Abstract
Here, we report a novel virus isolated from rice blast fungus, Magnaporthe oryzae, an important plant pathogen. This virus has an RNA genome of 3246 nucleotides. Its genome possesses two in-frame open reading frames (ORFs). The smaller ORF1 encodes a protein with significant similarity to a protein encoded by the ssRNA mycovirus Diaporthe ambigua RNA virus 1 (DaRV1). The larger ORF2 encodes a protein with similarity to RNA-dependent RNA polymerases (RdRp) of DaRV1 and other plant viruses of the family Tombusviridae. In silico analysis and comparisons with DaRV1 genome expression suggest that ORF2 is translated via a readthrough mechanism together with ORF1. Based upon results of this study, this virus, for which the provisional name Magnaporthe oryzae virus A (MoVA) is proposed, belongs to a new virus species. Furthermore, MoVA along with DaRV1 belong to a new taxon of mycoviruses that are evolutionarily related to plant viruses belonging to the family Tombusviridae.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Mycoviruses are widespread in filamentous fungi and yeast [1, 2]. Mycoviruses with dsRNA genomes are often encapsidated in isometric particles (members of the family Endornaviridae are an exception) and are currently classified into seven families: Totiviridae, Partitiviridae, Chrysoviridae, Reoviridae, Megabirnaviridae, Quadriviridae, and Endornaviridae (with genome segment numbers of 1, 2, 4, 11 or 12, 2, 4, and 1, respectively) [2]. Mycoviruses with single-stranded RNA genomes are classified into three major families: Hypoviridae, Narnaviridae, and Barnaviridae [3]. Many mycoviruses are inconspicuous, causing little or no obvious symptoms in their fungal hosts. However, some mycoviruses can induce phenotypic alterations, including hypovirulence and debilitation, that may be exploited for biological control, such as the successful use of Cryphonectria hypovirus 1 (CHV1) to control chestnut blight in Europe [4].
Magnaporthe oryzae (M. oryzae), formerly known as Magnaporthe grisea (anamorph: Pyricularia oryzae), is a filamentous heterothallic ascomycete that causes rice blast, one of the most prominent destructive fungal diseases of rice worldwide. M. oryzae is the first plant pathogenic fungus reported to contain virus particles [5–7]. Recently, complete sequences of two distinct viruses belonging to the family Totiviridae, named Magnaporthe oryzae virus 1 and 2 (MoV1 and MoV2), were determined [8, 9]. Moreover, Magnaporthe oryzae chrysovirus 1 (MoCV1), detected from a Vietnamese isolate, may negatively affect the growth of M. oryzae, as experimentally demonstrated by overexpression of the ORF4 in Saccharomyces cerevisiae [10, 11]. In addition, Urayama et al. [12] reported that infections by Magnaporthe oryzae chrysovirus 1-B (MoCV1-B) resulted in stronger symptoms displayed on M. oryzae compared to MoCV1-A. Thus, these findings indicate the presence of an array of mycoviruses in M. oryzae, some of which could be potentially manipulated as biological control agents. To date, no virus with a single-stranded RNA (ssRNA) genome has been reported from this fungus. In this paper, we report the detection and partial molecular characterization of a new mycovirus with a positive ssRNA genome, isolated from M. oryzae, which we have provisionally named Magnaporthe oryzae virus A (MoVA).
Provenance of the virus material
The M. oryzae strain, HN-018, was isolated from a rice leaf with rice blast disease in Hunan Province in China in 2012 and identified by rDNA-ITS sequencing (accession number: KP174726). dsRNAs were extracted as described by Morris and Dodds [13]. The dsRNA preparations were treated with RNase-free DNase I and S1 nuclease (TaKaRa, Dalian, China) in order to eliminate potential DNA and ssRNA molecules prior to gel electrophoresis and visualization (Fig. 1a). The cDNA library was constructed using the gel-purified dsRNAs and random hexanucleotide primers along with reverse transcriptase. Gaps in the genome sequence that were not covered by the cDNA library were filled by RT-PCR amplification using sequence-specific primers. To clone the terminal sequences of the dsRNA, adapter ligation and single-primer amplification were conducted [14]. All of the amplified cDNA products were cloned into the pMD18-T vector (TaKaRa) and sequenced in both orientations in at least three independent experiments. The assembled cDNA sequence was deposited in the GenBank database (accession number: KP174727). A multiple sequence alignment was performed using Clustal X [15]. A phylogenetic tree was constructed using the neighbor-joining (NJ) method in MEGA 6 with a bootstrap test of 1,000 re-samplings [16]. A transmembrane helix was predicted with TMHMM Server v. 2.0, and potential RNA secondary structures and motifs were predicted using the program RNA structure v. 5.6.
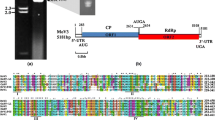
(a) dsRNA banding pattern of M. oryzae strain HN-018, separated on a 1 % agarose gel. (b) Genomic organization of MoVA. (c) Amino acid sequence alignment of the viral RdRps of MoVA and related myco- and plant viruses was carried out using the program Clustal X and highlighted using the GeneDoc application. Virus names are shown in the text or below. Tomato bushy stunt virus, TBSV; grapevine Algerian latent virus, GAlV
Sequence properties
The complete genome sequence of MoVA is 3246 nt in length with two consecutive, in-frame open reading frames (ORFs), spaced by 130 nt and with 5’- and 3’-end untranslated regions (UTRs) of 386 nt and 448 nt, respectively (Fig. 1a and b).
ORF1 was predicted to encode a 265-aa protein with a calculated molecular mass of 29.2 kDa. A BLASTP search showed that the deduced amino acid sequence of the 29.2-kDa protein has a maximum identity of 39 % (E-value: 7e-23; query cover: 67 %) to the protein encoded by ORF1 of DaRV1, whose function is unknown. As reported previously, the replication of positive-stranded RNA virus genomes often takes place in close association with membranes [17]. In the case of some tombusviruses, the targets are peroxisomal or mitochondrial membranes, and for the unencapsidated hypovirus Cryphonectria hypovirus 1 (CHV1), the association is with fungal vesicles of the trans-Golgi network (TGN) [18, 19]. In this study, a possible transmembrane helix was predicted at the N-terminus of the ORF1 protein (Fig. S1), suggesting a role in anchoring the viral protein to the host membrane, as proposed for DaRV1 [20]. Further experiments are required to investigate the localization of MoVA.
ORF2 (nt 1314 to 2798) encoded a 494-aa protein with a molecular mass of 54.6 kDa. A BLASTP search showed that the 54.6-kDa protein is closely related to the RdRps of two unclassified positive-strand ssRNA mycoviruses: DaRV1 and Sclerotinia sclerotiorum umbra-like virus 1 (Ssu-LV1) (48 % aa sequence identity with an E-value of 1e-125 and 92 % coverage for DaRV1; 48 % aa sequence identity with an E-value of 1e-95 and- 80 % coverage for Ssu-Lv1). Also, it has similarity to some plant viruses in the family Tombusviridae, including members of the genera Tombusvirus and Carmovirus, with aa sequence identities ranging from 28 % to 35 % (E-values from 2e-45 to 1e-125). In addition, a conserved domain database (CDD) search and multiple protein alignments confirmed that the 54.6-kDa protein shared a conserved viral RdRp domain in the subfamily RdRP_3 (pfam00998) (Fig. 1c). According to homology search results, we presumed that the MoVA dsRNA we isolated is the replicative intermediate of a mycovirus with a positive single-stranded RNA genome.
Since the two ORFs of MoVA are located in the same frame, they are likely to be expressed as a fusion protein via a readthrough strategy, as proposed for DaRV1. Additionally, a direct comparison of nucleotide sequences surrounding the stop codon revealed similarities to the corresponding part of the DaRV1 genome ([UAC-UAG-GGG] versus [UUU-UAG-GGA]) [21]. The readthrough requires a long-range RNA-RNA interaction between an extended stem-loop (SL) structure proximal to the readthrough site and a sequence in the 3’ untranslated region of the virus genome [22], and it occurs at a UAG stop codon followed by GGR [23], all of which are present in the MoVA genome. However, further experiments are needed to determine the expression mechanism and to identify the structures involved.
In the RdRp-based phylogenetic tree (Fig. 2), MoVA formed a clade with DaRV1 and SsU-LV1 that is evolutionarily related to members of the plant virus family Tombusviridae. Moreover, MoVA is closely related to DaRV1 in sequence similarity and genome organization. Each of the two viruses contains two main non-overlapping ORFs and shares significant sequence similarity in ORF1 and ORF2. Although the two mycoviruses (MoVA and DaRV1) are phylogenetically related to but in their genomic organization clearly differ from members of family Tombusviridae.
Phylogenic tree based on amino acid sequence alignment of viral RNA-dependent RNA polymerases, generated by the NJ method in the program MEGA 6.0. The numbers near the branches indicate the percentage of bootstrap replicates supporting the branch. The virus names and their respective GenBank accession numbers are as follows: carnation Italian ringspot virus (CAA59478.2), cowpea mottle virus (AAC54603.1), Cryphonectria hypovirus 3 (NP_05171 0.1), cymbidium ringspot virus (NP_613260.1), Diaporthe ambigua RNA virus 1(NP_037581.1), grapevine Algerian latent virus (AHZ12756.1), lettuce necrotic stunt virus (AFM91097.1), Moroccan pepper virus (BAN92400.1), Ophiostoma mitovirus 3a (NP_660176.1), pear latent virus (AAM49803.1), pelargonium line pattern virus (YP_238475.1), Sclerotinia homoeocarpa mitovirus (AAO21337.1), Sclerotinia sclerotiorum mitovirus 3(AGC24232.1), Sclerotinia sclerotiorum umbra-like virus 1 (AHE13862.1), tomato bushy stunt virus (CAB56480.1), turnip crinkle virus(NP_620720.3), Valsa ceratosperma hypovirus 1 (YP_005476604.1)
The MoVA RdRp contains the amino acid triplet GDN, which is frequently found in negative-sense RNA viruses instead of the GDD motif found in plant viruses. The same triplet was found in DaRV1 and Ssu-LV1 [24, 25]. Based on our results, we conclude that MoVA belongs to a putative new species of fungal viruses. Similarities in their genome organization and their close phylogenetic relationship to DaRV1 suggest that they both belong to a new taxon of mycoviruses with affinities to the extant members of the family Tombusviridae. Further studies will clarify if the virus causes any changes in the host phenotype as reported for DaRV1 in Diapothe ambigua and other Diaporthe species [20, 26, 27].
References
Ghabrial SA, Suzuki N (2009) Viruses of plant pathogenic fungi. Annu Rev Phytopathol 47:353–384
Pearson MN, Beever RE, Boine B, Arthur K (2009) Mycoviruses of filamentous fungi and their relevance to plant pathology. Mol Plant Pathol 10:115–128
King AMQ, Adams MJ, Carstens EB, Lefkowitz EJ (eds) (2012) Virus taxonomy: classification and nomenclature of viruses: ninth report of the international committee on taxonomy of viruses. Elsevier Academic Press, Amsterdam
Nuss DL (2005) Hypovirulence: mycoviruses at the fungal–plant interface. Nat Rev Microbiol 3:632–642
Yamashita S, Doi Y, Yora K (1971) A polyhedral virus found in rice blast fungus, Pyricularia oryzae Cavara. Ann Phytopathol Soc Japan 37:356–359
Chun SJ, Lee YH (1997) Inheritance of dsRNAs in the rice blast fungus, Magnaporthe grisea. FEMS Microbiol Lett 148:159–162
Hunst PL, Latterell FM, Rossi AE (1986) Variation in double-stranded RNA from isolates of Pyricularia oryzae. Phytopathology 76:674–678
Yokoi T, Yamashita S, Hibi T (2007) The nucleotide sequence and genome organization of Magnaporthe oryzae virus 1. Arch Virol 152:2265–2269
Maejima K, Himeno M, Komatsu K, Kakizawa S, Yamaji Y, Hamamoto H, Namba S (2008) Complete nucleotide sequence of a new double-stranded RNA virus from the rice blast fungus, Magnaporthe oryzae. Arch Virol 153:389–391
Urayama S, Kato S, Suzuki Y, Aoki N, Le MT, Arie T, Teraoka T, Fukuhara T, Moriyama H (2010) Mycoviruses related to chrysovirus affect vegetative growth in the rice blast fungus Magnaporthe oryzae. J Gen Virol 91:3085–3094
Urayama S, Ohta T, Onozuka N, Sakoda H, Fukuhara T, Arie T, Teraoka T, Moriyama H (2012) Characterization of Magnaporthe oryzae chrysovirus 1 structural proteins and their expression in Saccharomyces cerevisiae. J Virol 86:8287–8295
Urayama SI, Sakoda H, Takai R, Katoh Y, LeT Minh, Fukuhara T, Moriyama H (2014) A dsRNA mycovirus, Magnaporthe oryzae chrysovirus 1-B, suppresses vegetative growth and development of the rice blast fungus. Virology 448:265–273
Morris TJ, Dodds JA (1979) Isolation and analysis of double stranded RNA from virus-infected plant and fungal tissue. Phytopathology 69:854–858
Zhong J, Lei XH, Zhu JZ, Song G, Zhang YD, Chen Y, Gao BD (2014) Detection and sequence analysis of two novel co-infecting double-strand RNA mycoviruses in Ustilaginoidea virens. Arch Virol 159:3063–3070
Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Higgins DG (2007) Clustal W and Clustal X version 2.0. Bioinformatics 23:2947–2948
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 30:2725–2729
Schaad MC, Jensen PE, Carrington JC (1997) Formation of plant RNA virus replication complexes on membranes: role of an endoplasmic reticulum-targeted viral protein. EMBO J 16:4049–4059
Rubino L, Di Franco A, Russo M (2000) Expression of a plant virus non- structural protein in Saccharomyces cerevisiae causes membrane proliferation and altered mitochondrial morphology. J Gen Virol 81:279–286
Jacob-Wilk D, Turina M, Van Alfen NK (2006) Mycovirus Cryphonectria hypovirus 1 elements cofractionate with trans-Golgi network membranes of the fungal host Cryphonectria parasitica. J Virol 80:6588–6596
Preisig O, Moleleki N, Smit WA, Wingfield BD, Wingfield MJ (2000) A novel RNA mycovirus in a hypovirulent isolate of the plant pathogen Diaporthe ambigua? J Gen Virol 81:3107–3114
Skuzeski JM, Nichols LM, Gesteland RF, Atkins JF (1991) The signal for a leaky UAG stop codon in several plant viruses includes the two downstream codons. J Mol Biol 218:365–373
Newburn LR, Nicholson BL, Yosefi M, Cimino PA, White KA (2014) Translational readthrough in Tobacco necrosis virus-D. Virology 450:258–265
Firth AE, Wills NM, Gesteland RF, Atkins JF (2011) Stimulation of stop codon readthrough: frequent presence of an extended 3’ RNA structural element. Nuleic Acids Res 39:6679–6691
Poch O, Blumberg BM, Bougueleret L, Tordo N (1990) Sequence comparison of five polymerases (L proteins) or unsegmented negative-strand viruses: theoretical assignment of functional domains. J Gen Virol 71:1153–1162
Sleat DE, Banerjee AK (1993) Transcriptional activity and mutational analysis of recombinant vesicular stomatitis virus RNA polymerase. J Virol 67:1334–1339
Smit WA, Viljoen CD, Wingfield BD, Wingfield MJ, Calitz FJ (1996) A new canker disease of apple, pear and plum rootstocks caused by Diaporthe ambigua in South Africa. Plant Dis 80:1331–1335
Moleleki N, Wingfield MJ, Wingfield BD, Preisig O (2011) Effect of Diaporthe RNA virus 1 (DRV1) on growth and pathogenicity of different Diaporthe species. Eur J Plant Pathol 131:261–268
Acknowledgments
This study is funded by the National Key Technologies R & D Program of China (2012BAD15B04-1).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
This paper is in compliance with ethical standards for research. The authors declare no conflicts of interest.
Additional information
Y.-P. Ai and J. Zhong contributed equally to this work.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Ai, YP., Zhong, J., Chen, CY. et al. A novel single-stranded RNA virus isolated from the rice-pathogenic fungus Magnaporthe oryzae with similarity to members of the family Tombusviridae . Arch Virol 161, 725–729 (2016). https://doi.org/10.1007/s00705-015-2683-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00705-015-2683-9