Abstract
Breast cancer drug development costs nearly $610 million and 37 months in preclinical mouse model trials with minimal success rates. Despite these inefficiencies, there are still no consensus breast cancer preclinical models. Murine mammary adenocarcinoma 4T1-luc2 cells were implanted subcutaneous (SQ) or orthotopically percutaneous (OP) injection in the area of the nipple, or surgically into the chest 2nd mammary fat pad under direct vision (ODV) in Balb/c immunocompetent mice. Tumor progression was followed by in vivo bioluminescence and direct measurements, pathology and survival determined, and tumor gene expression analyzed by genome-wide microarrays. ODV produced less variable-sized tumors and was a reliable method of implantation. ODV implantation into the chest 2nd mammary pad rather than into the abdominal 4th mammary pad, the most common implantation site, better mimicked human breast cancer progression pattern, which correlated with bioluminescent tumor burden and survival. Compared to SQ, ODV produced tumors that differentially expressed genes whose interaction networks are of importance in cancer research. qPCR validation of 10 specific target genes of interest in ongoing clinical trials demonstrated significant differences in expression. ODV implantation into the chest 2nd mammary pad provides the most reliable model that mimics human breast cancer compared from subcutaneous implantation that produces tumors with different genome expression profiles of clinical significance. Increased understanding of the limitations of the different preclinical models in use will help guide new investigations and may improve the efficiency of breast cancer drug development .
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Breast cancer, the second leading cause of cancer death in women in the United States [1], has received hundreds of billions of U.S. dollars in research and drug development funding over the last 20 years and has produced thousands of peer-review publications [2]. The development of a single breast cancer drug costs approximately $610 million and requires on average 37 months of animal trials to screen for safety and efficacy [3]. Mouse models of metastatic breast cancer are the mainstay for screening for efficacy because of safety and cost, but too often drugs that pass animal screening fail in humans [4–6]. The number of projects terminated in phase III trials has increased from 36 in 2006–2007 to 53 in 2008–2010 and 45 in 2009–2011 [7]. Despite the central role of mouse models in breast cancer research and the great costs and inefficiencies of drug development, there is no standard mouse breast cancer metastasis model established as the optimal system for drug development [4–6, 8]. Moreover, in the era of gene targeted therapy where investigators increasingly focus on tumor genome profiles, little is known about the differences in the genetic signatures of the tumors produced by different models [9].
There has been a remarkable proliferation of publications using murine metastatic breast cancer models over the last 20 years, rising to more than 500 per year in the last 3 years. Simultaneously, there has been a divergence in the types of models used, from 3 in 1990 to 7 in 2008–2012, without a consensus model arising. In xenograft models, drugs are screened for efficacy against human tumors in immunodeficient mice. However, xenograft models obviate the anti-tumor immune response, confound tumor-host interactions important for cancer, obliterate the native mammary gland and associated vessel architecture and mammary microenvironment, and often produce false positive results [4–6]. Transgenic mouse models are useful to study tumorigenesis, but it usually takes months for the mice to develop tumors that are variable in timing and size, and can require more than a year to test one drug. Further, expensive diagnostic imaging modalities are required to monitor metastasis in vivo in these models, and unstable tumors can often lead to false positive results for novel therapeutics [5]. Syngeneic mouse models, such as murine mammary adenocarcinoma 4T1 cells tagged with firefly luciferase implanted in immune competent mice, can be used to screen for drug efficacy in vivo using bioluminescent technology. Such syngeneic models take into account the anti-tumor immune response, cancer-stromal interactions, and maintain and utilize the mammary microenvironment, and have been shown to produce more efficient cancer progression and metastasis than xenografts [4, 10, 11].
The choice of implantation site for xenografts, orthotopically directly by injection into the subcutaneous space (SQ) of the mammary gland or through a small incision (orthotopic implantation under direct vision, ODV) remains controversial, and critical analysis of the optimal approach for research and drug development is lacking [4, 5, 12–16]. Although the orthotopic implantation model was first described over 20 years ago [17], and the advantages of this model over other models were also described 15 years ago [18], ectopically implanted SQ model is widely used to date. In fact, it was also demonstrated utilizing high resolution fluorescent protein imaging as well as whole body fluorescent imaging in real time how this model can be utilized as an ideal in vivo system to study metastatic breast cancer [19–22]. However, the metastatic progression of each model, the feasibility of determining drug efficacy in mice with endpoints that are clinically important to humans, and differences in tumor genome profile have not been examined in enough detail to persuade the drug developers and critics to adopt it as the consensus model [4–6]. It has been argued that compared to SQ, ODV requires advanced surgical skill, and the tumors that are produced are difficult to monitor [4, 5, 12–16]. On the other hand, there is some evidence that ODV produces more efficient cancer progression and metastasis than SQ [4–6, 10, 23, 24].
The purpose of this study was to compare commonly used methods of implantation of breast cancer cells in syngeneic mice to determine which produces the most stable results and best mimics the progression of human disease. We also examined gene expression in tumors produced to evaluate the impact of implantation site on genetic targets important for cancer biology and therapy.
Materials and methods
Materials and animals used
Virginia Commonwealth University Institutional Animal Care and Use Committee approval was obtained for all experiments and all protocols were followed. Female Balb/c, nude, and C57Bl/6 mice, 12 weeks of age, weighing approximately 20 g were obtained from Jackson Laboratories. 4T1-luc2 murine mammary adenocarcinoma cell line genetically manipulated to overexpress the firefly luciferase gene was obtained from Caliper (Perkin Elmer). The cells were cultured in RPMI media, suspended at a concentration of 1 × 106 cells/100 µl, and 10 µl was then injected into the mice, unless specified otherwise.
Bioluminescent quantification of tumor burden
D-Luciferin (0.2 mL of 15 mg/mL stock; Perkin Elmer) was injected intraperitoneally (ip) into 4T1-luc2 tumor bearing mice and Xenogen IVIS 200 and Living Image software (Perkin Elmer) were used to quantify photons emitted by the cells as described [25, 26]. Metastasis was examined ex vivo 10 min after ip injection of luciferin.
Surgical techniques of cell implantation
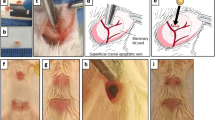
Subcutaneous implantation (SQ) Under anesthesia, the skin was tented up, and 10 µl of the 4T1-luc2 cell suspension was implanted into the subcutaneous space. Orthotopic Percutaneous mammary gland implantation (OP) Under anesthesia, 10 µl of the 4T1-luc2 suspension was implanted under the nipple into the mammary gland. Orthotopic mammary gland surgical implantation under Direct Vision (ODV) Under anesthesia, a 5-mm incision was made medial to the nipple, and a sterile cotton swab was used to expose the mammary gland. A 27G needle was used to implant 4T1-luc2 cells directly into the mammary gland under direct vision using x10 microscope magnification.
Gene expression profiling of 4T1 ODV and SQ tumors
Ten days after ODV or SQ (8 Balb/c mice per group, one implantation site each), the tumors were harvested and snap-frozen at −80 °C. Frozen tissue Sections (10 µm) were used for histopathological scoring after staining with hematoxylin and eosin and adjacent sections used for RNA isolation. All samples contained more than 70 % tumor. Total RNA was extracted, and the quality evaluated using a previously established method of sample processing [27]. Total RNA was extracted from multiple 10 µm frozen tissue sections using the MagMAX™-96 for Microarrays Total RNA Isolation Kit (Invitrogen Life Technologies, Carlsbad, CA), in an automated fashion using the MagMAX™ Express magnetic particle processor. RNA purity was judged by spectrophotometry at 260, 270, and 280 nm. RNA integrity was assessed by running 1 µl of every sample in RNA 6000 Nano LabChips on the 2100 Bioanalyzer (Agilent Technologies, Foster City, CA). The Affymetrix protocol (Affymetrix, Santa Clara, CA) has previously been described [27]. Briefly, starting with 500 ng of total RNA, cDNA synthesis and cRNA labeling were performed using the GeneChip® 3′ IVT Express Kit (Affymetrix). Ten µg of fragmented cRNA were hybridized to the GeneChip® Mouse Genome 430A 2.0 array for 16 h at 60 rpm in a 45 °C hybridization oven. This array provides a comprehensive coverage of the transcribed murine genome by including over 22,600 probe sets that analyze the expression level of over 14,000 well-characterized mouse transcripts. The arrays were washed and stained with streptavidin phycoerythrin (SAPE; Molecular Probes, Eugene, OR) in the Affymetrix fluidics workstation. Every chip was scanned at a high resolution, with pixelations ranging from 2.5 µm down to 0.51 µm by the Affymetrix GeneChip® Scanner 3000 according to the GeneChip Expression Analysis Technical Manual procedures (Affymetrix). After scanning, the raw intensities for every probe were stored in electronic files (in.DAT and.CEL formats) by the GeneChip Operating Software (GCODV v1.4, Affymetrix). The overall quality of each array was assessed by monitoring the 3′:5′ ratios for a housekeeping gene (GAPDH) and the percentage of “Present” genes (%P), with arrays exhibiting GAPDH 3′:5′ <3.0 and %p > 40 % considered good quality.
Quantitative real-time PCR
Real-time, quantitative reverse transcriptase polymerase chain reaction (QPCR) was used to assess gene expression levels of selected genes using the ABI Prism 7900 sequence detection system (Applied Biosystems) with probes and primer sets obtained from Applied Biosystems as previously described [28].
Statistical analysis
Student’s t test and Kaplan–Meier survival statistical analysis were utilized for photon emission and tumor surface area, and survival data, respectively. Standard statistical methods were utilized for microarray data analysis, including the Robust Multi-array Analysis (RMA) method [29], hierarchical cluster analyses, and false discovery rates [30].
Results
ODV generates larger and less variable tumors than OP and SQ
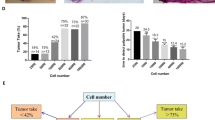
One of the challenges in the use of animal breast cancer models is the variability in size of the tumors between each animal after implantation. Higher variability requires a larger number of animals to reach statistical power resulting in soaring cost and efforts. Therefore, we compared three different implantation methods, ODV, OP, and SQ, commonly used in breast cancer research. Using luciferase-tagged 4T1-luc2 cells, we found that the ODV implantation method more consistently produced larger tumors than the OP or SQ methods, as determined by quantitative in vivo luminescence imaging and by qualitative imaging of the tumors (Fig. 1).
ODV produced greater cancer growth with less variability than OP or SQ. Four days after implantation of 1 × 105 cells by the respective methods in 4 separate locations in 5 mice in each group, bioluminescence was quantified, the black line represents the mean for each group of N = 20 (*p < 0.02, **p < 0.001)
Pathologic analysis demonstrated that cells implanted by OP, the most commonly used method of orthotopic implantation, were not uniformly distributed into the mammary gland, and cancer cells were observed all along the needle tract, from the skin to the mammary gland (Fig. 2a). This in part may explain why OP resulted in greater variability (Fig. 1). The OP method also can miss the targeted mammary fat pad entirely, resulting in intradermal, subcutaneous or intramuscular implantation. In contrast, ODV allows for accurate implantation into the mammary fat pad, as the procedure is performed under direct visualization of the target. Tumor size was determined by caliper measurements, where ODV produced larger tumors compared to SQ at day 28 after implantation (Fig. 2b). Pathologic analysis revealed a scar-like fibrotic capsule formed around the tumor after SQ, but not after ODV implantation (Fig. 2b). Mice implanted by the ODV method had a significantly shorter survival than SQ (Fig. 2c).
Pathological analyses demonstrate the possible cause of variabilities in OP or SQ. a H&E pathology demonstrated implantation along the needle tract (black arrow demonstrates skin implantation) and in the mammary gland (white arrow) after OP method. b ODV produced larger tumors than SQ (**p < 0.001; N = 8). H&E staining showed capsule formation around SQ but not ODV tumors. c Kaplan–Meier analysis demonstrated ODV produced shorter survival than Sq (p < 0.001)
Implantation of breast cancer cells into the chest 2nd mammary pad by ODV is a better mimic of human breast cancer progression
The vast majority of studies using orthotopic implantation as a metastatic breast cancer model implant cells into the 4th mammary pad, which is located at the abdomen. The rationale for this approach is that the 4th mammary pad is the largest and relatively easy to access. Therefore, it was of interest to examine metastasis to different organs after implantation of 4T1-luc2 cells into the abdominal 4th mammary pad compared to into the chest 2nd mammary pad. Twenty eight days after ODV implantation of 4T1-luc2 cells into the 2nd chest pad developed distant metastases, mostly to the lungs and to a lesser extent to the kidneys, liver and spleen detected by in vivo and ex vivo bioluminescence technology, (Fig. 3). This is consistent with our previous observation that ODV into the chest pad developed axially lymph node metastasis first and then lung metastasis, which is the same sequence as human breast cancer progression [25, 26]. In contrast, 28 days after ODV implantation into the 4th abdominal pad generated tumors that invaded through the abdominal wall, leading to peritoneal carcinomatosis, dissemination of cancer cells in the abdominal cavity, and much less lung metastasis was observed (Fig. 3). The majority of the tumor burden after ODV into abdominal pad was in the form of carcinomatosis, which is not seen in human breast cancer.
ODV implantation into the chest 2nd mammary pad better mimic human breast cancer progression than into the abdomen 4th mammary pad. 4T1-luc2 cells were implanted into either abdomen 4th mammary pad (Abdomen; closed bar) or chest 2nd mammary pad (Chest; open bar), and ex vivo IVIS imaging was obtained 28 days after implantation. Upper panel demonstrate the percentage of total photon counts of the metastatic lesions that had tumor in the respective organs, and their representative images are demonstrated below. Vast majority of the metastases from abdomen ODV was peritoneal carcinomatosis, whereas that of chest ODV was distributed among the distant organs. N = 5
ODV is not difficult to perform or learn
A review of the literature revealed that there was no uniformity in the orthotopic injection volume. The most commonly used injectate volumes, 10 and 100 µl, were compared (Fig. 4a–e). ODV implantation of isosulfan blue dye demonstrated greater spillage with 100 µl than with 10 µl (Fig. 4a, b). H&E staining revealed that ODV implantation of 4T1-luc2 cells in a volume of 100 µl (Fig. 4c) produced tissue disruption by dissection of the tissue plane between the fascia and subcutaneous fat and scattering of cancer cells in that layer, whereas in contrast, cells remained in the mammary pad after implantation of the same number of cells in a volume of 10 µl (Fig. 4d). Seven days after ODV implantation of cells in a 10 µl volume, larger tumors were produced than when the same number of cells were implanted in 100 µl determined by bioluminescence (Fig. 4e).
Less fluid is advantageous for ODV, and it is not difficult to learn. 100 µL ODV injection of isosulfan blue spilled out of the mammary gland (a), while 10 µL ODV stayed in the mammary gland (b). Implantation of the same number of cells, 100 versus 10 µL, cause greater tissue disruption, (c) versus (d), and less cancer growth (e). ODV required approximately 3 min longer to perform than other methods (f), and it only required 5 attempts for a researcher with no animal experience (closed diamond) to be proficient in this technique (g)
The major criticisms of ODV in favor of OP is that ODV is technically more demanding and requires more time to master. When the average time to perform these methods was directly compared, ODV required only 3 min more than SQ or OP (Fig. 4f). ODV learning curves were plotted for 3 investigators with no animal experience, 3 with some animal experience, and 3 professionally trained surgeons. As shown in Fig. 4g, even those with no animal experience were able to master the ODV method and were as swift as trained surgeons after 5 trials. In sum, ODV implantation of 10 µl minimized spillage out of the mammary gland and tissue disruption, optimized cancer growth, and was not burdensome to learn or perform.
Implantation of cancer cells in different sites produces tumors that differentially express genes important for breast cancer research and drug development
Because the tumor microenvironment plays a critical role in cancer biology [4, 5, 12–16], we sought to examine the effect of implantation of cancer cells into different microenvironments on gene expression profiles of the tumors that were produced. OP was not included in this analysis due to the limitations of this implantation method demonstrated above (Fig. 1). Initially, we compared the gene expression profiles of 4T1-luc2 cells in vitro (Cells) to ODV and SQ tumors using genome-wide microarrays. Unsupervised cluster analysis demonstrated that there was a significant difference in the clustering of the gene expression profiles between these groups (Fig. 5a). There were 10,350 genes (45.7 % of the entire transcriptome covered by the array) that were differentially expressed between the cells in vitro compared to the ODV and SQ tumors (FDR ≤ 1 %; p < 0.0001). Interestingly, genome-wide microarray analysis comparing SQ and ODV tumors showed that approximately 700 probe sets (3.1 % of the entire transcriptome covered by the array; lower white circle of Fig. 5a) were differentially expressed (FDR ≤ 15 %; p < 0.01). On the other hand, there were no significant differences in the gene expression profiles of tumors in different animals within each group. Supervised cluster analysis of ODV and SQ tumors demonstrated that 206 genes were differentially expressed by a 1.5-fold difference with p < 0.01 (Fig. 5b). A literature search identified 75 % of these 206 genes as either known targets of cancer therapy, cancer biomarkers, or tumor antigens (Fig. 5c). When the gene interaction networks of differentially expressed genes were analyzed for a significance of p ≤ 1 × 10−30, 8 networks were found with functions of known significance in cancer research and drug development (Table 1). Further analysis illustrated how these genes interact with pathways of interest to cancer research and targets of drug development (Fig. 6). These results were further validated by qPCR of 10 specific target genes of interest with significant differences in expression (p < 0.0001) in the development of targeted therapy including wnt5a (Fig. 7a), pdgfa (Fig. 7b), twist2 (Fig. 7c), s100a16 (Fig. 7d), cxcl14 (Fig. 7e), epha3 (Fig. 7f), hsp110 (Fig. 7g), cd59a (Fig. 7h), hdac1 (Fig. 7i), and notch4 (Fig. 7j). Although HDAC1 and NOTCH4 were differentially expressed, HDAC2 and NOTCH1 were not, as shown in Fig. 7i, j, respectively. Taken together, analysis of the differential expression of genetic pathways, gene interaction networks, and qPCR validation of specific targets suggest that the differences between the microenvironments in which SQ and ODV produce tumors differentially regulate gene profiles. Many of those are cancer biomarkers, know targets of cancer therapy, or genes that have been implicated tumor progression suggesting that the choice of the human breast cancer model can influence the development of new therapeutics in the era of targeted therapy.
ODV versus SQ: site of implantation produced tumors with different gene expression profiles. a Unsupervised cluster analysis using centered correlation and average linkage demonstrated that 10,350 genes were commonly different between Cells versus ODV and Cells versus SQ, which implicate the difference in gene expression profile between Cells and tumor in vivo is larger than ODV versus SQ. In total, 700 genes were differentially expressed between ODV and SQ tumors. b Supervised cluster analysis demonstrated the differential expression of 206 significant probe sets (3.1 % of entire transcriptome) by greater than 1.5-fold between ODV and Sq tumors, N = 8. c PubMed search of these 206 significant probe sets demonstrated that 75 % are known targets of therapy, cancer biomarkers, or cancer antigens, with 25 % of unknown clinical significance
ODV and SQ Differentially Expressed Genes Important for Cancer Research and Drug Development. With a significance of p < 0.0001, by genome-wide microarray and qPCR validation, there was a difference in the expression of Wnt5a (a), Cxcl14 (b), Pdgfa (c), S100a16 (d), Twist2 (e), Epha3 (f), Cd59a (g), Hsp110 (h), Hdac1 (i), and Notch4(j), but not Hdac2 (i) or Notch1 (j)
Discussion
Numerous mouse models have been used to study human breast cancer development and metastasis, ranging from spontaneous and carcinogen-induced models to genetically engineered and transplantation mouse models. Although it has been advocated that there should be a wider adoption of orthotopic implantation for cancer research instead of SQ [4, 10, 14, 15, 31–38], there has been a trend toward OP rather than ODV over the last 20 years. This trend has occurred despite the description of the orthotopic method over 20 years ago and of its advantages over other methods 15 years ago [17, 18].
Our results suggest that SQ and OP do not reliably form tumor into their target locations. These results support the reported variability of cancer cell growth and high false positive results in SQ [4–6, 10, 23, 24]. We also found that ODV produced greater tumor growth without a scar-like capsule and shorter survival than SQ. ODV implantation into the chest 2nd mammary pad produced a distribution of metastatic progression that modeled human breast cancer better than implantation into the abdominal 4th mammary pad, the most commonly used site of implantation. Implantation into the chest mammary pad produces metastasis to lung and other distant organs, while implantation into the 4th abdominal mammary pad mainly produced peritoneal disseminations secondary to direct invasion, which is almost never seen in human breast cancer patients. Despite the common misconception, we found that ODV implantation into the chest 2nd mammary pad is not difficult to perform or learn. We also found that a 10 µl volume of implantation was optimal and minimized disruption of the breast tissue. Considering that SQ and OP are the most commonly used methods, the inaccuracies and variable progression demonstrated in this study is an important consideration in favor of adopting ODV for preclinical testing of anti-cancer drug efficacy. These findings further support the multiple reports of Dr. Hoffman et al. that orthotopic models provide an optimal system for breast cancer research and drug development [17, 18].
Transcriptional profiling has been a common way to identify genes and signaling pathways important for carcinogenesis. However, rarely has there been a critical evaluation of how different methods and sites of implantation affect tumor genome-wide gene expression profiles. We have found that there was a significant difference in expression of hundreds of genes between the ODV and SQ tumors. Because the tumor microenvironment is critical to cancer biology, it is not surprising that implanting the same cancer cells into different microenvironments can alter tumor characteristics and gene expression. Strikingly, three quarters of these differentially expressed genes in ODV are of known importance in cancer cell biology and anti-cancer drug development. These differences might explain why SQ implantation models have failed to recapitulate the complexity of human breast cancer progression and failures of translation of positive preclinical trials to the clinic. Evaluation of gene interaction networks indicated that 8 of the most significant divergent networks upregulated in ODV tumors compared to SQ tumors have functions of importance for tumor development and are currently the focus of cancer research and drug development. Examples include Twist2, which is an epithelial-mesenchymal transition inducing E-cadherin repressor [39], and Epha3, which is a key player in tumor growth and survival [40]. These two genes are subjects of intense cancer biology research with the hope for future applications in therapy. Other examples include Wnt5a, an important player in breast epithelial cell migration and a marker of disease-free survival in breast cancer [41]; pdgf-alpha receptor which is correlated with breast cancer invasiveness and aggressiveness [42]; S100a16 (Fig. 7d) involved with cell motility, growth, differentiation, cell cycle progression, transcription, secretion, and a marker for circulating tumor cells, which is used as cancer diagnostic and prognostic markers [43, 44]; Cxcl14 promotes cell motility, which is associated with breast cancer development and progression, and its expression level is even associated with clinical time to breast cancer metastasis [45]. HSP110 (Fig. 7h) has been correlated with clinical stage in colorectal cancer and breast cancer, and is currently a target of cancer vaccines and molecular targeted chemotherapy [46, 47]. Antibodies to CD59a are currently under investigation to improve the efficacy of the chemotherapuetic agent rituximab [48]. HDAC1 is also currently under investigation as a target with a new class of inhibitors undergoing clinical trials in targeted cancer treatment. Interestingly, HDAC2 inhibitors showed promise in animals and failed in humans, and similarly HDAC2 demonstrated no significant differential expression between SQ and ODV, but HDAC1 which is thought to be a more appropriate target was differentially expressed [49]. NOTCH inhibitors provided a similar example: NOTCH 1 inhibitors demonstrated promise in xenograft mouse models, failed in humans, and recent findings suggest that NOTCH4 inhibitors instead offer greater promise [50]. These findings underscore the importance of characterizing the genomic profiles of the tumors produced by mouse models when screening for the efficacy of novel agents in the era of targeted therapy.
References
Siegel R, Ma J, Zou Z, Jemal A (2014) Cancer statistics, 2014. CA Cancer J Clin 64(1):9–29. doi:10.3322/caac.21208
Light DW (2009) Global drug discovery: Europe is ahead. Health Aff (Millwood) 28(5):w969–w977. doi:10.1377/hlthaff.28.5.w969
Adams CP, Brantner VV (2006) Estimating the cost of new drug development: is it really 802 million dollars? Health Aff (Millwood) 25(2):420–428. doi:10.1377/hlthaff.25.2.420
Schuh JC (2004) Trials, tribulations, and trends in tumor modeling in mice. Toxicol Pathol 32(Suppl 1):53–66
Ottewell PD, Coleman RE, Holen I (2006) From genetic abnormality to metastases: murine models of breast cancer and their use in the development of anticancer therapies. Breast Cancer Res Treat 96(2):101–113. doi:10.1007/s10549-005-9067-x
Gravekamp C, Sypniewska R, Gauntt S, Tarango M, Price P, Reddick R (2004) Behavior of metastatic and nonmetastatic breast tumors in old mice. Exp Biol Med (Maywood) 229(7):665–675
Mitka M (2012) Oncology drugs enter the US market faster than in Europe but cost more. JAMA, J Am Med Assoc 308(6):555. doi:10.1001/jama.2012.9535
Bloomston M, Zervos EE, Rosemurgy AS 2nd (2002) Matrix metalloproteinases and their role in pancreatic cancer: a review of preclinical studies and clinical trials. Ann Surg Oncol 9(7):668–674
Ding L, Ellis MJ, Li S, Larson DE, Chen K, Wallis JW, Harris CC, McLellan MD, Fulton RS, Fulton LL, Abbott RM, Hoog J, Dooling DJ, Koboldt DC, Schmidt H, Kalicki J, Zhang Q, Chen L, Lin L, Wendl MC, McMichael JF, Magrini VJ, Cook L, McGrath SD, Vickery TL, Appelbaum E, Deschryver K, Davies S, Guintoli T, Crowder R, Tao Y, Snider JE, Smith SM, Dukes AF, Sanderson GE, Pohl CS, Delehaunty KD, Fronick CC, Pape KA, Reed JS, Robinson JS, Hodges JS, Schierding W, Dees ND, Shen D, Locke DP, Wiechert ME, Eldred JM, Peck JB, Oberkfell BJ, Lolofie JT, Du F, Hawkins AE, O’Laughlin MD, Bernard KE, Cunningham M, Elliott G, Mason MD, Thompson DM, Jr., Ivanovich JL, Goodfellow PJ, Perou CM, Weinstock GM, Aft R, Watson M, Ley TJ, Wilson RK, Mardis ER (2010) Genome remodelling in a basal-like breast cancer metastasis and xenograft. Nature 464 (7291):999–1005. 10.1038/nature08989
Tao K, Fang M, Alroy J, Sahagian GG (2008) Imagable 4T1 model for the study of late stage breast cancer. BMC Cancer 8:228. doi:10.1186/1471-2407-8-228
Miller FR (1983) Tumor subpopulation interactions in metastasis. Invasion Metastasis 3(4):234–242
Hann B, Balmain A (2001) Building ‘validated’ mouse models of human cancer. Curr Opin Cell Biol 13(6):778–784
Le Devedec SE, van Roosmalen W, Maria N, Grimbergen M, Pont C, Lalai R, van de Water B (2009) An improved model to study tumor cell autonomous metastasis programs using MTLn3 cells and the Rag2(-/-) gammac (-/-) mouse. Clin Exp Metastasis 26(7):673–684. doi:10.1007/s10585-009-9267-6
Jonkers J, Derksen PW (2007) Modeling metastatic breast cancer in mice. J Mammary Gland Biol Neoplasia 12(2–3):191–203. doi:10.1007/s10911-007-9050-8
Teicher BA (2006) Tumor models for efficacy determination. Mol Cancer Ther 5(10):2435–2443. doi:10.1158/1535-7163.MCT-06-0391
Talmadge JE, Singh RK, Fidler IJ, Raz A (2007) Murine models to evaluate novel and conventional therapeutic strategies for cancer. Am J Pathol 170(3):793–804. doi:10.2353/ajpath.2007.060929
Fu X, Le P, Hoffman RM (1993) A metastatic orthotopic-transplant nude-mouse model of human patient breast cancer. Anticancer Res 13(4):901–904
Hoffman RM (1999) Orthotopic metastatic mouse models for anticancer drug discovery and evaluation: a bridge to the clinic. Invest New Drugs 17(4):343–359
Li X, Wang J, An Z, Yang M, Baranov E, Jiang P, Sun F, Moossa AR, Hoffman RM (2002) Optically imageable metastatic model of human breast cancer. Clin Exp Metastasis 19(4):347–350
Hoffman RM (2005) The multiple uses of fluorescent proteins to visualize cancer in vivo. Nat Rev Cancer 5(10):796–806. doi:10.1038/nrc1717
Hoffman RM, Yang M (2006) Subcellular imaging in the live mouse. Nat Protoc 1(2):775–782. doi:10.1038/nprot.2006.109
Hoffman RM, Yang M (2006) Whole-body imaging with fluorescent proteins. Nat Protoc 1(3):1429–1438. doi:10.1038/nprot.2006.223
Burger MM (2000) UICC study group on basic and clinical cancer research: animal models for the natural history of cancer. Meeting held at Woods Hole, MA (USA), June 21–23, 1999. International Union Against Cancer. Int J Cancer 85(3):303–305. doi:10.1002/(SICI)1097-0215(20000201)85:3<303:AID-IJC1>3.0.CO;2-2
Johnson JI, Decker S, Zaharevitz D, Rubinstein LV, Venditti JM, Schepartz S, Kalyandrug S, Christian M, Arbuck S, Hollingshead M, Sausville EA (2001) Relationships between drug activity in NCI preclinical in vitro and in vivo models and early clinical trials. Br J Cancer 84(10):1424–1431. doi:10.1054/bjoc.2001.1796
Rashid OM, Nagahashi M, Ramachandran S, Graham L, Yamada A, Spiegel S, Bear HD, Takabe K (2013) Resection of the primary tumor improves survival in metastatic breast cancer by reducing overall tumor burden. Surgery 153(6):771–778. doi:10.1016/j.surg.2013.02.002
Nagahashi M, Ramachandran S, Kim EY, Allegood JC, Rashid OM, Yamada A, Zhao R, Milstien S, Zhou H, Spiegel S, Takabe K (2012) Sphingosine-1-phosphate produced by sphingosine kinase 1 promotes breast cancer progression by stimulating angiogenesis and lymphangiogenesis. Cancer Res 72(3):726–735. doi:10.1158/0008-5472.CAN-11-2167
Dumur CI, Nasim S, Best AM, Archer KJ, Ladd AC, Mas VR, Wilkinson DS, Garrett CT, Ferreira-Gonzalez A (2004) Evaluation of quality-control criteria for microarray gene expression analysis. Clin Chem 50(11):1994–2002. doi:10.1373/clinchem.2004.033225
Hait NC, Allegood J, Maceyka M, Strub GM, Harikumar KB, Singh SK, Luo C, Marmorstein R, Kordula T, Milstien S, Spiegel S (2009) Regulation of histone acetylation in the nucleus by sphingosine-1-phosphate. Science 325(5945):1254–1257. doi:10.1126/science.1176709
Irizarry RA, Bolstad BM, Collin F, Cope LM, Hobbs B, Speed TP (2003) Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Res 31(4):e15
Storey J (2002) A direct approach to false discovery rates. J Royal Stat Soc 64(3):479–498. doi:10.1111/1467-9868.00346
Cabioglu N, Summy J, Miller C, Parikh NU, Sahin AA, Tuzlali S, Pumiglia K, Gallick GE, Price JE (2005) CXCL-12/stromal cell-derived factor-1alpha transactivates HER2-neu in breast cancer cells by a novel pathway involving Src kinase activation. Cancer Res 65(15):6493–6497. doi:10.1158/0008-5472.CAN-04-1303
Orimo A, Gupta PB, Sgroi DC, Arenzana-Seisdedos F, Delaunay T, Naeem R, Carey VJ, Richardson AL, Weinberg RA (2005) Stromal fibroblasts present in invasive human breast carcinomas promote tumor growth and angiogenesis through elevated SDF-1/CXCL12 secretion. Cell 121(3):335–348. doi:10.1016/j.cell.2005.02.034
Viola RJ, Provenzale JM, Li F, Li CY, Yuan H, Tashjian J, Dewhirst MW (2008) In vivo bioluminescence imaging monitoring of hypoxia-inducible factor 1alpha, a promoter that protects cells, in response to chemotherapy. AJR Am J Roentgenol 191(6):1779–1784. doi:10.2214/AJR.07.4060
Perou CM, Sorlie T, Eisen MB, van de Rijn M, Jeffrey SS, Rees CA, Pollack JR, Ross DT, Johnsen H, Akslen LA, Fluge O, Pergamenschikov A, Williams C, Zhu SX, Lonning PE, Borresen-Dale AL, Brown PO, Botstein D (2000) Molecular portraits of human breast tumours. Nature 406(6797):747–752. doi:10.1038/35021093
van’tVeer LJ, Dai H, van de Vijver MJ, He YD, Hart AA, Mao M, Peterse HL, van der Kooy K, Marton MJ, Witteveen AT, Schreiber GJ, Kerkhoven RM, Roberts C, Linsley PS, Bernards R, Friend SH (2002) Gene expression profiling predicts clinical outcome of breast cancer. Nature 415(6871):530–536. doi:10.1038/415530a
Wang Y, Klijn JG, Zhang Y, Sieuwerts AM, Look MP, Yang F, Talantov D, Timmermans M, Meijer-van Gelder ME, Yu J, Jatkoe T, Berns EM, Atkins D, Foekens JA (2005) Gene-expression profiles to predict distant metastasis of lymph-node-negative primary breast cancer. Lancet 365(9460):671–679. doi:10.1016/S0140-6736(05)17947-1
Minn AJ, Gupta GP, Siegel PM, Bos PD, Shu W, Giri DD, Viale A, Olshen AB, Gerald WL, Massague J (2005) Genes that mediate breast cancer metastasis to lung. Nature 436(7050):518–524. doi:10.1038/nature03799
Yang J, Mani SA, Donaher JL, Ramaswamy S, Itzykson RA, Come C, Savagner P, Gitelman I, Richardson A, Weinberg RA (2004) Twist, a master regulator of morphogenesis, plays an essential role in tumor metastasis. Cell 117(7):927–939. doi:10.1016/j.cell.2004.06.006
Tsuji T, Ibaragi S, Shima K, Hu MG, Katsurano M, Sasaki A, Hu GF (2008) Epithelial-mesenchymal transition induced by growth suppressor p12CDK2-AP1 promotes tumor cell local invasion but suppresses distant colony growth. Cancer Res 68(24):10377–10386. doi:10.1158/0008-5472.CAN-08-1444
Balakrishnan A, Bleeker FE, Lamba S, Rodolfo M, Daniotti M, Scarpa A, van Tilborg AA, Leenstra S, Zanon C, Bardelli A (2007) Novel somatic and germline mutations in cancer candidate genes in glioblastoma, melanoma, and pancreatic carcinoma. Cancer Res 67(8):3545–3550. doi:10.1158/0008-5472.CAN-07-0065
Hansen C, Howlin J, Tengholm A, Dyachok O, Vogel WF, Nairn AC, Greengard P, Andersson T (2009) Wnt-5a-induced phosphorylation of DARPP-32 inhibits breast cancer cell migration in a CREB-dependent manner. J Biol Chem 284(40):27533–27543. doi:10.1074/jbc.M109.048884
Carvalho I, Milanezi F, Martins A, Reis RM, Schmitt F (2005) Overexpression of platelet-derived growth factor receptor alpha in breast cancer is associated with tumour progression. BCR: Breast Cancer Res 7(5):R788–R795. doi:10.1186/bcr1304
Bianchi L, Canton C, Bini L, Orlandi R, Menard S, Armini A, Cattaneo M, Pallini V, Bernardi LR, Biunno I (2005) Protein profile changes in the human breast cancer cell line MCF-7 in response to SEL1L gene induction. Proteomics 5(9):2433–2442. doi:10.1002/pmic.200401283
Smirnov DA, Zweitzig DR, Foulk BW, Miller MC, Doyle GV, Pienta KJ, Meropol NJ, Weiner LM, Cohen SJ, Moreno JG, Connelly MC, Terstappen LW, O’Hara SM (2005) Global gene expression profiling of circulating tumor cells. Cancer Res 65(12):4993–4997. doi:10.1158/0008-5472.CAN-04-4330
Pelicano H, Lu W, Zhou Y, Zhang W, Chen Z, Hu Y, Huang P (2009) Mitochondrial dysfunction and reactive oxygen species imbalance promote breast cancer cell motility through a CXCL14-mediated mechanism. Cancer Res 69(6):2375–2383. doi:10.1158/0008-5472.CAN-08-3359
Wang XY, Chen X, Manjili MH, Repasky E, Henderson R, Subjeck JR (2003) Targeted immunotherapy using reconstituted chaperone complexes of heat shock protein 110 and melanoma-associated antigen gp100. Cancer Res 63(10):2553–2560
Lee SU, Kim BT, Min YK, Kim SH (2006) Protein profiling and transcript expression levels of heat shock proteins in 17beta-estradiol-treated human MCF-7 breast cancer cells. Cell Biol Int 30(12):983–991. doi:10.1016/j.cellbi.2006.07.005
Macor P, Tripodo C, Zorzet S, Piovan E, Bossi F, Marzari R, Amadori A, Tedesco F (2007) In vivo targeting of human neutralizing antibodies against CD55 and CD59 to lymphoma cells increases the antitumor activity of rituximab. Cancer Res 67(21):10556–10563. doi:10.1158/0008-5472.CAN-07-1811
Balasubramanian S, Verner E, Buggy JJ (2009) Isoform-specific histone deacetylase inhibitors: the next step? Cancer Lett 280(2):211–221. doi:10.1016/j.canlet.2009.02.013
Harrison H, Farnie G, Howell SJ, Rock RE, Stylianou S, Brennan KR, Bundred NJ, Clarke RB (2010) Regulation of breast cancer stem cell activity by signaling through the Notch4 receptor. Cancer Res 70(2):709–718. doi:10.1158/0008-5472.CAN-09-1681
Acknowledgments
The authors thank the Virginia Commonwealth University Health System Anatomic Pathology Research Services (APRS) Director Dr. Jorge A. Almenara and histotechnologists for technical assistance in the tissue processing, sectioning and staining. NIH grants (R01CA160688) and Susan G. Komen for the Cure Investigator Initiated Research Grant (IIR12222224) to KT, and NCI grant R01CA61774 to SS. MN is a Japan Society for the Promotion of Science Postdoctoral Fellow.
Conflicts of interest
All authors have no conflicts of interest in regards to the content of this manuscript.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Rashid, O.M., Nagahashi, M., Ramachandran, S. et al. An improved syngeneic orthotopic murine model of human breast cancer progression. Breast Cancer Res Treat 147, 501–512 (2014). https://doi.org/10.1007/s10549-014-3118-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10549-014-3118-0