Abstract
Background
Steroid-resistant nephrotic syndrome (SRNS) is a genetically heterogeneous disorder for which more than 25 single-gene hereditary causes have been identified.
Methods
Whole exome sequencing was performed in a 3-year-old girl with SRNS. We analyzed the expression of Crb2 and slit diaphragm molecules in the patient’s glomeruli, and compared it with that of controls or other nephrotic patients.
Results
Whole-exome analysis identified novel compound heterozygous mutations in exons 10 and 12 of CRB2 (p.Trp1086ArgfsX64 and p.Asn1184Thr, each from different parents; Asn1184 within extracellular 15th EGF repeat domain). Renal pathology showed focal segmental glomerulosclerosis with effaced podocyte foot processes in a small area, with significantly decreased Crb2 expression. Molecules critical for slit diaphragm were well-expressed in this patient’s podocytes. Crb2 expression was not altered in the other patients with congenital nephrotic syndrome with NPHS1 mutations.
Conclusions
These findings demonstrate that Crb2 abnormalities caused by these mutations are the mechanism of steroid-resistant NS. Although CRB2 mutations previously found in SRNS patients have been clustered within the extracellular tenth EGF-like domain of this protein, the present results expand the variation of CRB2 mutations that cause SRNS.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Idiopathic nephrotic syndrome (NS) is the most common form of NS in children [1]. Some 80–90 % of patients with childhood-onset NS are steroid-sensitive; this is called minimal change disease, and its renal biopsy findings show only diffuse foot process effacement on electron microscopy. Steroid-resistant NS (SRNS) is the second most common chronic kidney disease in the first three decades of life, and renal prognosis is generally poor with end-stage renal failure (ESRF) occurring in 30–40 % of patients [2]. The etiology and pathogenesis of SRNS have been a conundrum for decades; however, the single-gene cause can be found in some SRNS patients. To date, more than 25 single-gene causes of hereditary NS have been identified; they can display either dominant or recessive inheritance [3]. Very recently, next-generation sequencing technology discovered that a causative mutation can be detected in 30 % of all individuals who develop SRNS before 25 years of age [4]. As genetic alterations are associated with a poor response to immunosuppressive agents in children with SRNS [5, 6], genetic diagnosis of SRNS can offer a cause-based diagnosis and permit personalized treatment options for SRNS.
The family of Crumbs proteins, homologous to Drosophila Crumbs (Crb), contain large extracellular domains, including epidermal growth factor (EGF)-like and laminin G-like domains, a single transmembrane domain, and a cytoplasmic tail that contains single FERM and PDZ protein-binding motifs. The longest transcript of CRB2 (RefSeq NM_173689 [MIM 609720]) has 13 coding exons. In 2015, several CRB2 mutations were found in patients with SRNS [7]. Three of the four missense mutations (p.Cys620Ser, p.Arg628Cys, and p.Cys629Ser) identified in SRNS occur within exon 7 of CRB2, which encodes the extracellular tenth EGF-like domain of this protein [7]. Biallelic CRB2 mutations consistent with autosomal-recessive inheritance were also found in families who shared a phenotype comprising cerebral ventriculomegaly and echogenic kidneys with histopathological findings of congenital nephrosis [8].
Recent data resulting from morpholino-induced knockdown of zebrafish crb2b demonstrated some of the possible mechanisms of Crb2 in glomerular filtration. The crb2b −/− homozygotes resulted in the disappearance of slit diaphragms and decreased ZO-1 expression, which suggests that differentiation and protein trafficking of slit components are affected in crb2b mutants [7, 9]. However, pathological analysis of the morphological changes of podocytes or the expression pattern of Crb2 in patients with CRB2 mutations has not been reported, and the effect of pathogenic CRB2 mutations on the expression of other molecules in human podocytes is undetermined.
Here, we report a Japanese girl who presented with pneumococcal meningitis and SRNS in whom we found novel compound heterozygosity for two deleterious sequence variants in CRB2: a frameshift mutation and a missense mutation that predicted p.Asn1184Thr. Here we analyzed Crb2 expression and molecules at podocyte filtration slits in normal kidneys and the patient’s kidney. The data showed that Crb2 expression is specifically affected in these patients, indicating the pathogenicity of the mutations.
Materials and methods
Whole exome analysis and CRB2 gene analysis
Genomic DNA was extracted from peripheral white blood cells of the patient and parents using a QIAamp DNA Blood Midi Kit (Qiagen, Hilden, Germany). Exome sequences were enriched using a SureSelect v5+UTRs Kit (Agilent Technologies, Santa Clara, CA, USA) from 3 μg of genomic DNA, according to the manufacturer’s instructions. The captured DNA samples were subjected to massively parallel sequencing (100-bp paired-end reads) on an Illumina HiSeq2000 sequencing system (Illumina, Santiago, CA, USA). Short reads were aligned to the reference genome (hg19) using Burrows–Wheeler alignment (BWA) [10] with default parameter settings. Single-nucleotide variants and short insertions/deletions were called using SAMtools [11] with default parameter settings. Sanger sequencing was performed to detect CRB2 and to validate the presence of each variant detected by exome sequencing in the patient and her Japanese parents. The entire coding region and exon–intron boundaries of exons 10 and 12 of the CRB2 gene were amplified from the genomic DNA using polymerase chain reaction (PCR). The primers for exon 10 were 5′-GAGAACTTCACCGGCTGCTTG-3′ and 5′-GACTTCTCTGCCCCACCATA-3′. The primers for exon 12 were 5′-GGGACAGTGGATGGATAAGC -3′ and 5′- ATGACAGAGTGGCCCAGGAAC -3′.
Histological analysis
Tissue for light microscopy was collected and processed routinely whereas tissue for direct immunofluorescence was stained, utilizing fluorescein-tagged antibodies against immunoglobulin G (IgG), IgA, and IgM, and complement proteins C1q, C3, and C4. Biopsy tissue for electron microscopy was routinely fixed. Immunohistological analysis of podocyte protein expression was performed as follows. Paraffin-embedded samples from human renal biopsy samples were deparaffinized in xylene and rehydrated through an ethanol–H2O gradient, followed by heat-induced epitope retrieval by incubating in a target retrieval solution (S1699; Dako, Carpinteria, CA, USA) for 20 min at 121 °C. Sections were cooled to room temperature and incubated with the primary antibodies, followed by incubation with Alexa Fluor conjugated secondary antibodies (Invitrogen, Carlsbad, CA, USA). Images were obtained using a confocal microscope (model FV300; Olympus, Tokyo, Japan) and were processed using Adobe Photoshop CS6. The following antibodies were obtained commercially: rabbit polyclonal anti-CRB2 (Sigma-Aldrich, St Louis, MO, USA), mouse monoclonal anti-zonula occludens-1 (ZO-1; 33–9100; Invitrogen, Carlsbad, CA, USA), and rabbit polyclonal anti-synaptopodin (Meridian Life Science, Memphis, TN, USA). Rabbit polyclonal anti-nephrin IgG and rabbit polyclonal anti-podocin have been described previously [12, 13]. Control samples (donor kidney or biopsy samples from patients with minimal change disease obtained during remission periods) were stained at the same time. Kidney samples from patients with congenital nephrotic syndrome have also been described previously [13].
Preabsorption of Crb2 antibody
HEK293T cells were purchased from the ATCC (Manassas, VA, USA) and maintained in DMEM containing 10 % fetal bovine serum. Transfections were performed using Polyethylene-imine Max (PEI-Max) reagent (Polysciences, Warrington, PA, USA) following the manufacturer’s instructions. Cells were lyzed in a lysis buffer (20 mM Tris–HCl [pH 7.5], 150 mM NaCl, 1 mM EDTA, and 1 % NP-40) containing a protease inhibitor cocktail (Roche, Germany) for 15 min on ice. Lysates were clarified by centrifugation and incubated with agarose beads conjugated with anti-FLAG M2 antibody for 1 h at 4 °C. The diluted primary antibody for Crb2 was incubated with the immunoprecipitates from cells transfected with control vector or flag-Crb2 for antibody absorption.
Results
The patient was a 3-year-old girl with fever, edema, and macroscopic hematuria. She had an unremarkable birth and family history and no neurological or cardiac defects had been noted in her past regular medical checkups. On systemic examination, she had generalized edema and a depressed level of consciousness with the Glasgow Coma Scale of 10 (E3, V3, M4). Neck rigidity was present and Kernig’s sign was positive. Examination of the cerebrospinal fluid (CSF) revealed 2,384 cells/mm3; 97 % polymorphs, protein 189 mg/dL and sugar 0 mg/dL. CSF culture showed growth of Streptococcus pneumoniae. She had normal kidney function with blood urea nitrogen of 9.6 mg/dl and creatinine of 0.12 mg/dl. Urinalysis revealed proteinuria at 10.3 g/g creatinine, with urine sediments of 5–9 red blood cells per high power field. She was diagnosed with nephrotic syndrome with bacterial meningitis, and started on antibiotics, which successfully treated her meningitis. Thereafter, she received prednisolone (30 mg/day), but did not respond to a 4-week treatment. Thereafter, immunosuppressive therapy consisting of methylprednisolone pulse therapy and cyclosporine, and finally rituximab did not affect her massive proteinuria. She failed to achieve clinical remission after 5 months’ follow-up.
We hypothesized that this sporadic steroid-resistant NS was caused by mutations in known or novel nonsynonymous coding variants. To identify the causal variant, we then performed exome sequencing using next-generation sequencing for this patient. The mean sequencing coverage was 95 million reads, with 98.8 % of the total bases mapped to the reference genome, which encompassed 89.8 % of the targeted regions with coverage >10×. Among gene mutations reported to cause NS, two heterozygous mutations of CRB2 were found in exons 10 and 12 (Fig. 1). Direct sequencing of CRB2 confirmed that the patient had compound heterozygous mutations in CRB2 (Fig. 1a–c): c.3551A>C (predicting p.Asn1184Thr) was inherited from her healthy father and c.3256_3273del18insCGGCCCGGGGTGG (predicting p.Trp1086ArgfsX64) from her healthy mother. These mutations had not been registered in the database of single-nucleotide polymorphisms (SNPs) of the National Center for Biotechnology Information (dbSNP, www.ncbi.nlm.nih.gov), in the Exome Aggregation Consortium (ExAC) [14] or in the Japanese SNP control database established by the National Bioscience Data Base Center, which has genome-wide data for a million SNPs from 700 samples (http://gwas.biosciencedbc.jp/snpdb/snp_top.php). Furthermore, the absence of this DNA sequence abnormality in 1,240 alleles from 620 unrelated healthy Japanese individuals indicated that they are mutations and not polymorphisms. Asn1184 is within an extracellular 15th EGF repeat domain, and is highly conserved across various species (Fig. 1d). The functionality of p.Asn1184Thr was analyzed using the Sorting Intolerant From Tolerant (SIFT) web-based tool (http://sift.jcvi.org), the Polymorphism Phenotyping 2 (PolyPhen2) tool (http://genetics.bwh.harvard.edu/pph2), and Mutation Taster (http://www.mutationtaster.org) by homology modeling and threading. All three models predicted CRB2 p.Asn1184Thr to be “damaging.”
Genetic analysis of steroid-resistant nephrotic syndrome (SRNS) with CRB2 mutation. a Patient’s familial pedigree. Red: mutant alleles, WT wild-type allele. Sequence chromatograms for portions of b exon 10 and c 12 of CRB2 for the patient and her parents. The patient has compound heterozygous mutations c.3256_3273del18insCGGCCCGGGGTGG and c.3551A>C. d Protein alignment plot of Crb2 amino acid sequence for residues 1180–1188 in exon 12 of CRB2 (arrowhead), with complete conservation of the Asn1184 locus
A renal biopsy sample of the patient revealed focal segmental glomerulosclerosis (FSGS) by light microscopy (Fig. 2). Segmental sclerosis was evident in the perihilar area of one glomerulus, indicating FSGS, perihilar variant. No specific deposition of IgG, IgA or IgM, or of complement proteins C1q, C3, and C4, was detected by direct immunofluorescence. Electron microscopy revealed that the foot processes of podocytes were well retained, with the effacement of these processes in only a small area, indicating the secondary form of FSGS.
Kidney histopathology of the affected individual with biallelic CRB2 mutation. a On observation using light microscopy, the biopsy specimen included 21 glomeruli, many of which showed minor glomerular abnormalities (PAM stain, ×200). b Two glomeruli were obsolescent (PAM stain, ×600), and c three glomeruli showed expansion of mesangial areas with accumulation of matrix (PAS stain, ×600). d The perihilar type of segmental sclerosis was noted in one glomerulus (Masson stain, ×600), indicating the perihilar variant of focal segmental glomerulosclerosis. In podocytes, electron microscopy showed relatively well preserved foot processes, with the effacement of these processes in small areas (arrows in e, ×5,000, and f, ×8,000), indicating the secondary form of focal segmental glomerulosclerosis. Focal widening of subendothelial spaces was shown in the segmental capillary wall (arrowhead in e)
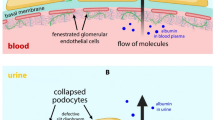
Next, we investigated Crb2 expression in human kidneys by using polyclonal antibodies against the extracellular part (amino acids 720–802) of human Crb2. Immunohistochemical staining of adult human kidney tissues (donor kidney) revealed intense signals for Crb2 in normal glomeruli along with glomerular capillary loops and parietal glomerular epithelial cells (Fig. 3a, b). These signals disappeared when antibody was pre-absorbed with anti-FLAG immunoprecipitates from cell lysates of HEK293 cells that express FLAG-tagged Crb2 (Fig. 3c–e), indicating the specificity of the signals. To determine the localization of Crb2 in glomeruli, paraffin-embedded sections of the adult human kidney were double-labeled with Crb2 and ZO-1, a podocyte marker. Intense signals for Crb2 in normal glomeruli were observed along with glomerular capillary loops, partially colocalizing with ZO-1 (Fig. 4). Notably, Crb2 staining was also observed outside the linear staining area marked by ZO-1. Next, we analyzed expressions of Crb2, ZO-1, synaptopodin, nephrin, and podocin in kidneys from the patient with Crb2 mutations and compared with those from control samples. Staining of Crb2 was undetected in glomeruli of the SRNS patient with CRB2 mutations (Fig. 4), but expressions of ZO-1, synaptopodin, and slit diaphragm proteins (nephrin and podocin) were retained in her podocytes (Figs. 4, 5). We also analyzed Crb2 expression in two congenital nephrotic syndrome (CNS) patients with NPHS1 mutations (CNS1: heterozygous missense mutation [P676R]; CNS2: homozygous mutation [Q839RfsX849]) [13]. Clear signals for Crb2 that demarcated glomerular capillaries were observed in glomeruli from the CNS patients with NPHS1 mutations (Fig. 6).
Crb2 expression in the kidney cortex. Human kidney sections were stained with anti-Crb2 antibody pre-adsorbed with anti-myc immunoprecipitates from lysates of HEK293T cells that expressed a, c control vector or b, d myc-tagged Crb2. Strong Crb2 signals were detected along glomerular capillary loops and in parietal epithelial cells (a and c), with specific signals detected in the vascular endothelium (e). Scale bar: 50 μm. e Lysates of HEK293T cells expressing control vector or Crb2 vector were immunoprecipitated with anti-FLAG antibody and analyzed by western blot with indicated antibodies. These immunoprecipitates were used for antibody absorption (a–d)
These results support the pathogenic role of CRB2 mutations in exons 10 and 12 in the patient, and imply that specific disruption of Crb2 expression causes podocyte dysfunction, which leads to massive proteinuria.
Discussion
In the present study, we identified novel CRB2 mutations in a girl with SRNS that was resistant to cyclosporine and rituximab. Immunohistochemical studies showed specific reduced expression of Crb2 in the patient’s podocytes, indicating the pathogenic role of the mutations. After identifying the genetic cause, immunosuppressive therapy was discontinued and supportive treatment was continued.
Recently, large international cohorts were studied for monogenic causes of childhood SRNS, in which single-gene mutations associated with SRNS were identified in ∼25–33 % of all families with SRNS [4, 5, 15, 16]. The earlier the age at onset, the more likely the causative mutations can be detected: single-gene causes of SRNS were detected in 61 % of the families of patients studied whose onset occurred in the first 12 months of life, compared with 25.3 % of families of patients whose disease began at age 1–6 years [4]. Although mutations in dominant genes such as INF2 and TRPC6 are more frequently found in early adulthood [4], mutations of recessive genes (NPHS1, LAMB2, and PLCE1) often manifest as SRNS in early childhood. Recessive mutations in CRB2 have been found in four different families affected by SRNS. Onset of SRNS due to CRB2 mutations was reported from the age 9 months to 6 years. Three of the four missense mutations (p.Cys620Ser, p.Arg628Cys, and p.Cys629Ser) previously found in CRB2-associated SRNS were clustered within the extracellular tenth EGF-like domain of this protein, and one mutation was in the cytoplasmic tail (p.Arg1249Gln) [7]. However, as p.Arg1249Gln has been reported in a homozygous state in two individuals who were not known to have renal or cerebral defects, the significance of this mutation is unclear [17]. One of the mutations found in the present case is in the 15th EGF-like domain, and the other was a frame-shift mutation. The present case demonstrated that even a mutation outside the tenth EGF-like domain can cause the SRNS phenotype; the association of mutations in the tenth EGF-like domain with SRNS was weakened by the present case.
Biallelic CRB2 mutations not only cause isolated early-onset SRNS [7], but also severe phenotypes that manifest congenital nephrotic syndrome, showing renal microcysts complicated with marked cerebral ventriculomegaly, gray matter heterotopias, and elevated maternal serum alpha-fetoprotein and amniotic fluid alpha-fetoprotein. Recent analysis also identified patients with obstructive congenital hydrocephalus, urinary tract anomalies and lung hypoplasia, features clinically consistent with a ciliopathy [18]. All the mutations reported in the present study and previous reports [7, 8, 17, 18] are shown in Fig. 7. The mechanisms leading to these different phenotypes caused by CRB2 mutations are unknown. With regard to genotype–phenotype correlation, the affected domain does not differentiate the phenotype per se. For example, homozygous missense mutations in the tenth EGF-like domain (C620S, C629S, R633W) can cause either SRNS or the CRB2-related syndrome/ciliopathy phenotype (Fig. 7). However, the missense mutations in SRNS or in pleiotropic phenotype reported so far are mutually exclusive (Fig. 7), raising the possibility of a genotype–phenotype relationship. Besides, the specificity or severity of the organ affected by CRB2 mutations seems to be complex because the severity of nephropathy is not necessarily parallel to extra-renal phenotypes. For example, a patient reported by Lamont et al. with compound heterozygous missense mutations of CRB2 (p.E643A and p.N800K) showed cerebral ventriculomegaly during the pregnancy, and a ventriculoperitoneal shunt was placed at 5 days of life to treat hydrocephalus [17]. Notably, she did not exhibit any detectable kidney abnormalities at 6 years of age. Future case series and in vitro or in vivo studies using knock-in mice or cell-specific knock-out models will be needed to determine the tissue-specific pathophysiological mechanisms of each mutation.
Schematic representation of CRB2 mutation positions. CRB2 contains large extracellular domains with 15 EGF-like domains (blue) and 3 laminin G-like domains (orange), a single transmembrane domain, and a cytoplasmic tail containing single FERM- and PDZ protein-binding motifs at the C-terminus. Mutations reported in patients with SRNS (above) or CRB2-related syndrome/ciliopathy phenotype (below) are shown. Double asterisk refers to this case, Ref indicates reference
Although crb2 expression in podocytes had been confirmed in zebrafish or rodents [7, 9], expression of Crb2 in human kidneys has not been reported. We found that Crb2 is exclusively expressed in podocytes and glomerular parietal epithelial cells in human glomeruli. Podocyte process patterning and cellular junction formation by assembling a protein complex that regulates actin cytoskeletal dynamics are influenced by slit diaphragm (SD) proteins such as nephrin and Neph1 [19]. By gene knock-down using morpholinos, Crb2b was found to be required for the morphological differentiation of podocyte foot processes [9, 20] as crb2b morphants had grossly disorganized the podocyte foot process architecture. While nephrin protein in zebrafish is present along the basal aspect of podocyte cell bodies, nephrin is apically mislocalized in podocytes in crb2b morphants, which suggests that crb2b might be required for the proper protein trafficking of nephrin [9]. In zebrafish morphants with stable heritable loss-of-function mutation in crb2b [7], expression of ZO-1, another component of slit diaphragm, is decreased, also suggesting that protein trafficking might be affected in crb2 mutants. In the present patient with decreased Crb2 expression, expression of nephrin, podocin, and ZO-1 was preserved. Furthermore, effacement of podocyte foot processes was observed in only a small area of the capillary wall. It should be noted that expression of Crb2 was maintained in the CNS patients with NPHS1 mutations. These data suggest that whereas Crb2 in zebrafish may participate in the upstream SD cascade by regulating expression and the proper targeting of SD proteins, the modality of regulation by Crb2 of the maintenance of the glomerular filtration barrier in human podocytes may differ from that of other species. There is also a possibility that the residual CRB2 mutant in the patient may cause differences in the expression of slit diaphragm molecules in podocytes in the patient and that in zebrafish crb2−/− mutants.
The specific loss of Crb2 expression in this patient’s podocytes indicates that SRNS of the present case is caused by novel compound heterozygous mutations of CRB2, expanding the genetic spectrum of CRB2-related diseases. The advance of the clinical and genetic characterization of the disease caused by CRB2 mutations will have important implications for the clinical evaluation and management of patients.
References
Eddy AA, Symons JM (2003) Nephrotic syndrome in childhood. Lancet 362:629–639
Benoit G, Machuca E, Antignac C (2010) Hereditary nephrotic syndrome: a systematic approach for genetic testing and a review of associated podocyte gene mutations. Pediatr Nephrol 25:1621–1632
Saleem MA (2013) New developments in steroid-resistant nephrotic syndrome. Pediatr Nephrol 28:699–709
Sadowski CE, Lovric S, Ashraf S, Pabst WL, Gee HY, Kohl S, Engelmann S, Vega-Warner V, Fang H, Halbritter J, Somers MJ, Tan W, Shril S, Fessi I, Lifton RP, Bockenhauer D, El-Desoky S, Kari JA, Zenker M, Kemper MJ, Mueller D, Fathy HM, Soliman NA, Hildebrandt F, Group SS (2015) A single-gene cause in 29.5% of cases of steroid-resistant nephrotic syndrome. J Am Soc Nephrol 26:1279–1289
Giglio S, Provenzano A, Mazzinghi B, Becherucci F, Giunti L, Sansavini G, Ravaglia F, Roperto RM, Farsetti S, Benetti E, Rotondi M, Murer L, Lazzeri E, Lasagni L, Materassi M, Romagnani P (2015) Heterogeneous genetic alterations in sporadic nephrotic syndrome associate with resistance to immunosuppression. J Am Soc Nephrol 26:230–236
Büscher AK, Beck BB, Melk A, Hoefele J, Kranz B, Bamborschke D, Baig S, Lange-Sperandio B, Jungraithmayr T, Weber LT, Kemper MJ, Tönshoff B, Hoyer PF, Konrad M, Weber S, (GPN) GPNA (2016) Rapid response to cyclosporin A and favorable renal outcome in nongenetic versus genetic steroid-resistant nephrotic syndrome. Clin J Am Soc Nephrol 11:245–253
Ebarasi L, Ashraf S, Bierzynska A, Gee HY, McCarthy HJ, Lovric S, Sadowski CE, Pabst W, Vega-Warner V, Fang H, Koziell A, Simpson MA, Dursun I, Serdaroglu E, Levy S, Saleem MA, Hildebrandt F, Majumdar A (2015) Defects of CRB2 cause steroid-resistant nephrotic syndrome. Am J Hum Genet 96:153–161
Slavotinek A, Kaylor J, Pierce H, Cahr M, DeWard SJ, Schneidman-Duhovny D, Alsadah A, Salem F, Schmajuk G, Mehta L (2015) CRB2 mutations produce a phenotype resembling congenital nephrosis, Finnish type, with cerebral ventriculomegaly and raised alpha-fetoprotein. Am J Hum Genet 96:162–169
Ebarasi L, He L, Hultenby K, Takemoto M, Betsholtz C, Tryggvason K, Majumdar A (2009) A reverse genetic screen in the zebrafish identifies crb2b as a regulator of the glomerular filtration barrier. Dev Biol 334:1–9
Li H, Durbin R (2009) Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25:1754–1760
Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, Subgroup GPDP (2009) The sequence alignment/map format and SAMtools. Bioinformatics 25:2078–2079
Harita Y, Kurihara H, Kosako H, Tezuka T, Sekine T, Igarashi T, Ohsawa I, Ohta S, Hattori S (2009) Phosphorylation of nephrin triggers Ca2+ signaling by recruitment and activation of phospholipase C-{gamma}1. J Biol Chem 284:8951–8962
Kajiho Y, Harita Y, Kurihara H, Horita S, Matsunaga A, Tsurumi H, Kanda S, Sugawara N, Miura K, Sekine T, Hattori S, Hattori M, Igarashi T (2012) SIRPalpha interacts with nephrin at the podocyte slit diaphragm. FEBS J 279:3010–3021
Lek M, Karczewski KJ, Minikel EV, Samocha KE, Banks E, Fennell T, O’Donnell-Luria AH, Ware JS, Hill AJ, Cummings BB, Tukiainen T, Birnbaum DP, Kosmicki JA, Duncan LE, Estrada K, Zhao F, Zou J, Pierce-Hoffman E, Berghout J, Cooper DN, Deflaux N, DePristo M, Do R, Flannick J, Fromer M, Gauthier L, Goldstein J, Gupta N, Howrigan D, Kiezun A, Kurki MI, Moonshine AL, Natarajan P, Orozco L, Peloso GM, Poplin R, Rivas MA, Ruano-Rubio V, Rose SA, Ruderfer DM, Shakir K, Stenson PD, Stevens C, Thomas BP, Tiao G, Tusie-Luna MT, Weisburd B, Won HH, Yu D, Altshuler DM, Ardissino D, Boehnke M, Danesh J, Donnelly S, Elosua R, Florez JC, Gabriel SB, Getz G, Glatt SJ, Hultman CM, Kathiresan S, Laakso M, McCarroll S, McCarthy MI, McGovern D, McPherson R, Neale BM, Palotie A, Purcell SM, Saleheen D, Scharf JM, Sklar P, Sullivan PF, Tuomilehto J, Tsuang MT, Watkins HC, Wilson JG, Daly MJ, MacArthur DG, Consortium EA (2016) Analysis of protein-coding genetic variation in 60,706 humans. Nature 536:285–291
Lovric S, Fang H, Vega-Warner V, Sadowski CE, Gee HY, Halbritter J, Ashraf S, Saisawat P, Soliman NA, Kari JA, Otto EA, Hildebrandt F (2014) Rapid detection of monogenic causes of childhood-onset steroid-resistant nephrotic syndrome. Clin J Am Soc Nephrol 9:1109–1116
McCarthy HJ, Bierzynska A, Wherlock M, Ognjanovic M, Kerecuk L, Hegde S, Feather S, Gilbert RD, Krischock L, Jones C, Sinha MD, Webb NJ, Christian M, Williams MM, Marks S, Koziell A, Welsh GI, Saleem MA, Group RtUSS (2013) Simultaneous sequencing of 24 genes associated with steroid-resistant nephrotic syndrome. Clin J Am Soc Nephrol 8:637–648
Lamont RE, Tan WH, Innes AM, Parboosingh JS, Schneidman-Duhovny D, Rajkovic A, Pappas J, Altschwager P, DeWard S, Fulton A, Gray KJ, Krall M, Mehta L, Rodan LH, Saller DN, Steele D, Stein D, Yatsenko SA, Bernier FP, Slavotinek AM (2016) Expansion of phenotype and genotypic data in CRB2-related syndrome. Eur J Hum Genet 24:1436–1444
Jaron R, Rosenfeld N, Zahdeh F, Carmi S, Beni-Adani L, Doviner V, Picard E, Segel R, Zeligson S, Carmel L, Renbaum P, Levy-Lahad E (2016) Expanding the phenotype of CRB2 mutations—a new ciliopathy syndrome? Clin Genet. doi:10.1111/cge.12764
Garg P, Holzman LB (2012) Podocytes: gaining a foothold. Exp Cell Res 318:955–963
Dreyer SD, Morello R, German MS, Zabel B, Winterpacht A, Lunstrum GP, Horton WA, Oberg KC, Lee B (2000) LMX1B transactivation and expression in nail-patella syndrome. Hum Mol Genet 9:1067–1074
Acknowledgments
The authors thank Dr. Masaki Nishimura (Shiga University of Medical Science) for providing Crb2 plasmid constructs.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Informed consent for DNA analysis was obtained from the parents. The study was performed with approval from the Ethics Committee of the University of Tokyo.
Conflicts of interest
The authors declare that they have no conflicts of interest.
Additional information
Tomohiro Udagawa and Tohaku Jo contributed equally to this work
Rights and permissions
About this article
Cite this article
Udagawa, T., Jo, T., Yanagihara, T. et al. Altered expression of Crb2 in podocytes expands a variation of CRB2 mutations in steroid-resistant nephrotic syndrome. Pediatr Nephrol 32, 801–809 (2017). https://doi.org/10.1007/s00467-016-3549-4
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00467-016-3549-4