Abstract
Background
Steroid-resistant nephrotic syndrome (SRNS) is a common cause of end-stage renal disease in children but also occurs as an adult-onset condition. In a subset of SRNS patients, pathogenic variants are found in genes coding for podocyte foot process proteins. The aim of this study was to define the role of pathogenic variants in Finnish patients with familial and sporadic SRNS.
Methods
We analyzed SRNS-related genes NPHS1, NPHS2, NEPH1, ACTN4, TRPC6, INF2, WT1, CD2AP, LAMB2, and PLCE1 for disease-causing variants using direct sequencing of exons and intron/exon boundaries in all members of a family with dominant SRNS with early onset and slow progression to end-stage renal disease. We carried out a whole genome sequencing in two affected and two healthy family members. The function of found podocin variant was studied using co-immunoprecipitation and immunohistochemistry. Podocyte gene sequences were analyzed in a cohort of Finnish non-familial SRNS patients.
Results
A heterozygous de novo deletion, c.988_989delCT in NPHS2, was found in all affected family members and in none of their healthy relatives, non-familial patients or controls. No other SRNS-related gene variant, coding or non-coding co-segregated with the disease phenotype in the family. While the truncated podocin remained able to bind nephrin, the expression of nephrin was fragmented and podocin expression reduced. The gene analysis of the non-familial SRNS patients revealed few variants.
Conclusion
The role of podocin variants in nephrotic syndrome may be more varied than previously thought.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Steroid-resistant nephrotic syndrome (SRNS) manifesting during the first year of life is a rare disorder often caused by variants in genes encoding glomerular podocyte proteins, especially nephrin (NPHS1) and podocin (NPHS2) [1–5]. SRNS starting later in childhood is in most cases multifactorial with unknown etiology, although the causative factors can include genetic variants. Familial forms of SRNS exist with recessive and dominant inheritance patterns. Variants in NPHS1 and NPHS2 are linked to the autosomal recessive inheritance, whereas variants in α-actinin-4 (ACTN4), transient receptor potential cation channel subfamily C member 6 (TRPC6) and inverted formin 2 (INF2), associate with the autosomal dominant pattern [6]. Altogether, variants in over 30 genes have been associated with SRNS, most of them in exceedingly rare cases or in association with extra-renal symptoms [7].
In this work, we analyzed the genes encoding for the three interacting podocyte proteins nephrin, podocin and neph1 in a cohort of Finnish pediatric SRNS patients including a family with newborn onset of proteinuria and exceptionally slow progression to renal failure. We also analyzed the sequences of ACTN4, TRPC6 and INF2 in the affected family members and carried out whole genome sequencing in two affected and two healthy family members to discover variants in the additional SRNS-related genes CD2AP, WT1 and PLCE1 as well as find all non-exonic variants in these SRNS-related genes. The familial disorder followed dominant inheritance pattern and was associated with a heterozygous, de novo mutation in NPHS2, encoding podocin.
Materials and methods
Patients
Two pediatric siblings with SRNS (Family AM, Fig. 1) were the starting point of this study. Seven other family members in three previous generations were recruited. Of these, three were inflicted with SRNS. In addition to Family AM, the study cohort included twelve pediatric patients with SRNS who were diagnosed at the University Hospitals in Finland after 1989. Nine of the twelve patients showed FSGS histology and MCNS. The clinical course of the disease of these patients was variable. Two had neurological symptoms.
Clinical data from patients and their families were gathered from the hospital records. Blood samples were collected during routine checkups, or on request in their local health centers.
Genetic analyses
Genomic DNA was isolated from frozen or fresh peripheral blood samples using the conventional molecular biology technique (Puregene EPTM DNA Purification Kit, Gentra Systems). The analyses of protein coding regions of NPHS1 (NM_004646), NPHS2 (NM_014625), NEPH1 (NM_018240), and ACTN4 (NM_004924) as well as exons 2 and 4 of INF2 (NM_022489) were performed using direct sequencing. Exons were amplified by PCR with flanking intronic primers. The reactions were performed in total volumes of 25 μl as previously described [8]. Occasionally, the denaturation temperature was raised up to 98 °C, or betaine was added to the reaction mixture. PCR products were subjected to automated sequence analysis by BigDye-terminator chemistry (v.3.1) on Genetic Analyzer 3730 (Applied Biosystems). The sequences were analyzed with GeneComposer version 1.1.0.1051 (www.GeneComposer.com) or 4Peaks version 1.7.2 (by A. Griekspoor and Tom Groothuis, mekentosj.com.).
When a nucleotide change resulting in an amino acid change was spotted, the significance of the change was studied by sequencing the same exon from 23 to 51 healthy controls except in the case of the deletion carried by Family AM which was checked from 101 healthy controls. In addition, the healthy members of Family AM were sequenced as controls.
In addition, Blueprint Genetics was commissioned to carry out a Nephrotic Syndrome sequencing panel of nine genes (http://blueprintgenetics.com/nephrotic-syndrome-panel/) to four healthy and three affected members of Family AM. While the panel included NPHS1, NPHS2, ACTN4 and TRPC6 that were previously sequenced it also contained all the rest of the INF2 exons as well as genes CD2AP, LAMB2, PLCE1 and WT1. The panel targets all exons, intron/exon boundaries and known mutations outside these regions.
The effects of amino acid changes were evaluated using online bioinformatics tools Predict Protein server [9], Prosite database [10] and PolyPhen [11].
Whole genome sequencing (WGS) was carried out in two affected (the first affected in the family, II-1 and her grand daughter, IV-2) and two healthy (I-1 and II-2) members of the family AM in the Finnish Institute of Molecular Medicine (www.fimm.fi, University of Helsinki, Biomedicum, Helsinki Finland). The aim was to look for sequence variants where the genotypes of the affected differ from the healthy in accordance with dominant inheritance pattern. The analysis revealed over 100,000 different type of variants spread across all autosomal chromosomes and chromosome X. We concentrated on variants in the coding and non-coding areas of genes associated with SRNS in the literature. As the disease in Family AM shows no extra-renal expression we focused our efforts on genes causing isolated SRNS including NPHS2, NPHS1, INF2, CD2AP, WT1, TRPC6, ACTN4, PLCE1, MYH9, Myo1E and ARHGAP24. The WGS data analysis was conducted using Integrative Genome Viewer (IGV) (www.broadinstitute.org/igv/).
Plasmids
Wild-type (WT) and c.988_989delCT-mutated NPHS2 sequences were inserted into pCMV-Myc–vector (Clontech, Palo Alto, CA, USA) to give them N-terminal Myc-tag. The WT-NPHS1 was inserted into pcDNA3.1–vector (Invitrogen, Carlsbad, CA, USA). Plasmids were amplified using JM109 competent E. coli cells, QIAGEN Plasmid Maxi Kit (QIAGEN inc. Valencia, CA, USA) and the protocol provided by the kit.
Co-immunoprecipitation of nephrin and podocin
HEK293FT cells were transiently transfected with two plasmids: one carrying WT-nephrin, the other carrying either WT-podocin or c.988_989delCT-mutated podocin. The transfection was carried out using FuGENE 6–protocol (Roche, Basel, Switzerland). After incubating for 24 h, the cells were washed in PBS and lysed in non-denaturing lysis buffer containing 20 mM TrisHCl pH8, 139 mM NaCl, 10 % glycerol, 1 % NP40, 2 mM EDTA, and phosphatase and proteinase inhibitors. Cell lysates were incubated with c-Myc (A-14) antibody (Santa Cruz Biotechnology Inc., Santa Cruz, CA, USA) over night at +4 °C on gentle agitation. After this, 50 μl 50 % rec Protein G-Sepharose beads (Invitrogen, Carlsbad, CA, USA) were added to the antibody–protein complex mix and incubated over night at +4 °C on gentle agitation to bind the antibody to the beads. Beads were washed with the lysis buffer and fractioned by SDS-PAGE. Western blot analysis was performed using nephrin antibody (Santa Cruz Biotechnology Inc., Santa Cruz, CA, USA) followed by the incubation with horseradish peroxidase-coupled anti-goat antibody. Immobilized antibodies were detected using chemiluminescence.
Immunofluorescence staining
Antibodies used for immunofluorescence staining of cryosections of the kidney tissue samples were goat polyclonal antibody against the C terminus of the human podocin C-18 (sc-22296, Santa Cruz Biotechnology, Santa Cruz, CA), goat polyclonal antibody against the N terminus of the human podocin N-21 (sc-22294, Santa Cruz Biotechnology, Santa Cruz, CA), and rabbit polyclonal antibody against human nephrin (UP3) (by courtesy of Karl Tryggvason lab, Karolinska Institute, Stockholm, Sweden). The staining was carried out in the traditional method using snap-frozen, acetone-fixed cryosections (5 μm) of kidney tissue. Samples included thin needle biopsy of affected family member (IV-1 in Fig. 1), nephrectomised tissue from congenital nephrotic syndrome of the Finnish type (CNF) patient and normal human kidney from adult donors as control. Light microscopy was performed with a standard Leica DM RX light microscope equipped with an Olympus DP70 digital camera.
The binding of the antibodies C-18 and N-21 to truncated podocin was confirmed using Western blot analysis (Fig. 2a).
Analysis of wild-type and c.988_989delCT podocin of patient IV-1. a Binding of antibodies recognizing the N terminus (N21) and C terminus (C18) of podocin to WT and c.988_989delCT podocin in transfected HEK293FT lysates. a WT-podocin. b c.988_989delCT podocin. b Co-Immunoprecipitation of podocin and nephrin visualized with α-nephrin antibody. IP Immunoprecipitation. TP Total protein. a Lysates of HEK 293FT. b Lysates of HEK 293FT cells transfected with WT-podocin. c Lysates of HEK 293FT cells transfected with c.988_989delCT podocin. d Lysates of HEK 293FT cells transfected with nephrin. e Lysates of HEK 293FT cells transfected with both WT-podocin and nephrin. f Lysates of HEK 293FT cells transfected with both mutated podocin and nephrin
Results
Clinical findings in the familial SRNS
Clinical findings of the affected members of Family AM are cataloged in Table 1. In four of the five affected family members, mild proteinuria appeared during early childhood. The fifth affected (II-1 in Fig. 1) was diagnosed in a routine checkup in the age of 18 years, as her proteinuria had been symptomless. In two of the five affected (II-1, III-2), the proteinuria slowly increased leading to ESRD in adulthood. The kidney function of third affected (III-3) has been worsening but he has yet to reach ESRD, and the other two affected (IV-1, IV-2) are still children. In biopsy samples taken during childhood or when the proteinuria was still mild (II-1, III-2, IV-1), the histology of the affected shows only MCNS while biopsies taken after the kidney function has started to deteriorate show FSGS (II-1, III-3). All patients were given corticosteroid medication with little effect. Other members of Family AM (I-1, I-2, II-2, III-1) are healthy with no signs of kidney disease.
NPHS2 in the familial NS
Genetic analysis was performed on five cases and four healthy family members (Fig. 1). Sequencing of the podocin gene (NPHS2) revealed a heterozygous de novo deletion in exon 8 (c.988_989delCT) in all affected cases and in none of the healthy family members or in the unrelated controls. To see if a second mutation would lie outside the analyzed exons and exon/intron boundaries, NS panel of nine genes was commissioned. In NPHS2, an intronic variant (rs115778946) was discovered in all three affected family members included in the analysis and in one healthy family member. However, the first affected family member (II-1) has inherited this invariant from her mother and passes it on to her children together with c.988_989delCT. The father of her affected children (II-2) and the mother of her affected grandchildren (III-1) do not carry it. Thus, it appears that both found variants lie in the same allele of the gene and all affected have one allele of NPHS2 with no disease-causing variants. The WGS of two affected and non-affected family members was carried out to reveal variants that are unreachable for the gene panel. Only a few non-coding variants were found and these did not follow such inheritance pattern that could explain the phenotype.
c.988_989delCT results in a 343 aa polypeptide instead of the 382 aa wild type. The Prosite database calculated that the mutated sequence lacked one predicted N-glycosylation site (from aa 355: NRTQ), two protein kinase C phosphorylation sites (from aa 332: TEK and from aa 354: SNR) and a cAMP- and cGMP-dependant protein kinase phosphorylation site (from aa 377: KKDS).
Co-immunoprecipitation of normal and c.988_989delCT–podocin and nephrin showed that the truncated podocin bound to nephrin similar to the normal podocin or slightly more strongly (Fig. 2b). Immunohistochemistry for nephrin in biopsy sample of IV-1 showed that while the protein was expressed in kidney glomeruli, the staining pattern was uneven and discontinuous. The staining of podocin was done using two antibodies, one staining the N terminus of the protein and the other the C terminus carrying the variant. The epitope recognized by the C-terminal antibody C-18 is partially disrupted by the c.988_989delCT variant, and thus the ability of C-18 to bind mutated podocin was tested with Western blot analysis from the transfected HEK293 lysates. Figure 2 A shows that the C-18 binding of c988_989delCT podocin is similar to WT-podocin. The immunostaining of podocin shows that while the C-terminal antibody stains slightly less than the N-terminal antibody in c.988_989delCT mutated glomeruli, the staining of both is greatly reduced when compared to the glomeruli of normal kidney tissue (Fig. 3).
Immunofluorescence staining of podocin and nephrin. Images magnified ×20. a Podocin staining in normal human kidney by N21 antibody. b Podocin staining in CNF kidney by N21 antibody. c Podocin staining in c.988_989delCT-mutated kidney of patient IV-1 by N21 antibody. d Podocin staining in normal human kidney by C18 antibody. e Podocin staining in CNF kidney by C18 antibody. f Podocin staining in c.988_989delCT-mutated kidney of patient IV-1 by C18 antibody. g Nephrin staining in normal human kidney. h Nephrin staining in CNF. i Nephrin staining in c.988_989delCT-mutated kidney
Other podocyte genes in the familial NS
Sequencing of the podocyte protein coding genes NPHS1, Neph1 and ACTN4 uncovered only known polymorphisms. Similarly, none of the patients had variants leading to aa changes in INF2 exons 2 and 4, where majority of the mutations leading to dominant FSGS are clustered [12].
The NS panel of Blueprint Genetics did not find any variant in any gene that would have followed the disease phenotype. The WGS revealed one variant that was shared by the two analyzed affected family members (II-1 and IV-2) but not the two healthy ones. Direct sequencing of this non-coding variant, a nucleotide deletion (rs76500597) in WT1, in rest of the family members revealed no shared genotype between the affected individuals: one affected did not carry the deletion while two healthy family members did (one as a homozygote). All sequence variants of Family AM in SRNS-associated genes can be viewed in Table 2.
Genes in non-familial cases of SRNS
One of the twelve sporadic SRNS cases in the study had aa changing variants in NPHS2, p.P341R in one allele and p.R229Q in two alleles. SRNS was diagnosed in this patient at the age of six and she is now waiting for the kidney transplant at the age of 19 years.
In the NPHS1 gene, six of the eleven patients had one or more of three known amino acid changing polymorphisms (p.E117K, p.R402Q, p.N1077S), which were also displayed in the healthy controls in similar frequencies. No patient carried amino acid changing variants in the NEPH1 gene. A few polymorphisms, which did not lead to amino acid changes, were detected in all three genes.
Discussion
We describe a family with an exceptional clinical course of NS. In the affected family members, proteinuria started in the newborn period but remained moderate up to two decades of life until escalating in association with progressive renal failure. ESRD was reached at the age of 25–40 years. While the early kidney biopsies showed MCNS, FSGS histology became evident after proteinuria worsened. All affected patients had a novel heterozygous deletion c.988_989delCT in the podocin gene (NPHS2), which was not present in their healthy family members, in controls or in a cohort of non-familial Finnish SRNS patients. No other mutation was found in NPHS2 (including non-coding regions) or in other SRNS-related genes.
The NPHS2 variants cause autosomal recessive form of SRNS [13, 14]. Patients with two NPHS2 variants usually develop SRNS from birth to 6 years of life and reach ESRD before the end of the first decade [15]. However, phenotypic variability is associated with particular NPHS2 variants. In patients with p.R229Q in one allele and another disease-causing variant in the other allele, SRNS often starts after the age of 17 years [13, 15, 16]. In cases where only one heterozygous alteration has been identified the disease has started later [2]. It has been assumed the second variant is intronic or in the promoter, and thus escapes detection in the sequencing of the exons.
This seems not to be the case in the family presented here. The disease follows dominant inheritance pattern in three consecutive generations. Undetected variant or variants would have to come into the family from three unrelated sources. The c.988_989delCT is de novo variant in II-1 that occurs in the family at the same time as the disease phenotype appears and then co-segregates perfectly through generations. This suggests that it has a significant role in the NS in the family.
In the literature, other small frameshift variations in NPHS2 exon 8 have been described [2, 17–21]. Most of these appear as heterozygotes or compound heterozygotes and thus their specific function is difficult to determine. McCarthy et al. [20] reported two FSGS patients with heterozygous exon 8 variant F344fs as well as heterozygous R229Q, and the course of their disease shares similarities with the course of the disease in patients from Family AM with early onset (11 months and 2 years) and only moderate reduction in kidney function after over a decade long follow-up time (13 and 11 years). The effect of R229Q to the phenotype is unclear as its pathogenicity escapes precise definition. Interestingly, Bouchireb et al. [21] described a novel small deletion at almost the same location as c.988_989delCT. The deletion c.989_992del (p.Leu330ProfsX17) was found in one patient as a heterozygote and it is the only variant this patient is reported to have [Leiden Open Variation Database (LOVD 3.0:www.lovd.nl/NPHS2)]. As clinical data concerning the course of disease of this patient are not reported, it is impossible to ascertain if this patient expresses a similar phenotype to the patients in Family AM.
The NPHS2 deletion c.988_989delCT results in frameshift after amino acid 329. The altered protein gains a novel fourteen amino sequence before a stop codon appears truncating the protein to 343 amino acids instead of the 384 amino acids of the WT-podocin. The truncated podocin had quite normal or slightly elevated affinity to the major slit diaphragm protein nephrin but, in immunofluorescence staining of glomeruli of an affected family member, the expression of nephrin was affected both quantitatively and qualitatively. The Predict Protein revealed that the two- and three-dimensional structure of the podocin was not drastically altered by c.988_989delCT. The protein–protein interactions are often facilitated by amino acid residue proline and c.988_989delCT erases nine proline residues. The truncated podocin may have difficulty to interact with other proteins the same way the WT does.
It is probable that the c.988_989delCT is a gain-of-function variant. These have different mechanisms: hypermorphs increase the activity of the mutated protein, as is the case in ACTN4 and its disease-causing variants, neomorphs give the mutated protein a new function separate from the WT-protein, and antimorphs tweak the proteins’ function to be antagonistic to the WT. The antimorphs are also known as the dominant negative mutations.
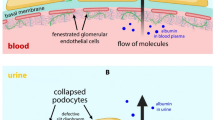
The immunofluorescent staining of podocin was notably reduced in glomeruli of the affected tissue compared to WT. This was seen with both N- and C-terminal antibodies. The amount of reduction leads to assume that it is not only due to c.988_989delCT podocin failing to express properly but that the expression of the WT copy is also hampered. This would suggest that the variant has a dominant negative effect, and it could also explain the altered staining of nephrin, as podocin plays a role in anchoring the protein to the cell membrane. However, the underlying mechanism is unknown and requires more studying and, what the effect of c.988_989delCT may be, its negative impact on the glomerular filtration barrier is likely to be a modest one, leading to the exceptionally slow progression of the disease.
In dominantly inherited SRNS, candidate genes include ACTN4, TRPC6, INF2, WT1, CD2AP and ARHGAP [22–27]. We explored the possibility that the causative mutation would be found in these. None of the variants in these genes followed the disease phenotype.
Besides NPHS2, we also included other genes that cause recessive form of NS to the study. No disease phenotype following variant, homozygous or compound heterozygous, was found in NPHS1, PLCE1, LAMB2, Myo1E or MYH9. As the disease in all affected members of Family AM was unambiguously limited to the kidney, we did not look for variants in genes causing disease characterized by extrarenal manifestations. As genome-wide sequencing was a method used in this study, it should be noted that the weakness of this method is its difficulty in identifying large deletions, insertions and complex rearrangements. It is possible these play a role in the genetic background of NS in this family.
The analysis of NPHS1, NPHS2 and NEPH1 in the cohort of Finnish pediatric patients with sporadic SRNS showed very little genetic variation. While Neph1 is associated to proteinuria only in animal models [28] and NPHS1 mutations are mainly found in congenital NS, previous studies have found frequency of disease-causing NPHS2 variants in sporadic SRNS to be 10–28 % [3, 19]. In the cohort included to this study, there was only one patient with potentially disease-causing variants, homozygous p.R229Q and novel variant p.P341R in NPHS2. This is slightly less than anticipated. Frequencies of disease-causing variants in the genes behind SRNS are known to differ between populations [1, 2, 29]. SRNS is relatively rare in Finland and it is possible that lack of NPHS2 variation is the reason behind this. However, the cohort size in this study is insufficient to draw conclusions, and larger scale study must be conducted to clarify the matter. The analysis of NPHS2 variants in sporadic SRNS does, however, act as a type control for the familial SRNS discussed in this article, underlying the rareness of their variant.
This paper outlines an unusual phenotype of familial NS. Although the disease mechanism is not clear we offer evidence that suggests that a small heterozygous deletion in the NPHS2 gene plays a significant role in its pathogenesis. It seems that not only the altered gene but also the nature of the alteration itself may have an effect on the character of the disease.
References
Hinkes BG, Mucha B, Vlangos CN, et al. Nephrotic syndrome in the first year of life: two thirds of cases are caused by mutations in 4 genes (NPHS1, NPHS2, WT1, and LAMB2). Pediatrics. 2004;119:e907–19.
Hinkes B, Vlangos C, Heeringa S, et al. Specific podocin mutations correlate with age of onset in steroid-resistant nephrotic syndrome. J Am Soc Nephrol. 2008;19:365–71.
Karle SM, Uetz B, Ronner V, et al. Novel mutations in NPHS2 detected in both familial and sporadic steroid-resistant nephrotic syndrome. J Am Soc Nephrol. 2002;13:388–93.
Kestilä M, Lenkkeri U, Männikkö M, et al. Positionally cloned gene for a novel glomerular protein–nephrin—is mutated in congenital nephrotic syndrome. Mol Cell. 1998;1:575–82.
Philippe A, Nevo F, Esquivel EL, et al. Nephrin mutations can cause childhood-onset steroid-resistant nephrotic syndrome. J Am Soc Nephrol. 2008;19:1871–8.
Santín S, Bullich G, Tazón-Vega B, et al. Clinical utility of genetic testing in children and adults with steroid-resistant nephrotic syndrome. Clin J Am Soc Nephrol. 2011;6:1139–48.
Bierzynska A, Soderquest K, Koziell A. Genes and podocytes—new insights into mechanisms of podocytopathy. Front Endocrinol (Lausanne). 2015;5:226.
Lenkkeri U, Männikkö M, McCready P, et al. Structure of the gene for congenital nephrotic syndrome of the finnish type (NPHS1) and characterization of mutations. Am J Hum Genet. 1999;64:51–61.
Rost B, Yachdav G, Liu J. The PredictProtein server. Nucleic Acids Res. 2004;1(32):W321–6.
Bairoch A, Bucher P, Hofmann K. The PROSITE database, its status in 1997. Nucleic Acids Res. 1997;1(25):217–21.
Adzhubei IA, Schmidt S, Peshkin L, et al. A method and server for predicting damaging missense mutations. Nat Methods. 2007;7:248–9.
Gbadegesin R, Hinkes B, Vlangos C, et al. Mutational analysis of NPHS2 and WT1 in frequently relapsing and steroid-dependent nephrotic syndrome. Pediatr Nephrol. 2007;22:509–13.
Tonna SJ, Needham A, Polu K, et al. NPHS2 variation in focal and segmental glomerulosclerosis. BMC Nephrol. 2008;29:9–13.
Relle M, Cash H, Brochhausen C, et al. New perspectives on the renal slit diaphragm protein podocin. Mod Pathol. 2011;24:1101–10.
Machuca E, Hummel A, Nevo F, et al. Clinical and epidemiological assessment of steroid-resistant nephrotic syndrome associated with the NPHS2 R229Q variant. Kidney Int. 2009;75:727–35.
Tsukaguchi H, Sudhakar A, Le TC, et al. NPHS2 mutations in late-onset focal segmental glomerulosclerosis: R229Q is a common disease-associated allele. J Clin Invest. 2002;110:1659–66.
Berdeli A, Mir S, Yavascan O, et al. NPHS2 (podocin) mutations in Turkish children with idiopathic nephrotic syndrome. Pediatr Nephrol. 2007;22:2031–40.
Mbarek IB, Abroug S, Omezzine A. Novel mutations in steroid-resistant nephrotic syndrome diagnosed in Tunisian children. Pediatr Nephrol. 2011;26:241–9.
Santín S, Tazón-Vega B, Silva I, et al. Clinical value of NPHS2 analysis in early- and adult-onset steroid-resistant nephrotic syndrome. Clin J Am Soc Nephrol. 2011;6:344–54.
McCarthy HJ, Bierzynska A, Wherlock M, et al. Simultaneous sequencing of 24 genes associated with steroid-resistant nephrotic syndrome. Clin J Am Soc Nephrol. 2013;8:637–48.
Bouchireb K, Boyer O, Gribouval O, et al. NPHS2 mutations in steroid-resistant nephrotic syndrome: a mutation update and the associated phenotypic spectrum. Hum Mutat. 2014;35:178–86.
Kaplan JM, Kim SH, North KN, et al. Mutations in ACTN4, encoding alpha-actinin-4, cause familial focal segmental glomerulosclerosis. Nat Genet. 2000;24:251–6.
Winn MP, Conlon PJ, Lynn KL, et al. A mutation in the TRPC6 cation channel causes familial focal segmental glomerulosclerosis. Science. 2005;308:1801–4.
Brown EJ, Schlöndorff JS, Becker DJ, et al. Mutations in the formin gene INF2 cause focal segmental glomerulosclerosis. Nat Genet. 2010;42:72–6.
Kim JM, Wu H, Green G, et al. CD2-associated protein haploinsufficiency is linked to glomerular disease susceptibility. Science. 2003;23(300):1298–300.
Gbadegesin RA, Lavin PJ, Hall G, et al. Inverted formin 2 mutations with variable expression in patients with sporadic and hereditary focal and segmental glomerulosclerosis. Kidney Int. 2012;81:94–9.
Akilesh S, Suleiman H, Yu H, et al. Arhgap24 inactivates Rac1 in mouse podocytes, and a mutant form is associated with familial focal segmental glomerulosclerosis. J Clin Invest. 2011;121:4127–37.
Donoviel DB, Freed DD, Vogel H, et al. Proteinuria and perinatal lethality in mice lacking NEPH1, a novel protein with homology to NEPHRIN. Mol Cell Biol. 2001;21:4829–36.
Franceschini N, North KE, Kopp JB, et al. NPHS2 gene, nephrotic syndrome and focal segmental glomerulosclerosis: a HuGE review. Genet Med. 2006;8:63–75.
Acknowledgments
We thank Tuike Helmiö for excellent technical assistance.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
This work was supported by grants from the Sigrid Juselius Foundation, the Pediatric Research Foundation and Helsinki University Central Hospital Research Fund.
Ethical approval
All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee at which the studies were conducted (IRB: HUS 509/E7/05) with the 1964 Helsinki declaration and its later amendments or comparable ethical standards.
Informed consent
Informed consent was obtained from all individual participants (or their parents) included in the study.
Conflict of interest
The authors have declared that no conflict of interest exists.
About this article
Cite this article
Suvanto, M., Patrakka, J., Jahnukainen, T. et al. Novel NPHS2 variant in patients with familial steroid-resistant nephrotic syndrome with early onset, slow progression and dominant inheritance pattern. Clin Exp Nephrol 21, 677–684 (2017). https://doi.org/10.1007/s10157-016-1331-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10157-016-1331-3