Abstract
Nigrospora oryzae is a pathogen that can infect plants of various species. Here, we report the isolation of a novel mycovirus from N. oryzae infecting rice, as well as the complete genome sequence and genomic organization of this virus, which we have named “Nigrospora oryzae nonsegmented RNA virus 1” (NoNRV1). The genome of NoNRV1 contained two non-overlapping open reading frames (ORF1 and ORF2) potentially encoding a protein with an unknown function in ORF1 and a putative RNA-dependent RNA polymerase (RdRp) in ORF2. Homology and phylogenetic analysis revealed that NoNRV1 was most similar to the Ustilaginoidea virens nonsegmented virus 1 (UvNV-1) and distantly related to members of the virus family Partitiviridae. It is proposed that NoNRV1, together with UvNV-1 and other related viruses, might represent a novel virus taxon of mycoviruses belonging to a partitivirus-like lineage.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Mycoviruses, or fungal viruses, have been described in major fungal groups, including yeasts, oomycetes, and filamentous fungi, and most of them have been found in plant-pathogenic fungi [1–3]. Most mycoviruses have double-stranded (ds) RNA genomes and belong to one of six families: Totiviridae, Partitiviridae, Chrysoviridae, Reoviridae, Megabirnaviridae, Quadriviridae and the proposed family “Alternaviridae” [4]. However, the classification is being updated, with an increasing number of novel mycoviruses having been reported that may be phylogenetically related to plant viruses or animal viruses but cannot yet be conclusively assigned to a particular family. Generally, most mycoviruses do not produce discernible symptoms in their fungal hosts [1]. However, some mycoviruses can induce phenotypic aberrations or alterations in their host, including hypovirulence and debilitation. Such mycoviruses may be manipulated as biological control agents against fungal diseases. This capacity has been demonstrated for the control of chest blight disease in Europe, by the use of Cryphonectria hypovirus 1 as a biocontrol agent [5]. Moreover, mycoviruses have expanded our understanding of fungal and viral pathogenesis, virus evolution, the viral life cycle, and interaction with their fungal hosts. Additionally, newly discovered and unassigned mycoviruses will promote progress in the advanced study of mycovirology. In recent years, the establishment of a virus family named “Amalgaviridae” has been proposed to accommodate a group of new viruses whose genomes are composed of an undivided, relatively small dsRNA segment [6]. In addition, several unclassified dsRNA viruses that infect fungal hosts were recently found to have a similar genomic composition [7, 8].
N. oryzae is a known pathogen that is able to infect maize, rice, sorghum, cotton, weeds, cotton (Gossypium hirsutum), Kentucky bluegrass (Poa pratensis), brown mustard (Brassica juncea), Aloe vera, Dendrobium candidum, and various other plants [9–13]. To date, no mycovirus has been identified from N. oryzae. Here, we describe a new RNA virus isolated from the pathogenic fungus N. oryzae and provisionally name it “Nigrospora oryzae nonsegmented RNA virus 1” (NoNRV1).
Provenance of the virus material
N. oryzae isolate HN-21, which was isolated from a diseased rice leaf in Hunan Province, China, in 2015, was cultured on potato dextrose agar. Its identity was determined by rDNA-ITS sequencing (accession number: KT266531). Extraction of dsRNA from mycelia was done as described by Morris and Dodds [14]. The dsRNA nature of these extracts was ascertained by digestion with RNase-free DNase I and S1 nuclease (TaKaRa, Dalian, China), which was used to eliminate potential DNA and ssRNA molecules, and by the cellulose (CF-11) column chromatography extraction method (with 16 % ethanol concentration). The dsRNA sample electrophoresed on a 1 % (w/v) agarose gel, stained with ethidium bromide, and viewed on a UV transilluminator. A fragment of approximately 3 kbp was purified and used as a template for reverse transcription and cDNA library synthesis, using random hexanucleotide primers and reverse transcriptase. Sequence gaps not covered by the cDNA library in the initial round of sequencing were filled by RT-PCR amplification using primers that were designed based on the cDNA sequences obtained. To clone the ends of the dsRNA, adapter ligation and amplification were carried out as described previously [15]. All of the amplified cDNA products were cloned into the pMD18-T vector (TaKaRa) and sequenced, with each base having been determined independently at least three times. The final contigs were assembled and deposited in the GenBank database under accession number KT258976. Homology searches and multiple sequence alignment were performed against the National Center for Biotechnology Information (NCBI) databases (http://www.ncbi.nlm.nih.gov/genomes) using the programs BLAST and ClustalX [16]. Based on sequence alignment, a molecular phylogenetic tree was constructed by the neighbor-joining (NJ) method, with 1,000 bootstrap re-samplings in MEGA 6 [17].
Sequence analysis
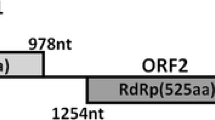
The full-length cDNA of NoNRV1 was 2857 nt in length with a G+C composition of 57.9 %. Two putative open reading frames (ORF1 and ORF2), spaced 58 nt apart and preceded and followed by untranslated regions (UTRs) of 34 nt and 56 nt respectively, were found in the coding strand of NoNRV1. A schematic representation of the genomic organization of NoNRV1 is shown in Fig. 1. We also detected another ORF (from nt 667 to 1188), located within ORF2. However, the potentially encoded product of this ORF displayed no detectable similarity to any proteins in the NCBI database, and no putative conserved domain was detected. The functions of this ORF and its encoded product are unknown.
Genomic organization of NoNRV1, Alternaria longipes dsRNA virus 1 (AlRV1), southern tomato virus (STV), and Heterobasidion RNA virus 6. The open reading frames (ORFs) and untranslated regions of these viruses are indicated by shaded boxes and black lines, respectively. The positions of the initiation and termination codons of the respective ORFs are indicated by numbers above solid lines. The genomes of NoNRV1, AlRV1 and STV (2857 nt and 3415 nt, respectively) contain two nonoverlapping ORFs. STV is 3437 nt in length, containing two overlapping ORFs, and HRV6 (1884 nt in length) has one ORF
ORF1 encodes a putative 174-aa protein with an approximate molecular mass of 19.3 kDa. A BLASTp search using the deduced aa sequence of ORF1 showed no significant similarity to any of the viral sequences present in the NCBI protein databases, with the exception of a hypothetical protein encoded by Ustilaginoidea virens nonsegmented virus 1 (UvNV1) showing 37 % aa sequence identity (E-value: 1e-24; query cover: 95 %). The fact that no putative conserved domain was detected might suggest an unknown function of this ORF1 protein. ORF2, from nt position 618 to 2801, was predicted to encode a 82.8-kDa polyprotein consisting of 727 aa. A Blastp search of the NCBI protein database showed that the 82.8-kDa protein has a degree of sequence similarity to the RdRps of some unclassified dsRNA viruses. The UvNV1 is the most closely related one, with 31 % aa sequence identity (E-value: 6e-43; query cover: 63 %), followed by Bryopsis mitochondria-associated dsRNA (BDRM), which has 26 % aa sequence identity of (E-value: 1e-19; query cover: 45 %) [18, 19]. The ORF2-encoded protein also appears to be distantly related to the RdRps of viruses in the family Partitiviridae, but with relatively low coverage. Alignment of multiple protein sequences and comparison to known viral sequences, using a conserved domain search, demonstrated that the ORF2-encoded protein of NoNRV1 shared a conserved viral RdRp domain (pfam00680) containing conserved motifs that are characteristic of the RdRps of dsRNA mycoviruses.
A phylogenetic tree was constructed based on multiple alignments of the selected aa sequences of viral RdRps (Fig. 2). This revealed that NoNRV1, as expected from the homology search results, formed an independent clade together with UvNV-1 and BDRM in the unclassified family. This NoNRV1 clade was distantly related to members of the family Partitiviridae but distinct from the viruses in the family Totiviridae.
Phylogenic analysis of NoNRV1. The phylogenic tree was constructed, using MEGA 6, by the neighbor-joining method (1,000 bootstrap replicates) based on a multiple aa sequence alignment of the viral RdRps of NoNRV1 and other selected viruses. The values near the branches indicate the percentage of bootstrap replicates supporting the branch. The virus names and GenBank accession numbers are as follows: Alternaria longipes dsRNA virus 1 (YP_009052469.1), Aspergillus fumigatus partitivirus 1 (CAY25801.2), Atkinsonella hypoxylon virus (L39125.1), Beauveria bassiana RNA virus 1 (AKC57301.1), beet cryptic virus 3 (AAB27624.1), blueberry latent virus (YP_003934623.1), Botryotinia fuckeliana partitivirus 1 (AM491609), Bryopsis mitochondria-associated dsRNA (BAA25883.1), Ceratocystis resinifera partitivirus (AY603052.1), Colletotrichum acutatum RNA virus 1 (AGL42312.1), Discula destructiva virus 1 (NP_116716.1), Fragaria chiloensis cryptic virus (YP_001274391.1), Fusarium graminearum dsRNA mycovirus-4 (YP_003288790.1), Gremmeniella abietina RNA virus 6 (AIU98624.1), Helminthosporium victoriae virus 190S (U41345), Heterobasidion RNA virus 6 (AHA82556.1), pepper cryptic virus 1 (AEJ07890.1), Raphanus sativus cryptic virus 2 (ABB04855.1), rhododendron virus A (ADM36020.1), Saccharomyces cerevisiae virus L-A (AAA50321.1), Sclerotinia sclerotiorum partitivirus S (GQ280377.1), southern tomato virus (YP_002321509.1), Ustilaginoidea virens nonsegmented virus 1 (AIE77248.1), Ustilaginoidea virens partitivirus 2 (AGR45851), Ustilaginoidea virens partitivirus 4 (AGJ03719), Ustilaginoidea virens unassigned RNA virus HNND-1 (YP_009154709), white clover cryptic virus 1 (AY705784.1)
Members of the newly established family Amalgaviridae, which includes a new group of plant-infecting dsRNA viruses such as southern tomato virus, blueberry latent virus, and rhododendron virus A, share a nonsegmented genome and a totiviruses-like organization [6]. These viruses form a sister phylogenetic clade based on RdRp sequences when compared to viruses in the family Partitiviridae. Investigators have consequently proposed the amalgaviruses to be an evolutionary nexus between the families Partitiviridae and Totiviridae [20]. Our phylogenetic analysis suggested that the BbV1-related viruses (such as Alternaria longipes dsRNA virus 1 and UvNV1) and a group of unclassified dsRNA viruses (as represented by Heterobasidion RNA virus 6 [HRV6] or Fusarium graminearum dsRNA mycovirus 4 [FgV4]) were phylogenetically grouped and formed a dominant cluster together with the amalgaviruses. This result was accordant with the conclusion proposed by Koloniuk et al. [21] that these unclassified dsRNA viruses, which infect fungal hosts, constitute a distinct lineage (called amalgamycoviruses) and can be classified as members of the family Amalgaviridae. However, although they share the similarities of being nonsegmented and having a totiviruses-like genome organization, these viruses also exhibit differences in their genome organization. The viruses of the BbV1-related clade have two non-overlapping ORFs, whereas the plant-infecting amalgaviruses have two overlapping ORFs (Fig. 1). The viruses in the HRV6-related clade have only one large ORF in the RdRp-encoding strand, and the genome is not confined to one dsRNA segment, as is seen, for example, in FgV4. NoNRV1, despite exhibiting similarities in genome organization to a BbV1-related virus that is closely related to amalgaviruses as well as members of the family Totiviridae, this virus, along with UvNV-1 and BDRM, was phylogenetically separate from members of the family Amalgaviridae, and it seems to have a closer evolutionary relationship to the members in the family Partitiviridae, consistent with the RdRp-based Blastp search results.
In conclusion, NoNRV1, UvNV-1, and BDRM may be considered members of a novel taxon accommodating unassigned mycoviruses in a partitivirus-like lineage that is distinct from the members of amalgaviruses and other amalgavirus-like mycoviruses. As far as we are aware, this article is the first written report of a mycovirus identified from the phytopathogenic fungus Nigrospora oryzae.
References
Ghabrial SA, Suzuki N (2009) Viruses of plant pathogenic fungi. Annu Rev Phytopathol 47:353–384
Pearson MN, Beever RE, Boine B, Arthur K (2009) Mycoviruses of filamentous fungi and their relevance to plant pathology. Mol Plant Pathol 10:115–128
Xie JT, Jiang DH (2014) New Insights into mycoviruses and exploration for the biological control of crop fungal diseases. Annu Rev Phytopathol 52:45–68
Ghabrial SA, Castón JR, Jiang D, Nibert ML, Suzuki N (2015) 50-plus years of fungal viruses. Virology 479:356–368
Nuss DL (2005) Hypovirulence: mycoviruses at the fungal–plant interface. Nat Rev Microbiol 3:632–642
Nibert ML, Ghabrial SA, Maiss E, Lesker T, Vainio EJ, Jiang D, Suzuki N (2014) Taxonomic reorganization of family Partitiviridae and other recent progress in partitivirus research. Virus Res 188:128–141
Lin Y, Zhang H, Zhao C, Liu S, Guo L (2014) The complete genome sequence of a novel mycovirus from Alternaria longipes strain HN28. Arch Virol 160:577–580
Kotta-Loizou I, Sipkova J, Coutts RH (2015) Identification and sequence determination of a novel double-stranded RNA mycovirus from the entomopathogenic fungus Beauveria bassiana. Arch Virol 160:873–875
Zheng L, Shi F, Kelly D, Hsiang T (2012) First Report of Leaf Spot of Kentucky Bluegrass (Poa pratensis) Caused by Nigrospora oryzae in Ontario. Plant Dis 96:909
Sharma P, Meena PD, Chauhan JS (2013) First Report of Nigrospora oryzae (Berk. & Broome) Petch Causing Stem Blight on Brassica juncea in India. J Phytopathol 161:439–441
Zhai LF, Liu J, Zhang MX, Hong N, Wang GP, Wang LP (2013) The First Report of Leaf Spots in Aloe vera Caused by Nigrospora oryzae in China. Plant Dis 97:1256
Wu JB, Zhang CL, Mao PP, Qian YS, Wang HZ (2014) First report of leaf spot caused by Nigrospora oryzae on Dendrobium candidum in China. Plant Dis 98:996
Zhang LX, Li SS, Tan GJ, Shen JT, He T (2012) First Report of Nigrospora oryzae causing leaf spot of cotton in China. Plant Dis 96:1379
Morris TJ, Dodds JA (1979) Isolation and analysis of double stranded RNA from virus-infected plant and fungal tissue. Phytopathology 69:854–858
Zhong J, Lei XH, Zhu JZ, Song G, Zhang YD, Chen Y, Gao BD (2014) Detection and sequence analysis of two novel co-infecting double-strand RNA mycoviruses in Ustilaginoidea virens. Arch Virol 159:3063–3070
Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Higgins DG (2007) Clustal W and Clustal X version 2.0. Bioinformatics 23:2947–2948
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 30:2725–2729
Zhang T, Jiang Y, Dong W (2014) A novel monopartite dsRNA virus isolated from the phytopathogenic fungus Ustilaginoidea virens and ancestrally related to a mitochondria-associated dsRNA in the green alga Bryopsis. Virology 462:227–235
Koga R, Fukuhara T, Nitta T (1998) Molecular characterization of a single mitochondria-associated double-stranded RNA in the green alga Bryopsis. Plant Mol Biol 36:717–724
Sabanadzovic S, Valverde RA, Brown JK, Martin RR, Tzanetakis IE (2009) Southern tomato virus: the link between the families Totiviridae and Partitiviridae. Virus Res 140:130–137
Koloniuk I, Hrabáková L, Petrzik K (2015) Molecular characterization of a novel amalgavirus from the entomopathogenic fungus Beauveria bassiana. Arch Virol 160:1585–1588
Acknowledgments
This study was supported by Hunan Provincial Innovation Foundation for Postgraduates (grant number CX2014B291).
Author information
Authors and Affiliations
Corresponding author
Additional information
Q. Zhou and J. Zhong contributed equally to this work.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Zhou, Q., Zhong, J., Hu, Y. et al. A novel nonsegmented double-stranded RNA mycovirus identified in the phytopathogenic fungus Nigrospora oryzae shows similarity to partitivirus-like viruses. Arch Virol 161, 229–232 (2016). https://doi.org/10.1007/s00705-015-2644-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00705-015-2644-3