Abstract
Identification of molecular regulators of hepatocellular carcinoma (HCC) initiation and progression is not well understood. We chemically induced HCC in male Wistar rats by administration of diethyl nitrosamine (DEN) and 2-acetylaminofluorene (2-AFF). Using 2D-electrophoresis and MALDI-TOF–MS/MS analyses, we characterized differentially expressed proteins in liver tissues at early stage of HCC progression. Using RT-PCR analysis, we quantified the mRNA expression of the characterized proteins and validated the transcript expression with tumor tissues of clinically confirmed HCC patients. Using bioinformatic tools, we analyzed a network among the introduced proteins that identified their interacting partners and analyzed the molecular mechanisms associated with signaling pathways during HCC progression. We characterized a protein, namely, pre-mRNA splicing factor 1 homolog (ISY1), which is upregulated at both transcriptome and proteome levels at HCC initiation, progression, and tumor stages. We analyzed the interacting partners of ISY1, namely, APOA-1, SYNE1, MMP10, and MTG1. Real-time PCR analysis confirmed elevated expression of APOA-1 mRNA at HCC initiation, progression, and tumor stages in animals undergoing tumorigenesis. The mRNA expression of the interacting partners was validated with tumor tissues of clinically confirmed liver cancer patients; the analysis revealed significant elevation in expression of transcripts. The transcriptome and proteome analyses complement each other and dysregulation in mRNA and protein expression of these regulators may play critical role in HCC initiation and progression.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Liver cancer is the second leading cause of cancer-related mortality and accounts for the fifth highest cancer incidence annually (Ranjpour et al. 2018b). Hepatocellular carcinoma (HCC) is the most common primary liver cancer. It is highly aggressive and is often detected at a late stage; thus, new biomarkers are urgently needed for its early diagnosis (Faraj Shaglouf et al. 2019; Ranjpour et al. 2021). HCC is an asymptomatic disease developed in up to 90% of the patients with cirrhotic background as one of the main reasons that obstructs its early detection leading to inadequate treatment and poor survival rate (Ferrín et al. 2015). Infection with chronic hepatitis B and C (Li and Lan 2016), obesity, type 2 diabetes, nonalcoholic fatty liver disease (Marengo et al. 2016), and high amount of alcohol consumption (Vieira-Castro and Oliveira 2017) are important causes leading to progression of HCC. Recent developments in gene expression profiling techniques have allowed HCC to be classified into defined subclasses that provide a strong ground for further research of prospective mechanisms underlying the development of HCC and novel biomarkers for its early diagnosis (Chiou and Lee 2016). Signaling cascades of Ras/Raf/MEK/ERK play important role in transmission of signals involved in apoptosis and components of these pathways are aberrantly expressed in different human cancers, especially in HCC (Chen et al. 2011). Despite the fact that precise mechanism that leads to progression of HCC remains unclear, a number of studies indicate that progressive gathering of mutations and genetic changes in hepatocytes may lead to HCC development (Aravalli et al. 2013; Ranjpour et al. 2021).
The current study was carried out to get deeper insight into the molecular changes that may lead to HCC progression. The total cellular proteins of control and tumor-bearing animals were resolved on 2D gels, the differentially expressed proteins were analyzed by PD Quest analysis of the 2D gels, and the selected proteins were characterized using MALDI-TOF–MS/MS analysis. Real-time PCR analysis relatively quantified the transcript expression of the target proteins. Further, the obtained results have been validated with the tumor tissues of clinically confirmed HCC patients. We analyzed a specific HCC protein network that revealed significant interrelationship among the query protein and its interacting partners some of which were earlier characterized by us using the same animal model (Faraj Shaglouf et al. 2019; Malik et al. 2013b; Ranjpour et al. 2020), showing cross-talk among various components of signaling pathways associated to HCC development.
Methodology
Ethical statement
The present research has been carried out using liver tissue samples of an animal model which is specifically induced to study HCC at early stage of cancer initiation (Ranjpour et al. 2018b). The data from the animal model, present (ISY1 homolog) and previous (nesprin-1 isoform X6: SYNE1; stromelysin-2: MMP10; mitochondrial ribosome-associated GTPase 1 isoform X2: MTG1; apolipoprotein A-1: APOA-1) (Faraj Shaglouf et al. 2019; Ranjpour et al. 2018a) studies has been validated with tumors of clinically confirmed HCC patients. The ethical permission for the study on animal and human samples was granted by Jamia Hamdard Institutional Ethics Committee where experiments were performed and Ethics Committee from Pushpawati Singhania Research Institute (PSRI) Hospital, New Delhi, India, where the human samples were obtained. Consent was obtained from patients whose samples were used for these studies.
The rodent model for study of HCC has been developed by our group (Malik et al. 2013a). Briefly, rats were randomly divided into two groups, namely, carcinogen-treated and control. Chemical carcinogens diethyl nitrosamine (DEN) and 2-acetylaminofluorene (2-AAF) were used to induce HCC and promote its progression in male Wistar rats. Animals in carcinogen-treated groups were administered one high intravenous (I.V.) dose of DEN (200 mg/kg body weight) followed by oral doses of 2-AAF (150 mg/kg body weight) dissolved in 1% CMC, on alternative days of the first week of each month for 4 months. Based on the carcinogen dose and time period of its administration, the carcinogen-treated animals were subdivided into three groups 1 M (sacrificed 1 month after carcinogen treatment), 2 M (sacrificed 2 months after carcinogen treatment), and 4 M (sacrificed 4 months after carcinogen treatment). Therefore, a total of three, six, and twelve doses of 2-AAF were administered to the animals in treated groups 1 M, 2 M, and 4 M, respectively. Animals in control group were sham fed with normal saline at the same schedule. Rats in groups 1 M, 2 M, and 4 M were anesthetized with chloroform and were sacrificed at the end of 1, 2, and 4 months after carcinogen treatment. The liver samples were collected from control and carcinogen-treated groups and kept at − 80 °C, for future proteomic studies and quantification of mRNA expression of the target genes (Ranjpour et al. 2018a).
Proteomic studies
The preserved liver tissues (100–150 mg) were homogenized in urea lysis buffer (urea: 8 M, CHAPS: 65 mM, DTT: 65 mM, thiourea: 2 M, Tris: 33 mM, and PMSF: 6 mM) using a Polytron PT3100 homogenizer. The homogenate was centrifuged at 10,000 rpm for 10 min (Chaudhary et al. 2013; Faraj Shaglouf et al. 2019). The protein concentration in supernatant was estimated by Bradford’s method (Bradford 1976). The extracted proteins (120 μg) were diluted in rehydration buffer containing 8 M urea, 2% CHAPS, 0.25% (v/v; pH 3–10) ampholytes, 30 mM DTT, and 0.1% bromophenol blue (BPB) to make a final volume of 200 μl. The aliquots of control and carcinogen-treated samples were loaded on 11-cm IPG strips (immobilized linear pH gradient, pH 4–7, GE Healthcare) and allowed to rehydrate overnight. Soon after rehydration, IPG strips containing protein samples were focused on an EttanIPGphor II isoelectric focusing system (GE Healthcare) for 500 V-1 h, 1000 V-1 h, 6000 V-2 h, and 6000 V for a total of 35,000 V-hour. The electro-focused IPG strips were sealed on the top of 12% gradient SDS polyacrylamide gel electrophoresis, and the proteins were allowed to be fractionated for 2D analysis. The gels were silver stained and analyzed by PD Quest software (Bio-Rad). The protein of interest was characterized by MALDI-TOF–MS/MS analysis as described earlier (Faraj Shaglouf et al. 2019; Ranjpour et al. 2018b).
Characterization of protein using MALDI-TOF–MS/MS analysis
2D gels of control and carcinogen-treated group (1 M and 4 M) were analyzed by PD Quest software. The spot of differentially expressed protein was excised from gel, de-stained in potassium ferricyanide (15 mM), then sodium thiosulfate (50 Mm), three times for 10 min each, thoroughly dehydrated with acetonitrile and dried. The gel piece was rehydrated with DTT by incubation with iodoacetamide for 45 min; it was then incubated with ammonium bicarbonate, completely dehydrated with acetonitrile, and dried. Trypsin (150–200 ng) was used to digest the protein in presence of 50 mM ammonium bicarbonate for 16 h at 37 °C. The resulting peptides were extracted with the peptide extraction solution and re-suspended in tris–acetate buffer. Finally, the protein sample was analyzed by MALDI TOF/TOF Ultraflex III instrument version 3.2. The peptide mass fingerprints were submitted to the Mascot server for peptide mass fingerprint search in the NCBInr “Rattus norvegicus” database for protein characterization (Faraj Shaglouf et al. 2019; Ranjpour et al. 2018a).
Protein network analysis
The STRING v6 software was used to analyze the interacting partners of the characterized protein (ISY1 homolog) and to understand the functional involvement of these proteins with etiopathogenesis of HCC (Szklarczyk et al. 2011). We have earlier characterized several proteins associated with HCC initiation and progression by inducing the same HCC model (Faraj Shaglouf et al. 2019; Malik et al. 2013b; P Katare et al. 2018; Ranjpour et al. 2018a; Ranjpour et al. 2018b; Ranjpour et al. 2020; Ranjpour et al. 2021); in order to better understand the interrelationship among those proteins with the characterized protein in the present study (ISY1 homolog) during HCC development, we have added the Uniprot ID of all these proteins during network analysis, in the current study. Further, to understand a better insight of molecular mechanism of HCC initiation, some global proteins showing vital role in HCC pathogenesis have also been added (Table 1).
Quantification of transcript expression by real-time PCR analysis
Fifty milligrams of liver tissue samples was used to isolate RNA using Tri reagent method combined with RNA isolation kit (Mini Sure Spin total RNA isolation kit, Nucleopore, Genetix Biotech Asia, India). The RNA was quantified by NanoDrop 2000c spectrophotometer 56 (Thermo Fisher Scientific Incorporation, USA) at A260/A280. Using Verso cDNA Synthesis Kit (Thermo Fisher Scientific, USA), 1000 ng of normalized RNA was reverse transcribed to synthesize cDNA. The conventional PCR was carried out to check the integrity of cDNA using GAPDH specific primers. Further, the gene specific primers were designed to amplify the specific amplicons using T100™ thermal cycler PCR, Bio-Rad System, USA. The detailed information of the primers has been mentioned in Table 2.
PCR mixture was composed of 10 μl of 2 × PCR master mix, 0.5 μl of cDNA (500 ng), 10 μM of each forward and reverse primers, and nuclease-free water. Further the relative transcript quantification was carried out using Roche lightcycler 480 Real Time PCR System (Thermo Fisher Scientific, USA) in presence of SYBR green dye chemistry. Quantitative PCR was carried out in total volume of 25 µl using 12.5 µl of 2 × SYBR green master mix (Maxima), 0.4 µM of forward and reverse primers, 1 µl of 5 times diluted cDNA, and 9.5 µl of nuclease free water. All reactions were performed in triplicates. The gapdh gene was used as reference normalizer and control samples were taken as calibrator. The constant threshold value was used to obtain the Ct values. The relative gene expression was quantified with comparative ΔCt(2−ΔΔCT) method of Livak and Schmittgen (2001).
The statistical analysis
The cDNA samples were run in triplicates. One-way ANOVA and post hoc Dunnett test were carried out by using GraphPad Prism (version 8); the difference between treated and control groups was considered to be significant at P ˂ 0.05. On the other hand, unpaired t-test was used to analyze RT-PCR data of human samples using GraphPad Prism software (version 8).
Results
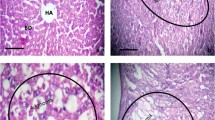
Histological evaluation revealed HCC initiation at 1 month and tumorigenesis at 4 months after carcinogen treatment (Fig. 1). The analysis revealed presence of nodules in liver tissues of carcinogen-treated animals stating that the tumors are of field cancerization.
Histopathology evaluation. (a top) 100 × and (a bottom) 400 × magnification of liver from animals in control group showing the centrizonal area and normal arrangement of hepatocytes in the liver lobule. (b top) 100 × and (b bottom) 400 × magnification of liver from animal in 1 M group showing mild centrizonal sinusoidal dilatation; (c top) 100 × and (c bottom) 400 × magnification of liver from animals in 2 M group showing normal arrangement of hepatocytes in the liver lobule and the centrizonal area with mild dilatation of sinusoids. (d top) 100 × and (d bottom) 400 × magnification of liver from animals in 4 M group showing adenocarcinoma formation in the form of abnormal clustering of vacuolated hepatocytes in the liver lobule; the figure shows the edge of the adenocarcinoma with vacuolated hepatocytes having large hyperchromatic nuclei at the right and normal hepatocytes with smaller nuclei at the left side. PT, portal triad; CV, central vein; VH, vacuolated hepatocytes; NH, normal hepatocytes; 1 M, 1 month; 2 M, 2 months; 4 M, 4 months post-carcinogen treatment
Identification of the differentially expressed proteins
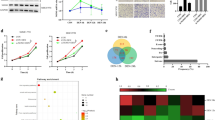
Total liver proteins of control and carcinogen-treated groups 1 (1 M) and 4 (4 M) months after carcinogen treatment were resolved on 2D gels; PD Quest analysis of the gels was performed and compared the protein profiles at HCC initiation (1 M) and tumor (4 M) stages. Based on differences in protein intensities, differentially expressed proteins were analyzed. A protein spot was selected for further analysis which showed elevation in its expression at 1 (1 M) and 4 months (4 M) after carcinogen treatment. The target spot was assigned a unique spot number (SSP) 0019 by the PD Quest software (Fig. 2a (c–f)). The MALDI-TOF–MS/MS characterization of the target protein spot was carried out that introduced the protein as pre-mRNA splicing factor 1 (ISY1) homolog (Fig. 2b (g and h)). Detailed information of peptide sequence analysis of ISY1 homolog has been represented in Table 3.
PD Quest analysis of the target protein and its characterization by MALDI-TOF–MS/MS analysis. (a) PD Quest analysis of 2D gels. (a-c) Master gel represents all protein spots of control and treated groups 1 M and 4 M. The analysis gave unique SSP number 0019 to the target protein (ISY1) and compared its expression level based on its intensity during HCC development. The pop up graphs showed the protein expression pattern and revealed upregulation in expression of the target protein at 1 (HCC initiation) and 4 (tumor stage) months after carcinogen treatment (a–c: master gel, a–d: control, a–e: 1 month, and a–f: 4 months after carcinogen treatment). (b) MALDI-TOF–MS/MS analysis of the target spot: b (g) spectra of ISY1 characterization. b (h) Mascot search result of ISY1 protein. Protein score is − 10*Log (P), where P is the probability that the observed match is a random event. Protein score greater than 52 is significant (P < 0.05)
HCC specific network involving interactions of ISY1 homolog and its interacting partners
The analysis introduced the interacting partners of ISY1 homolog, namely, mitochondrial ribosome-associated GTPase 1 isoform X2 (MTG1), stromelysin-2 (MMP10), nesprin-1 isoform X6 (SYNE1), apolipoprotein A1 (APOA-1), transthyretin (TTR), complement C3 (C3), serine/threonine protein kinase A-Raf (ARAF), cytosolic phospholipase A 2 delta (PLA2G4D), and fatty acid 2-hydroxylase (FA2H). Previous to this, we had characterized these proteins at early stage of HCC initiation using the same animal model (Faraj Shaglouf et al. 2019; Malik et al. 2013a; Malik et al. 2013b; P Katare et al. 2018; Ranjpour et al. 2018b; Ranjpour et al. 2020). The network revealed interrelationship among these proteins through their neighboring partners, namely, MAPK1, lamin B (LMNB1), EGFR/FOS/JUN, pre-mRNA processing factor 6 homolog (PRPF6), pre-mRNA processing factor 31 homolog (PRPF31), ribosomal RNA-processing protein 1B (RRP1B), splicing factor 3B subunit 3 (SF3B3), phytanoyl-CoA dioxygenase peroxisomal (PHYH), and peptidyl-prolyl cis–trans isomerase (PPIH) during HCC progression (Fig. 3a and b). The following observations were obtained through the network analysis.
-
1-
ISY1 is inter-connected to MTG1 through interactions among SF3B3 and PRPF31.
-
2-
ISY1 is connected to FA2H through PPIH and PHYH interactions.
-
3-
The interaction among ISY1 and MMP10 has been mediated through interactions among SF3B3, PRPF6, ARAF, MAPK1, and EGFR/FOS/JUN.
-
4-
The interaction among ISY1 and APOA-1/TTR/C3 has been mediated through interactions between SF3B3 PRPF6 and ARAF.
-
5-
ISY1 is interacted with ARAF through interactions among SF3B3 and PRPF6; further MAPK1 mediates interactions among ISY1 and SYNE1 through LMNB1.
-
6-
The interaction between ISY1 and PLA2G4D has been mediated through interactions among SF3B3, PRPF6, ARAF, and MAPK1.
In silico analysis of protein network during HCC progression. (a) Master protein network in HCC. (b) HCC specific protein network involving interactions of the experimentally characterized proteins. The green color proteins are ISY1 and its interacting partners, namely, APOA-1, SYNE1, MMP10, and MTG1; the gray color proteins are HCC-related proteins reported by our group, previously (ARAF, TTR, C3, FA2H, and PLA2G4D). The network reveals the interrelationship among the characterized proteins and their interacting partners associated with signaling pathways during HCC development. These proteins have vital regulatory roles during HCC progression
Transcriptomic studies
The quantitative PCR analysis of ISY1 homolog was performed and revealed significant elevation (P value: 0.0015) in expression of the transcript at initiation, progression, and tumor stages of HCC. The levels of its transcript expression continued to increase with progression of cancer stating 1.4-fold elevation at 1 month (HCC initiation), 2.5-fold increase at 2 months (HCC progression stage), and 3.8-fold increase at 4 months (tumor stage) after carcinogen treatment. Similarly, we quantified the mRNA expression level of one of the interacting partners of ISY1 homolog, APOA-1 in the animal model. The analysis showed significant elevation (P value: 0.005) in its mRNA expression during HCC progression; the analysis showed 2.47-fold increase at 1 month and 4.5-fold elevation at 2 months post-carcinogen treatment; its expression tends to decrease during development of tumors and showed 2.05-fold increase at 4 months post-carcinogen treatment suggesting its critical regulatory role at initiation and very early progression stages of HCC. The representative data of real-time PCR analysis of ISY1 homolog and APOA-1 transcripts (rat liver samples) has been shown in Fig. 4a.
Real-time PCR analysis of the target transcripts in Rattus norvegicus and human. (a) The analysis revealed significant increase in ISY1 (**P value = 0.0015) expression during HCC progression in animals undergoing tumorigenesis as compared to controls. A significant increase in levels of APOA-1 expression (**P value = 0.005) was observed during HCC development in animals undergoing tumorigenesis as compared to controls; however, there was a slight decrease in the level of mRNA expression of APOA-1 at tumor stage (4 months) as compared to HCC initiation (1 month). One-way ANOVA and post hoc Dunnett test were carried out by using GraphPad Prism. The data has been represented as mean ± SEM (n = 3) and consider significant at (*P < 0.05). (b) We observed a significant increase in expression of ISY1 *P value (0.0308), APOA-1 **P value (0.0022), SYNE1 **P value (0.0032), MMP10 **P value (0.0011), and MTG1 ***P value (0.0004) in liver tumors of clinically confirmed HCC patients as compared to the adjacent tissues. Unpaired t-test was used to analyze RT-PCR data using GraphPad Prism software (version 8). HCC P-1, HCC patient 1; HCC P-2, HCC patient-2; HCC P-3, HCC patient-3
Further, we quantified the mRNA expression of ISY1 homolog, APO-A1, SYNE1, MMP10, and MTG1 in tumor tissues of clinically confirmed HCC patients. The PCR analysis showed significant increase in transcript expression of ISY1, APOA-1, SYNE1, MMP10, and MTG1 in liver tumors of HCC patients as compared to adjacent non-tumor tissues (controls). The analysis of transcript expression in human and animal livers complements each other. Unpaired t-test was used to analyze RT-PCR data using GraphPad Prism software (version 8). The mRNA analysis of ISY1 showed 2.27-fold significant increase in HCC patient 1 (HCC P-1), 1.94-fold significant increase in HCC patient 2 (HCC P-2), and 1.58-fold significant increase in HCC patient 3 (HCC P-3) vis-à-vis controls (P value = 0.0308). The analysis of transcripts for APOA-1 reported 2.28-fold significant increase in HCC P-1, 1.85-fold significant elevation in HCC P-2, and 2.74-fold significant increase in HCC P-3 (P value = 0.0022). On the other hand, mRNA expression of SYNE1 had relatively less pronounced change in HCC patients as compared to controls. We observed 1.32-, 1.47-, and 1.14-fold significant increase (P value = 0.0032) in the levels of the mRNA expression in HCC P-1, HCC P-2, and HCC P-3, respectively. Besides, MMP10 transcript expression was significantly upregulated (P value = 0.0011) in HCC patients and revealed 3.39-fold increase in HCC-P1, 2.49-fold increase in HCC-P2, and 1.75-fold increase in HCC-P3. However, the MTG1 mRNA expression showed the highest level of significant elevation (P value = 0.0004) among these transcripts stating 2.71-, 12.64-, and 3.59-fold increase in HCC P-1, HCC P-2, and HCC P-3, respectively (Fig. 4b).
Discussion
The animal model of HCC was chemically induced in male Wistar rats. We used tissues from carcinogen-treated and placebo-treated animals as the experimental and control groups. DEN and 2-AAF were used to induce HCC. Rats were randomly divided into two groups, namely, control and carcinogen-treated. Carcinogen-treated groups were further subdivided into three different groups, 1 M (sacrificed 1 month after carcinogen treatment), 2 M (sacrificed 2 months after carcinogen treatment), and 4 M (sacrificed 4 months after carcinogen treatment). The carcinogen-treated animals were given a single high I.P. dose of DEN; after the 7-day recovery period, rats were administered 2-AAF, orally. Three doses were administered on three alternative days of the first week of every month for 4 months. Therefore, a total of three, six, and twelve doses of 2-AAF were administered to the animals in treated groups 1 M, 2 M, and 4 M, respectively. Rats in control group received normal saline on the same schedule. Animals from all groups were kept in glass chambers containing cotton soaked with diethyl ether to be anesthetized and sacrificed at 1, 2, and 4 months after carcinogen treatment. At the time of sacrifice, rats were transcardially perfused with saline and after death were dissected to excise livers for proteomic and genomic analyses (Malik et al. 2013a; Ranjpour et al. 2020).
Histological analysis revealed HCC initiation, progression, and tumorigenesis at 1, 2, and 4 months post-carcinogen treatment, respectively. The tumors were developed as the result of field cancerization and were not of monoclonal origin. Similar field cancerization may be taking place in case of human liver cancer (Ranjpour et al. 2018a, 2021). Total proteins of liver tissues were resolved on 2D gels and differentially expressed proteins were analyzed by PD Quest analysis of the gels. The analysis showed significant changes in expression of several proteins at HCC initiation and tumor stages. Based on protein intensity, a protein spot was selected which showed significant elevation in its expression associated with HCC pathogenesis at cancer initiation and tumor stages. MALDI-TOF–MS/MS analysis characterized the protein as pre-mRNA splicing factor ISY1 homolog. The quantitative analysis of its transcript expression also revealed upregulation in its expression at HCC initiation (1 M), during its progression (2 M), and at tumor (4 M) stages. Thus, the transcriptomic and proteomic analyses complement each other. ISY1 homolog is a splicing factor which is poorly characterized (Du et al. 2015). It controls biogenesis of pro-miRNA (Anczuków and Krainer 2016; Du et al. 2015), modulating expression of a vast group of target genes (Anczuków and Krainer 2016). Most splicing factors are involved in the progression of the cell cycle. In presence of mutation, cell cycles get arrested at G2/M phase (inter phase) in which DNA replication takes place (Du et al. 2018). Besides, the expression of highly dynamic ISY1 homolog controls poised pluripotency through mRNA and miRNAs to control the fate of embryonic stem cells (Du et al. 2015). DNA damage induces the expression of ISY1 homolog (Jaiswal et al. 2020). The DEN and 2-AAF used to induce HCC are known to damage DNA (Malik et al. 2013a) and thus may be one of the factors lead to elevated expression of ISY1 in HCC bearing animals as a result of administrating the carcinogen. Dysregulation in alternative splicing pathway plays a critical role in HCC progression (Liu et al. 2014). Cancer cells exhibit modified splicing activity as compared to ordinary cells. The generation of pro-tumorigenic isoforms is primarily related to alternate splicing event which is implicated in cancer progression stages including cell proliferation, metastasis, and apoptosis (Anczuków and Krainer 2016). We have recently reported elevated expression of two different isoforms of two proteins, namely, SYNE1 and MTG1. The association of these proteins with HCC initiation and progression has been elucidated (Faraj Shaglouf et al. 2019). Alternative splicing is an important process taking place in all species including human and is a significant factor to proteomic complexity (Singh and Cooper 2012); it is a complex nuclear process and a natural origin of errors of cancer causing gene expression (Venables 2004). This information also supports our findings that dysregulation in expression of different isoforms of proteins or/and genes has been documented in HCC and may be affected by aberrant expression of some splicing factors. The data obtained from our study is in agreement with previous studies.
On the other hand, we analyzed a HCC specific protein network associated with several signaling pathways by incorporating the experimentally identified proteins introduced by our group in the present study and earlier. Using the reported animal model specific for HCC, our group recently characterized several differentially expressed proteins, namely, SYNE1, MTG1, MMP10, APOA-1, TTR, C3, ARAF, cytosolic phospholipase A2 delta, and FA2H and reported significant elevation in their expression at HCC initiation, progression, and tumor stages (Faraj Shaglouf et al. 2019; Malik et al. 2013b; P Katare et al. 2018; Ranjpour et al. 2018a; Ranjpour et al. 2018b; Ranjpour et al. 2020). The network reveals correlation among ISY1 and these proteins and represents the interacting partners of ISY1 associated with HCC pathogenesis. The network states interrelationship among key regulators of lipid metabolism, EGFR signaling, MAPK, and splicing pathways including APO-A1, TTR, C3, cytosolic phospholipase A2 delta, FA2H, EGFR/JUN/FOS, MAPK1, ARAF, MMP10, SYNE1, ISY1, and MTG1 during HCC progression. The network reveals interactions among ISY1 and MTG1 through PRPF31 and SF3B3. On the other hand, ISY1 is connected to FA2H through PPIH and PHYH. MTG1 is connected to RRPIB that is a member of metastasis susceptibility genes (Kinoshita et al. 2017) and its increased expression level has been reported in HCC (Kinoshita et al. 2017). PRPF31 has been introduced as a novel drug target for HBV treatment (Kinoshita et al. 2017). SF3B3 is essential for pre-mRNA splicing. Mutations in sf3b3 gene have been reported in different cancers (Cretu et al. 2016). The sf3b3 gene is differentially expressed in HCC and plays critical role in HCC development supporting cell proliferation and invasion of cancer cells involved in endocytosis (Choi et al. 2017; Naboulsi et al. 2016). Moreover, ppih gene is involved in post-translational modification, protein folding, endocrine system development and function (Rashid et al. 2014) and its expression is downregulated in cancer (Ibáñez et al. 2014). PPIH is a RNA binding protein associated with pre-mRNA splicing and translation machinery in tumorigenesis promoting cancer progression (Zhang et al. 2018). The phyh gene is downregulated in liver tumors of zebra fish; it has regulatory role in cellular metabolic processes and is regulated by hyperactive Raf–MEK signaling in zebra fish liver cells (He et al. 2011). The direct interactions among APOA-1 with ARAF, TTR, and C3 have been reported by the analyzed network. We previously reported the regulatory role of APOA-1, TTR, and C3 in lipid metabolism during HCC progression (Malik et al. 2013b; P Katare et al. 2018; Ranjpour et al. 2018a). ARAF mediates the interactions of ISY1 with APOA-1 and MAPK1 through PRPF6 and SF3B3 interactions; it also mediates interactions among APOA-1 and SYNE1 through interactions of MAPK1 and LMNB1. ARAF is a member of MAPK pathway and is associated with HCC progression (Ranjpour et al. 2018a, 2018b). MAPK pathway has critical role in HCC progression (Chen et al. 2019). MAPK is important mediator of EGFR signaling in liver (Kannangai et al. 2006). The HCC proliferation and metastasis are regulated through EGF-EGFR signaling pathways (Huang et al. 2014). The network shows direct interaction among MAPK1 and PLA2G4D. We earlier reported the elevated expression of PLA2G4D and demonstrated its regulatory role in lipid metabolism associated with HCC progression. PLA2G4D regulates TGF-β signaling pathway that controls growth of primary hepatocytes and hepatoma cells (Ranjpour et al. 2020). The network also shows the interrelationship among MAPK1 and SYNE1 through LMNB1. LMNB1 is a member of lamin family having different isoforms controlling DNA repair, gene expression, cell differentiation, and apoptosis participating in cancer development (Sakthivel and Sehgal 2016). LMNB1 is over expressed in HCC and is considered as a potential biomarker for diagnosis of cirrhotic liver and HCC. The regulatory role of lamins in MAPK signaling pathway was previously reported (Sakthivel and Sehgal 2016).
We quantified the mRNA expression of ISY1 homolog, APOA-1, MMP10, MTG1, and SYNE1 in liver tumors of clinically confirmed HCC patients by employing real-time PCR analysis. The quantification of mRNA expression for these proteins revealed significant upregulation in the transcript expression that validated our experimental findings from animal studies and are complementary to these observations. Besides, we quantified the mRNA expression of APOA-1 in animals undergoing tumorigenesis that revealed significant upregulation in its expression at HCC initiation, progression, and tumor stages vis-a-vis controls. However, we observed a slight decrease in its transcript expression at tumor stage as compared to HCC initiation and progression stages. We earlier reported elevated expression of APOA-1 in sera of rats undergoing tumorigenesis at HCC initiation, during its progression, and at tumor stages (Ranjpour et al. 2018a). There has been a report showing low levels of APOA-1 of liver cancer patients with circulating tumor cells at multiple tumor stages and during tumor recurrence (Ma et al. 2016). It may be due to extensive damage to liver during metastasis as well as in case of tumor recurrence where loss of liver function may have taken place during chemo- and/or radio-therapy and surgical intervention. This can explain the apparent discrepancy between this report and our study showing elevated levels of APOA-1, at both mRNA and protein levels, at HCC initiation, its progression, and tumor stages. APOA-1 is involved in lipid metabolism and molecular transport and is regulated by transcription factor HNF4αa (Dillon et al. 2013). It has been implicated as potential serum biomarker for HCC (Kang et al. 2010).
Taken together, we state that ISY1, APOA-1, SYNE1, MMP10, and MTG have critical regulatory role at HCC initiation and progression stages. The network analysis reveals interrelationship among these proteins and their interacting partners playing critical regulatory role associated with lipid metabolism, EGFR signaling, MAPK, and splicing pathways during HCC progression.
Conclusion
Altogether, we reported elevation in expression of ISY1, SYNE1, MMP10, MTG1, and APOA-1 in HCC initiation and during its progression. The correlation among these proteins and their interacting neighbors, namely, EGFR, JUN, FOS, MAPK, LMNB1, PPIH, PHYH, LMNB1, SF3B3, and HGF play critical role(s) in modulating cell proliferation, metastasis, apoptosis, DNA repair, protein folding, post-transitional modifications, cell structure and nuclear integrity associated with lipid metabolism, EGFR signaling, MAPK, and splicing pathways during HCC development. Dysregulation in expression of the members of these pathways may affect the normal activity of hepatocytes and result in abnormal cell growth and proliferation.
The findings of the current study elucidate the possible functional role of ISY1, APOA-1, SYNE1, MMP10, and MTG1 in HCC pathogenesis. The interrelationship among these proteins plays critical role(s) associated with signaling pathways during HCC progression. The validation with large cohort of human HCC samples and further mechanistic studies are required to explore the exact functional role of these proteins and their corresponding genes during HCC initiation and progression.
Abbreviations
- HCC:
-
Hepatocellular carcinoma
- TTR:
-
Transthyretin
- DEN:
-
Diethylnitrosamine
- 2-AAF:
-
2-Aminoacetylfluorine
- APOA1:
-
Apolipoprotein A-1
- BD:
-
Bile duct
- HA:
-
Hepatic artery
- 2DE:
-
2D-electrophoresis
- ISY1:
-
Pre-mRNA splicing factor 1 homolog
- SYNE1:
-
Nesprin-1 isoform X6
- MMP10:
-
Stromelysin-2
- MTG1:
-
Mitochondrial ribosome-associated GTPase 1 isoform X2
- PLA2G4D:
-
Cytosolic phospholipase A 2 delta
- LMNB11:
-
Lamin B
- PRPF6:
-
Pre-mRNA processing factor 6 homolog
- PRPF31:
-
Pre-mRNA processing factor 31 homolog
- RRP1B:
-
Ribosomal RNA-processing protein 1B
- SF3B3:
-
Splicing factor 3B subunit 3
- PHYH:
-
Phytanoyl-CoA dioxygenase, peroxisomal
- PPIH:
-
Peptidyl-prolyl cis–trans isomerase
References
Anczuków O, Krainer AR (2016) Splicing-factor alterations in cancers. RNA (new York, NY) 22:1285–1301. https://doi.org/10.1261/rna.057919.116
Aravalli RN, Cressman EN, Steer CJ (2013) Cellular and molecular mechanisms of hepatocellular carcinoma: an update. Arch Toxicol 87:227–247. https://doi.org/10.1007/s00204-012-0931-2
Bakiri L, Hamacher R, Graña O, Guío-Carrión A, Campos-Olivas R (2017) Liver Carcinogenesis by FOS-Dependent Inflammation and Cholesterol Dysregulation. J Exp Med 214:1387–1409. https://doi.org/10.1084/jem.20160935
Bradford MM (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72:248–254. https://doi.org/10.1006/abio.1976.9999
Chaudhary N, Bhatnagar S, Malik S, Katare DP, Jain SK (2013) Proteomic analysis of differentially expressed proteins in lung cancer in Wistar rats using NNK as an inducer. Chem Biol Interact 204:125–134. https://doi.org/10.1016/j.cbi.2013.05.004
Chen L, Shi Y, Jiang CY, Wei LX, Wang YL, Dai GH (2011) Expression and prognostic role of pan-Ras, Raf-1, pMEK1 and pERK1/2 in patients with hepatocellular carcinoma. Eur J Surg Oncol 37:513–520. https://doi.org/10.1016/j.ejso.2011.01.023
Chen J, Ji T, Wu D, Jiang S, Zhao J, Lin H, Cai X (2019) Human mesenchymal stem cells promote tumor growth via MAPK pathway and metastasis by epithelial mesenchymal transition and integrin α5 in hepatocellular carcinoma. Cell Death Dis 10:425. https://doi.org/10.1038/s41419-019-1622-1
Chiou SH, Lee KT (2016) Proteomic analysis and translational perspective of hepatocellular carcinoma: identification of diagnostic protein biomarkers by an onco-proteogenomics approach. Kaohsiung J Med Sci 32:535–544. https://doi.org/10.1016/j.kjms.2016.09.002
Choi J-H et al (2017) Mutations acquired by hepatocellular carcinoma recurrence give rise to an aggressive phenotype. Oncotarget 8:22903–22916. https://doi.org/10.18632/oncotarget.14248
Cretu C et al (2016) Molecular architecture of SF3b and structural consequences of its cancer-related mutations. Mol Cell 64:307–319. https://doi.org/10.1016/j.molcel.2016.08.036
Dillon ST, Bhasin MK, Feng X, Koh DW, Daoud SS (2013) Quantitative proteomic analysis in HCV-induced HCC reveals sets of proteins with potential significance for racial disparity. J Transl Med 11:239. https://doi.org/10.1186/1479-5876-11-239
Du P, Wang L, Sliz P, Gregory Richard I (2015) A biogenesis step upstream of microprocessor controls miR-17∼92 expression. Cell 162:885–899. https://doi.org/10.1016/j.cell.2015.07.008
Du P et al. (2018) An intermediate pluripotent state controlled by microRNAs is required for the naive-to-primed stem cell transition. Cell Stem Cell 22https://doi.org/10.1016/j.stem.2018.04.021
Faraj Shaglouf LH, Ranjpour M, Wajid S, Jain SK (2019) Elevated expression of cellular SYNE1, MMP10, and GTPase1 and their regulatory role in hepatocellular carcinoma progression. Protoplasma. https://doi.org/10.1007/s00709-019-01423-w
Ferrín G et al (2015) Plasma protein biomarkers of hepatocellular carcinoma in HCV-infected alcoholic patients with cirrhosis. PLoS ONE 10:e0118527. https://doi.org/10.1371/journal.pone.0118527
He S, Krens SG, Zhan H, Gong Z, Hogendoorn PC, Spaink HP, Snaar-Jagalska BE (2011) A ΔRaf1-ER-inducible oncogenic zebrafish liver cell model identifies hepatocellular carcinoma signatures. J Pathol 225:19–28. https://doi.org/10.1002/path.2936
Huang P, Xu X, Wang L, Zhu B, Wang X, Xia J (2014) The role of EGF-EGFR signalling pathway in hepatocellular carcinoma inflammatory microenvironment. J Cell Mol Med 18:218–230. https://doi.org/10.1111/jcmm.12153
Ibáñez K, Boullosa C, Tabarés-Seisdedos R, Baudot A, Valencia A (2014) Molecular evidence for the inverse comorbidity between central nervous system disorders and cancers detected by transcriptomic meta-analyses. PLoS Genet 10:e1004173. https://doi.org/10.1371/journal.pgen.1004173
Jaiswal AS et al (2020) The splicing component ISY1 regulates APE1 in base excision repair. DNA Repair 86:102769. https://doi.org/10.1016/j.dnarep.2019.102769
Kang X, Sun L, Guo K, Shu H, Yao J, Qin X, Liu Y (2010) Serum protein biomarkers screening in HCC patients with liver cirrhosis by ICAT-LC-MS/MS. J Cancer Res Clin Oncol 136:1151–1159. https://doi.org/10.1007/s00432-010-0762-6
Kannangai R, Sahin F, Torbenson MS (2006) EGFR is phosphorylated at Ty845 in hepatocellular carcinoma. Mod Pathol 19:1456–1461. https://doi.org/10.1038/modpathol.3800665
Katare DP, Malik S, Mani RJ, Ranjpour M (2018) Novel mutations in transthyretin gene associated with hepatocellular carcinoma. Mol Carcinog 57:70–77. https://doi.org/10.1002/mc.22732
Katare DP, Malik S, Mani RJ, Ranjpour M, Jain SK (2018) Novel mutations in transthyretin gene associated with hepatocellular carcinoma. Mol Carcinog 57:70–77. https://doi.org/10.1002/mc.22732
Kinoshita W, Ogura N, Watashi K, Wakita T (2017) Host factor PRPF31 is involved in cccDNA production in HBV-replicating cells. Biochem Biophys Res Commun 482:638–644. https://doi.org/10.1016/j.bbrc.2016.11.085
Li L, Lan X (2016) Association between hepatitis B virus/hepatitis C virus infection and primary hepatocellular carcinoma risk: a meta-analysis based on Chinese population. J Cancer Res Ther 12:C284-c287. https://doi.org/10.4103/0973-1482.200763
Liu L, Xie S, Zhang C, Zhu F (2014) Aberrant regulation of alternative pre-mRNA splicing in hepatocellular carcinoma. Crit Rev Eukaryot Gene Expr 24:133–149. https://doi.org/10.1615/critreveukaryotgeneexpr.2014007702
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(-delta delta C(T)) method. Methods (san Diego, Calif) 25:402–408. https://doi.org/10.1006/meth.2001.1262
Ma XL et al (2016) Apolipoprotein A1: a novel serum biomarker for predicting the prognosis of hepatocellular carcinoma after curative resection. Oncotarget 7:70654–70668. https://doi.org/10.18632/oncotarget.12203
Malik S, Bhatnagar S, Chaudhary N, Katare DP, Jain S (2013a) DEN+ 2-AAF-induced multistep hepatotumorigenesis in Wistar rats: supportive evidence and insights. Protoplasma 250:175–183
Marengo A, Rosso C, Bugianesi E (2016) Liver cancer: connections with obesity, fatty liver, and cirrhosis. Annu Rev Med 67:103–117. https://doi.org/10.1146/annurev-med-090514-013832
Malik S, Pande Katare D, Jain S (2013b) Proteomic profiling of hepatocellular carcinoma: elevated circulating complement C3 protein level displays role for potential early cancer biomarker vol 56. doi:https://doi.org/10.1016/j.molimm.2013b.05.126
Naboulsi W et al (2016) Quantitative proteome analysis reveals the correlation between endocytosis-associated proteins and hepatocellular carcinoma dedifferentiation. Biochimica et Biophysica Acta 1864:1579–1585. https://doi.org/10.1016/j.bbapap.2016.08.005
Paterlini-Bréchot P et al (2003) Hepatitis B virus-related insertional mutagenesis occurs frequently in human liver cancers and recurrently targets human telomerase gene. Oncogene 22:3911–3916. https://doi.org/10.1038/sj.onc.1206492
Ranjpour M, Katare DP, Wajid S, Jain SK (2018a) HCC specific protein network involving interactions of EGFR with A-Raf and transthyretin: experimental analysis and computational biology correlates. Anticancer Agents Med Chem. https://doi.org/10.2174/1871520618666180507141632
Ranjpour M, Wajid S, Jain SK (2018b) Elevated expression of A-Raf and FA2H in hepatocellular carcinoma is associated with lipid metabolism dysregulation and cancer progression. Anticancer Agents Med Chem. https://doi.org/10.2174/1871520618666181015142810
Ranjpour M, Wajid S, Jain SK (2020) Elevated expression of cytosolic phospholipase A2 delta is associated with lipid metabolism dysregulation during hepatocellular carcinoma progression. Cell J 22:17–22. https://doi.org/10.22074/cellj.2020.6527
Ranjpour M, Wajid S, Jain SK (2021) Elevated expression of sepiapterin reductase, regulator of G protein signaling 1, hypothetical protein CXorf58 homolog, and zinc finger and BTB domain–containing protein 21 isoform X2 is associated with progression of hepatocellular carcinoma. Protoplasma. https://doi.org/10.1007/s00709-021-01632-2
Rashid OM et al (2014) An improved syngeneic orthotopic murine model of human breast cancer progression. Breast Cancer Res Treat 147:501–512. https://doi.org/10.1007/s10549-014-3118-0
Sakthivel KM, Sehgal P (2016) A novel role of lamins from genetic disease to cancer biomarkers. Oncol Rev 10:309–309. https://doi.org/10.4081/oncol.2016.309
Singh RK, Cooper TA (2012) Pre-mRNA splicing in disease and therapeutics. Trends Mol Med 18:472–482. https://doi.org/10.1016/j.molmed.2012.06.006
Szklarczyk D et al (2011) The STRING database in 2011: functional interaction networks of proteins, globally integrated and scored. Nucleic Acids Res 39:D561-568. https://doi.org/10.1093/nar/gkq973
Venables JP (2004) Aberrant and alternative splicing in cancer. Cancer Res 64:7647–7654. https://doi.org/10.1158/0008-5472.can-04-1910
Vieira-Castro ACM, Oliveira LCM (2017) Impact of alcohol consumption among patients in hepatitis C virus treatment. Arq Gastroenterol 54:232–237. https://doi.org/10.1590/s0004-2803.201700000-33
Yu XT, Zhu SN, Xu ZD, Hu XQ, Zhu TF, Chen JQ, Lu SL (2007) Roles of EGFR-Stat3 signal pathway in carcinogenesis of experimental hepatoma in rats. J Cancer Res Clin Oncol 133:145–152. https://doi.org/10.1007/s00432-006-0139-z
Yuen MF, Wu PC, Lai VC, Lau JY, Lai CL (2001) Expression of c-Myc, c-Fos, and c-jun in hepatocellular carcinoma. Cancer 91:106–112. https://doi.org/10.1002/1097-0142(20010101)91:1%3c106::aid-cncr14%3e3.0.co;2-2
Zou C-D et al (2016) MicroRNA-107: a novel promoter of tumor progression that targets the CPEB3/EGFR axis in human hepatocellular carcinoma. Oncotarget 7:266–278. https://doi.org/10.18632/oncotarget.5689
Zhang B et al. 2018 Pan-cancer analysis of RNA binding proteins (RBPs) reveals the involvement of the core pre-mRNA splicing and translation machinery in tumorigenesis. doi:https://doi.org/10.1101/341263
Acknowledgements
M. R. is grateful to international council for cultural relation (ICCR), Govt. of India, for their financial support [No.# 2-261/2013-14/ISD-II].
Funding
The work was supported by a MRP grant from university grant commission (UGC) to S. K. Jain [Grant # 40–155/2011 (SR)].
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Communicated by Handling Editor: Jörn Bullerdiek.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Shaglouf, L.H.F., Ranjpour, M., Wajid, S. et al. Elevated expression of ISY1, APOA-1, SYNE1, MTG1, and MMP10 at HCC initiation: HCC specific protein network involving interactions of key regulators of lipid metabolism, EGFR signaling, MAPK, and splicing pathways. Protoplasma 260, 651–662 (2023). https://doi.org/10.1007/s00709-022-01796-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00709-022-01796-5