991) Influence of proline residues on protein conformation. J Mol Biol 218:397–412
Man Z, Xu M, Rao Z, Guo J, Yang T, Zhang X, Xu Z (2016) Systems pathway engineering of Corynebacterium crenatum for improved l-arginine production. Sci Rep 6:28629. doi:10.1038/srep28629
Marco-Marı́n C, Ramón-Maiques S, Tavárez S, Rubio V (2003) Site-directed mutagenesis of Escherichia coli acetylglutamate kinase and aspartokinase III probes the catalytic and substrate-binding mechanisms of these amino acid kinase family enzymes and allows three-dimensional modelling of aspartokinase. J Mol Biol 334:459–476. doi:10.1016/j.jmb.2003.09.038
Park JH, Lee SY (2008) Towards systems metabolic engineering of microorganisms for amino acid production. Curr Opin Biotechnol 19:454–460. doi:10.1016/j.copbio.2008.08.007
Park SH, Kim HU, Kim TY, Park JS, Kim SS, Lee SY (2014) Metabolic engineering of Corynebacterium glutamicum for l-arginine production. Nat Commun 5:4618. doi:10.1038/ncomms5618
Ramon-Maiques S, Fernandez-Murga ML, Gil-Ortiz F, Vagin A, Fita I, Rubio V (2006) Structural bases of feed-back control of arginine biosynthesis, revealed by the structures of two hexameric N-acetylglutamate kinases, from Thermotoga maritima and Pseudomonas aeruginosa. J Mol Biol 356:695–713. doi:10.1016/j.jmb.2005.11.079
Schendzielorz G, Dippong M, Grunberger A, Kohlheyer D, Yoshida A, Binder S, Nishiyama C, Nishiyama M, Bott M, Eggeling L (2014) Taking control over control: use of product sensing in single cells to remove flux control at key enzymes in biosynthesis pathways. ACS Synth Biol 3:21–29. doi:10.1021/sb400059y
Schmidt AE, Sun MF, Ogawa T, Bajaj SP, Gailani D (2008) Functional role of residue 193 (chymotrypsin numbering) in serine proteases: influence of side chain length and beta-branching on the catalytic activity of blood coagulation factor XIa. Biochemistry 47:1326–1335. doi:10.1021/bi701594j
Schneider J, Niermann K, Wendisch VF (2011) Production of the amino acids l-glutamate, l-lysine, l-ornithine and l-arginine from arabinose by recombinant Corynebacterium glutamicum. J Biotechnol 154:191–198. doi:10.1016/j.jbiotec.2010.07.009
Shin JH, Lee SY (2014) Metabolic engineering of microorganisms for the production of l-arginine and its derivatives. Microb Cell Fact 13:166. doi:10.1186/s12934-014-0166-4
Tauch A, Kirchner O, Loffler B, Gotker S, Puhler A, Kalinowski J (2002) Efficient electrotransformation of corynebacterium diphtheriae with a mini-replicon derived from the Corynebacterium glutamicum plasmid pGA1. Curr Microbiol 45:362–367. doi:10.1007/s00284-002-3728-3
Xu H, Dou W, Xu H, Zhang X, Rao Z, Shi Z, Xu Z (2009) A two-stage oxygen supply strategy for enhanced l-arginine production by Corynebacterium crenatum based on metabolic fluxes analysis. Biochem Eng J 43:41–51. doi:10.1016/j.bej.2008.08.007
Xu M, Rao Z, Dou W, Jin J, Xu Z (2012) Site-directed mutagenesis studies on the l-arginine-binding sites of feedback inhibition in N-acetyl-l-glutamate kinase (NAGK) from Corynebacterium glutamicum. Curr Microbiol 64:164–172. doi:10.1007/s00284-011-0042-y
Xu M, Rao Z, Dou W, Yang J, Jin J, Xu Z (2012) Site-directed mutagenesis and feedback-resistant N-acetyl-l-glutamate kinase (NAGK) increase Corynebacterium crenatum l-arginine production. Amino Acids 43:255–266. doi:10.1007/s00726-011-1069-x
Yu P, Xu M (2012) Enhancing the enzymatic activity of the endochitinase by the directed evolution and its enzymatic property evaluation. Process Biochem 47:1089–1094. doi:10.1016/j.procbio.2012.03.015
Zhang B, Wan F, Qiu YL, Chen XL, Tang L, Chen JC, Xiong YH (2015) Increased l-arginine production by site-directed mutagenesis of N-acetyl-l-glutamate kinase and proB gene deletion in Corynebacterium crenatum. Biomed Environ Sci 28:864–874. doi:10.3967/bes2015.120
Zhao Q, Luo Y, Dou W, Zhang X, Zhang X, Zhang W, Xu M, Geng Y, Rao Z, Xu Z (2016) Controlling the transcription levels of argGH redistributed l-arginine metabolic flux in N-acetylglutamate kinase and ArgR-deregulated Corynebacterium crenatum. J Ind Microbiol Biotechnol 43:55–66. doi:10.1007/s10295-015-1692-8
Acknowledgements
This work was supported by the High-tech Research and Development Programs of China (2015AA021004), the National Natural Science Foundation of China (31300028), the Jiangsu Provincial National Basic Research Program (BK20150002, BK20130137), the Research Project of Chinese Ministry of Education (113033A), the Program Funded by China Scholarship Council, the Project Funded by the Priority Academic Program Development of Jiangsu Higher Education Institutions, the 111 Project (No. 111-2-06), and the Jiangsu province “Collaborative Innovation Center for Advanced Industrial Fermentation” industry development program.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare no financial or commercial conflict of interest.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Zhang, J., Xu, M., Ge, X. et al. Reengineering of the feedback-inhibition enzyme N-acetyl-l-glutamate kinase to enhance l-arginine production in Corynebacterium crenatum . J Ind Microbiol Biotechnol 44, 271–283 (2017). https://doi.org/10.1007/s10295-016-1885-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10295-016-1885-9