Abstract
Despite the strong ecological importance of ectomycorrhizal (ECM) fungi, their vertical distribution remains poorly understood. To our knowledge, ECM structures associated with trees have never been reported in depths below 2 meters. In this study, fine roots and ECM root tips were sampled down to 4-m depth during the digging of two independent pits differing by their water availability. A meta-barcoding approach based on Illumina sequencing of internal transcribed spacers (ITS1 and ITS2) was carried out on DNA extracted from root samples (fine roots and ECM root tips separately). ECM fungi dominated the root-associated fungal community, with more than 90% of sequences assigned to the genus Pisolithus. The morphological and barcoding results demonstrated, for the first time, the presence of ECM symbiosis down to 4-m. The molecular diversity of Pisolithus spp. was strongly dependent on depth, with soil pH and soil water content as primary drivers of the Pisolithus spp. structure. Altogether, our results highlight the importance to consider the ECM symbiosis in deep soil layers to improve our understanding of fine roots functioning in tropical soils.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Ectomycorrhizal (ECM) fungi represent a significant fraction of the microbial biomass in forest ecosystems (Churchland and Grayston 2014; Clemmensen et al. 2013; Hogberg and Hogberg 2002; Pagano and Lugo 2019). They establish symbiotic associations with roots and play a major role in tree functioning notably by improving mineral nutrition and water capture for their hosts (Johnson and Gehring 2007; Lehto and Zwiazek 2011). ECM fungi are commonly believed to occur in the surface horizon because of higher fine root densities (Tedersoo et al. 2003; Rosling et al. 2003). However, recent studies in deep soils highlighted the abundance and diversity of microbial communities down to 10-m depth (Gocke et al. 2017; Li et al. 2014; Pereira et al. 2017; Zheng et al. 2017). These results suggest that soil microorganism in deep soil horizons could play an important role in supplying ecosystem services, especially for carbon storage (Churchland and Grayston 2014).
A number of studies on ectomycorrhizas have reported vertical differentiation of ECM fungi through the soil horizons but, in general, has only compared organic with mineral soil layers (Dickie et al. 2002; Genney et al. 2006; Moyersoen et al. 1998; Rosling et al. 2003; Lindahl et al. 2007). Unfortunately, despite the strong ecological importance of ECM fungi, their vertical distribution remains poorly understood and rarely investigated in very deep soil layers (Bakker et al. 2006; Clemmensen et al. 2015; Trocha et al. 2017). In particular ecosystems, such as thin soils with a deep granite layer, few ECM root tips have been observed down to a depth of 4-m in bedrock fissures (Bornyasz et al. 2005; Querejeta et al. 2007). However, to date, there is no study reporting clearly ECM structures associated with tree roots in soil layers deeper than 2-m, neither their diversity.
The area of eucalypt plantations in Brazil was around 5.6 million ha in 2014 (IBÁ 2015), representing around 70% of the planted forests in the country. Among the species used for afforestation, Eucalyptus grandis W. Hill ex Maiden is a suitable model for studying deep roots and their associated mycorrhizal fungi because this fast-growing tree explores a huge volume of soil with functional specialization of fine roots in deep soil layers to take up cations (Bordron et al. 2019; da Silva et al. 2011; Pradier et al. 2017). Although fine root densities decrease rapidly with depth, the density of fine deep roots on the entire soil profile could represent an important part of the fine root biomass (Laclau et al. 2013), and very deep roots can play a key role in tree survival during droughts in eucalypt plantations (Christina et al. 2015, 2017). A better understanding of deep root functioning (including fungal symbiosis) and their role in water capture is very important as drought events are likely to become more frequent with global climate change (Phillips et al. 2016).
A recent study using minirhizotrons reported ECM structures associated with deep roots down to 6-m in a Brazilian eucalypt plantation (Lambais et al. 2017). However, even though this methodology is well suited for root observations and has already been used to study the dynamics of ECM fungi (Allen and Kitajima 2013; McCormack et al. 2017), the method itself could affect mycorrhization, especially in deep soil layers. Spores or mycelium could be transported during the digging process and the minirhizotron tube setup. Moreover, the environmental disturbance near the tube (e.g., changes in oxygen or water content) might create a favorable environment for fungal growth, leading to overestimating mycorrhizal colonization of very deep roots.
Here, we examined whether or not the ectomycorrhizal association occurs in deep tree roots (> 2 m) in Eucalyptus grandis plantations in Brazil. To assess the effects of depth on ECM fungi, we sampled fine roots and ECM root tips developed in ten soil layers during the digging of two large pits down to a depth of 4-m in two eucalypt plots differing by their water availability. We determined the fungal community composition by meta-barcoding targeting the fungal internal transcribed spacers (ITS1 and ITS2) of sampled roots. We hypothesized that the diversity of ECM species associated with eucalypt roots would decrease with depth and that the community structure would also differ, according to soil physical and chemical properties (pH, water content, C and nutrient contents) measured in the soil layers.
Materials and methods
Study site and soil properties
The experiment was conducted at the Itatinga research station of the University of São Paulo in Brazil (23° 02′ S, 48° 38′ W). The climate is subtropical humid with a mean annual rainfall of 1,360 mm and a mean temperature of 19°C (with a cold dry season between June and September). The soil is a deep Ferralsol (water table at 17-m) developed on Cretaceous sandstone. Soil texture, mineralogy, and physical and chemical properties have been described previously (Battie-Laclau et al. 2013; Laclau et al. 2010; Pradier et al. 2017). This highly weathered soil is dominated by the sand fraction, with a clay content ranging from 15% on the first 50 cm to 20–25% in deep soil layers. The soil is acidic and the nutrient contents are low. The main physical and chemical properties of soil are presented in the supplementary Table S1 from results published in Pradier et al. (2017). Briefly, independently of the plot and only for the depth 0–20 cm, the soil has a pH in CaCl2 of 4.17, total C and N contents of 0.69 and 0.04 % respectively, and resin P and K concentrations of 10.22 and 10.58 mg kg−1 respectively.
Experimental design
A split-plot experimental design was set up in 2010 with a highly productive Eucalyptus grandis clone (provided by the Suzano Company, Brazil). The experiment was described in detail by Battie-Laclau et al. (2013). Six treatments (three fertilization regimes × two throughfall exclusion regimes) were applied in three blocks. The area of the individual subplots was 864 m2, with 144 trees at a spacing of 2 × 3 m. Our study was carried out only for one fertilization rate treatment (non-limiting for tree growth), which was representative of the silvicultural practices in commercial eucalypt plantations (12 g N m−2, 3.3 g P m−2, 17.5 g K m−2, 200 g m−2 of dolomitic lime and trace elements applied the first 3 months after planting). Two pits were dug in the same block, one in the throughfall exclusion plot (W−) and the other one in the undisturbed throughfall plot (W+). The W− plot had two 40-cm-wide transparent plastic sheets in each inter-row that were shaped as gutters to exclude 37% of the throughfalls. We decided to carry out careful time-consuming sampling in one pit for each water regime rather than less cautiously sampling in several pits per plot. This method also provided enough deep fine roots and ECM root tips to be collected for analysis, but was extremely time-consuming below 1-m. Other studies in this experimental setup have shown that soil properties (Maquere 2008), tree growth (Christina et al. 2018), and fine root densities (B. Bordron et al., unpublished) were similar in the three blocks for a given combination of treatments.
The soil water content (SWC) was measured from planting to soil sampling at half-hourly intervals, with CS616 probes (Campbell Scientific Inc., Logan, UT, USA), installed at 6 depths down to 4-m (0.15, 0.5, 1, 2, 3, and 4 m; three CS616 probes for each soil depth). The soil profile was dryer and SWC was more uniform in W− than in W+ around the sampling period (Supplementary Fig. S1).
Fine roots and ECM root tips sampling
Samples were collected between May and June 2015. The trees were 5 years old with a mean tree height of 20 m and a root front depth down to 17 m (Christina et al. 2011). A 1.5-m square pit was dug in the center of the 3-m-wide inter-row of both the exclusion (W−) and control (W+) plots, between four neighbor trees. Fine roots (< 2 mm) and ECM root tips were very carefully and manually sampled, layer by layer, while the pit was being dug in order to prevent any contamination between two adjacent soil layers and to collect the maximum biomass of fine roots in each soil layer. Ten soil layers were sampled from 0 to 4-m depth (0–20, 20–40, 40–60, 60–100, 100–150, 150–200, 200–250, 250–300, 300–350, and 350–400 cm) as described in Pradier et al. (2017). The deeper soil layers were thicker than the upper layers to obtain enough fine roots and ECM root tips quantities for analysis.
Two dominant ECM morphotypes were separated during the collection of root tips and photographed in the laboratory in the days following the sampling using an Olympus TG2 camera (sensor BSI CMOS 12 Mpx, 1/2.3″; lens 4× 25–100 mm f/2-4.9, sensitivity 100–6400 ISO). The fine roots and ECM root tips were washed with tap water and stored at − 20 °C until analysis. All the samples were ground in liquid nitrogen before DNA extraction. For each depth, three subsamples of fine roots and ECM root tips were used to constitute pseudo-replicates (n = 3).
DNA extraction, ITS amplification, and Illumina Miseq sequencing
DNA was extracted from two separate samples, fine roots or ECM root tips (80 mg each), using the FastDNA SPIN Kit (MP Biomedicals Santa Ana, CA, USA) according to the manufacturer’s recommendations with some modifications. Samples were homogenized in 800 μL of CLS-VF buffer and 200 μL of PPS by vortexing for 10 min, followed by incubation for 15 min at room temperature. The samples were centrifuged (14,000g, 10 min), and a binding matrix equal in volume to the volume of supernatant was added, and agitated for 5 min at room temperature. After a centrifugation (14,000g, 10 min), a guanidine wash step was added as described by Tournier et al. (2015) to improve the purity of DNA extract. The DNA binding matrix was resuspended in 500 μL of guanidine thiocyanate (5.5 M), transferred to a spin filter column, and centrifuged at 14,000g for 1 min. This step was repeated and followed by two SEWS-M washes. Finally, the DNA was eluted in 100 μL of DES. For all samples, extractions were made in triplicate and stored at − 20°C before analysis.
The internal transcribed spacers ITS1 and ITS2 of the nuclear ribosomal RNA were amplified using the primers ITS1FI2 (5′-GAACCWGCGGARGGATCA-3′) and ITS2 (5′-GCTGCGTTCTTCATCGATGC-3′) for the ITS1 region (Schmidt et al. 2013) and the primers ITS86F (5′-GTGAATCATCGAATCTTTGAA-3′) and ITS4 (5′-TCCTCCGCTTATTGATATGC-3′) for the ITS2 region (De Beeck et al. 2014). The forward and reverse primers were modified prior to amplification by adding the adaptor sequences 5′-CTTTCCCTACACGACGCTCTTCCGATCT-3′ and 5′-GGAGTTCAGACGTGTGCTCTTCCGATCT-3′, respectively, which are needed for multiplexing the PCR products before sequencing. The amplification reaction was performed in a final volume of 25 μL with the modified primers (0.6 μM each), 2 μL of DNA extract, 200 μM of each dNTP, 200 ng/mL BSA, GoTaq® DNA Polymerase (2 units), and 1× Green GoTaq® Reaction Buffer (Promega, Charbonnieres, France) at 94°C for 3 min, 30 cycles of 95°C for 45 s, 52C (ITS1 region) or 55°C (ITS2 region) for 45 s, 72°C for 45 s with a final elongation step at 72°C for 10 min. To improve recovery and limit PCR bias, three PCR replicates per sample were pooled, evaporated using a vacuum concentrator and resuspended in 50 μL of sterile water. All amplicon products were analyzed using paired-end Illumina MiSeq sequencing (2 × 250 bp) by Get-PlaGe (Genotoul, Castanet-Tolosan, France) with single multiplexing using a home-made 6 bp index, which was added to the reverse primers for a second PCR with 12 cycles, using the forward primer (AATGATACGGCGACCACCGAGATCTACACTCTTTCCCTACACGAC) and the reverse primer (CAAGCAGAAGACGGCATACGAGAT-index-GTGACTGGAGTTCAGA-CGTGT).
The resulting PCR products were purified and loaded onto the Illumina MiSeq cartridge according to the manufacturer’s instructions. The quality of the run was checked internally using PhiX (20%), and then each pair-end sequence was assigned to the samples with the help of the home-made 6 bp index.
Data processing
The data analyses were conducted as described in Maghnia et al. (2017). Illumina sequencing, base calling, and demultiplexing were carried out using RTA v1.18.54, MCS 2.6 and bcl2fastq2.17. Paired Illumina MiSeq reads were merged with vsearch v2.3.0 (Rognes et al. 2016) using the command fastq_mergepairs and the option fastq_allowmergestagger. Primer clipping was performed with cutadapt v1.9 (Martin 2011) forcing a full-length match for sample tags and allowing a 2/3-length partial match for forward and reverse primers. Only reads containing both primers were retained. For each trimmed read, the expected error was estimated with vsearch’s command fastq_filter and the option eeout. Each sample was then dereplicated, i.e., strictly identical reads were merged, using vsearch’s command derep_fulllength, and converted to FASTA format.
To prepare for clustering, the samples were pooled and processed by another round of dereplication with vsearch. Files containing expected error estimates were also dereplicated to retain only the lowest expected error for each unique sequence. Clustering was performed with swarm v2.1.9 (Mahé et al. 2014), using a local threshold of one difference and the fastidious option. Operational taxonomic unit (OTU) representative sequences were then searched for chimeras with vsearch’s command uchime_denovo (Edgar et al. 2011). In parallel, representative sequences were assigned using the stampa pipeline (https://github.com/frederic-mahe/stampa/) and a custom version of the fungal reference database UNITE v7 (https://unite.ut.ee/) (Koljalg et al. 2013).
Clustering results, expected error values, taxonomic assignments, and chimera detection results were used to build a raw OTU table. Up to that point, reads that could not be merged, reads without tags or primers, reads shorter than 32 nucleotides, and reads with uncalled bases (“N”) had been eliminated. To create the “cleaned” OTU table, additional filters were applied to keep only non-chimeric OTUs, OTUs with an expected error per nucleotide below 0.0002, OTUs containing more than three reads or seen in two samples. All codes and representative OTU sequences can be found in HTML format (Supplementary File S1), and the raw data are available under the bioproject PRJEB27622 (https://www.ebi.ac.uk/ena/data/view/PRJEB27622).
Statistics
Tables were transformed using the R tidyverse package version 1.1.1 (Wickham 2017), and the plots were generated using the R ggplot2 package version 2.2.1 (Wickham 2009). According to the analyzed compartments, two independent rarefactions were performed with R vegan package rrarefy function for downstream statistical tests (rarefaction on 25,267 or 41,551 reads per sample, for fine roots and ECM root tips, respectively).
The differences in Pisolithus sp. structure were estimated using the Bray-Curtis dissimilarity matrix (from the abundance of reads in each Pisolithus OTUs) and visualized using non-metric multi-dimensional scaling (NMDS) using the R vegan package metaMDS() function. For each of the W+ and W− pits, the significance of the differences in Pisolithus spp. structure was tested by permutational multivariate analysis of variance (PERMANOVA) using the R vegan package adonis() function. Multivariate dispersion was estimated using the R vegan package betadisper() and permutest() functions as this can affect PERMANOVA results.
Diversity (Shannon) and richness (number of OTUs) were estimated using the R vegan package. The impact of depths regarding the soil surface (0–20 cm) as a reference was estimated with generalized linear models using the R stats package and glm() function (R Core Team 2017). The Gaussian model showed the smallest Akaike information criterion (AIC).
For each of the W+ and W− pits, the relationship between the soil parameters and the variation of the Pisolithus spp. structure with the depth was analyzed by redundancy analysis (RDA) using the R vegan package rda() function. For the RDA analysis, only the 12 most abundant OTUs of Pisolithus spp. were used (OTUs with more than 50 sequences all over the depths).
Results
The amplicon sequencing for both fine roots and ECM root tips resulted in 10,807,477 reads (number of raw sequences at the output of the sequencer) with a median length of 304 bp (with 95% of the reads between 241 and 330 bp). After filtering, a total of 7,156,053 sequences was analyzed (25,267 and 45,551 sequences per samples for fine roots and ECM root tips, respectively).
Strong dominance of the genus Pisolithus in the ECM fungal community
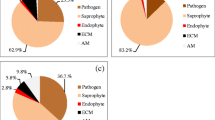
The total root-associated fungal community of E. grandis trees was dominated by ECM fungi with 84% of ECM fungal sequences in the W+ pit and 81% in the W− pit (Fig. 1). The genus Pisolithus, notably represented by P. microcarpus, was the most common with 98% (W+) and 93% (W−) of total ECM fungal sequences in the pits. Six other genera of ECM fungi were identified (Fig. 1), i.e., Tomentella, Laccaria, Thelephora, Scleroderma, Coltricia, and Rhizopogon, mainly represented by Laccaria alba, Thelephora palmata, Scleroderma citrinum, and Coltricia cinnamomea.
Fungal composition from fine roots from the W+ and W− pits after standardization and rarefaction of the data, for all soil layers from 0 to 4-m. Number of fungal ITS2 reads divided between none ECM fungal genera and ECM fungal genera (a, d); for ECM fungal genera only (b, e); for ECM fungal genera excluding Pisolithus (c, f)
Visual inspection of the root tips showed two main morphotypes, corresponding to ectomycorrhizas formed by Pisolithus spp. (yellow morphotype) and Scleroderma spp. (white morphotype), throughout the soil profiles (Fig. 2). The hyphal mantles were thick and had most often bright appearance. ECM root tips were generally ramified and could cover a large area of fine roots. Hyphae and rhizomorphs were observed at all depths. Fruiting bodies of these two ECM fungi were observed in the field (data not shown).
Variation in the Pisolithus spp. diversity with depth
As Pisolithus was strongly predominant in the ECM fungal community, its diversity was characterized in more detail by sequencing separately root tips that were typical of this ECM fungus (Fig. 2). Three hundred twenty-one OTUs of Pisolithus were identified, but the majority of these OTUs were represented by very small number of sequences by OTUs. Finally, we identified 12 OTUS with more than 50 sequences all over the depth (representing 58% of the analyzed sequences from ECM root tips).
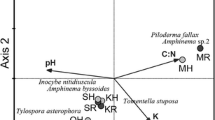
The NMDS analysis showed a very strong structuration of Pisolithus with the depth in the W+ pit, and to a lesser extent in the W− pit (Fig. 3). A similar observation was obtained with Pisolithus sequences retrieved from fine roots and using the ITS1 marker (data not shown). PERMANOVA revealed in each pit a significant effect of depth (P = 0.0001) on the Pisolithus spp. structure (R2 of 0.97 and 0.77, in W+ and W− pit respectively). The homogeneity of variance dispersion among depths was verified (HOMOVA, P = 0.449 and P = 0.634, in W+ and W− pit respectively).
Non-metric multidimensional scaling (NMDS) based on Bray-Curtis dissimilarities of Pisolithus spp. ITS2 data from root tips. Circles represent the W+ pit and triangles represent the W− pit. Each depth (n = 3) is identified using a color gradient with the lightest color assigned to the surface layer and the darkest color assigned to the deepest layer
The Pisolithus spp. diversity and, to a lesser extent, richness tented to be higher in W+ than in W− (Fig. 4). Linear model based on a Gaussian distribution showed significant impacts of depth on richness and diversity in comparison with the soil surface (0–20 cm) mostly in the W− pit (Fig. 4). The highest diversity (P < 0.01) and richness (P < 0.05) compared with the soil surface (0–20 cm) were observed between 200 and 350 cm in the W− pit. A significant lower diversity (P < 0.001) was also observed regarding the soil surface (0–20 cm) between 40 and 200 cm deep.
Diversity indices calculated from Pisolithus spp. ITS2 data from the W+ and W− pits as a function of depth (from root tips). Bars indicate standard errors (n = 3). Difference among the soil surface (0–20 cm) and the other depths were estimated using a generalized linear model based on a Gaussian distribution. Significance codes: ***P < 0.001; **P < 0.01; *P < 0.05
Relationships between the Pisolithus spp. structure and the soil physical and chemical properties
The relationships between the Pisolithus spp. structure (represented by the 12 most abundant OTUs) and the soil physical and chemical properties (described in Supplementary Table S1) were investigated using redundancy analysis (Fig. 5). The six physical and chemical properties (soil pH, soil humidity, C, N, P, K) explained 26.9% of the total variability in W+ and 21.5% in W−.
Redundancy analysis (RDA) for the W+ and W− pits using the 12 most abundant OTUs of Pisolithus spp. from root tips and 6 soil physical and chemical properties (pH, humidity, carbon and nitrogen (%), phosphorus and potassium (mg kg−1). The OTUs are shown in black and the soil physical and chemical properties by the red arrows. Each depth (n = 3) is identified using a color gradient with the lightest color assigned to the surface layer and the darkest color assigned to the deepest layer
In W+, OTUs 4, 52, 63, 112, 1067, and 2603 were predominant in the upper soil layers (from 0–20 cm to 60–100 cm) and covariate positively with soil C, N, K, and P contents. OTUs 2, 15, 19, and 1579 were predominant in deep layers (from 150–200 cm to 350–400 cm) and covariate positively with soil pH and soil moisture. As observed with the NMDS analysis, Pisolithus spp. structuration with depth was weaker in W− than in W+. In W−, OTU 1 was predominant in the upper soil layers (from 0–20 cm to 150–200 cm) and covariated positively with soil C, N, P, and K concentrations. The soil layers from 200–250 cm to 350–400 cm were associated with higher soil pH and soil moisture, along with higher relative density of all OTUs (except OTU 1).
Discussion
The current study presents the first confirmation of ECM symbiosis down to 4-m deph using both ECM root tips observations and meta-barcoding approach.
ECM fungi diversity associated with eucalypts
The ECM genera identified in this study were consistent with known ECM fungi associated with eucalypt roots, as reviewed in Sulzbacher et al. (2013), with notably a strong predominance of species belonging to the genus Pisolithus (> 90% of sequences). In most eucalypt plantations studied so far, a low level of ECM diversity is generally reported with most often a single genus (Ducousso et al. 2012). Pisolithus microcarpus was the most abundant species in our study, which is in agreement with previous observations in eucalypt plantations (Giachini et al. 2000). Secondary metabolites and phenol compounds found in eucalypt leaves or in eucalypt root exudates have been shown to promote mycorrhization by Pisolithus (Lagrange et al. 2001; Steffen et al. 2013) and could explain the predominance of this fungus. In addition, this fungus participates in tolerance to metals such as copper and aluminum (Egerton-Warburton 2015; Silva et al. 2013), which could be an ecological advantage in acidic tropical soils.
Pisolithus spp. associated with very deep eucalypt roots
One of the main challenges of our study was to demonstrate the presence of mycorrhizal structures down to 4-m depth and to evaluate changes of ECM fungal diversity through the vertical profile. Previous studies have shown fungal distributions in soil profiles suggesting vertical niche partitioning of the ECM fungi associated with roots (Dickie and Koide 2014; Dickie et al. 2002; Genney et al. 2006; Taylor and Bruns 1999), but those studies have only investigated soil layers down to 90 cm (Bakker et al. 2006; Clemmensen et al. 2015). Bakker et al. (2006) speculated that they did not sample deeply enough to determine the limits of ECM fine roots. The presence of ECM symbiosis associated with deep roots questioned about the dispersion of fungal spores in the deep layers of soil, which could be brought from the surface by water percolation, growing roots, and may be up to a certain depth limit, by macrofauna.
Although our sampling did not allow quantifying the mycorrhizal root tips precisely, field observations indicated that the abundance of symbiotic structures decreased with depth. Stone et al. (2014) recently showed that even though extracellular microbial activity decreases with increasing soil depth, specific enzyme activities (i.e., biomass-normalized) can increase with depth. This suggests the importance of considering more than just the abundance to gain insights into the role of biological processes in deep soil horizons.
Effect of physical and chemical properties on the structuration of Pisolithus spp. within depth
Soil water content and soil pH were the main factors correlated with the variations in ECM fungal composition with depth. These parameters have been previously shown to affect ECM fungal communities (Hogberg et al. 2007; Joergensen and Wichern 2008; Rousk et al. 2009; Scattolin et al. 2008). Nevertheless, the explained variation by the RDA is low (26.9% and 21.5%, in W+ and W− pit respectively) and indicates that other parameters not measured in our study, structure the Pisolithus with depth. The presence of symbiotic associations in deep soil layers also suggests a sufficient amount of oxygen for the germination of spores and saprotrophic hyphal growth. A recent work showed that young deep eucalypt roots have considerable amounts of aerenchyma (M. Gomide et al., unpublished). The roots themselves therefore may supply oxygen to ECM fungi in very deep soil layers in addition to oxygen diffusing from the soil surface through airspace in the soil.
Impact of rainfall reduction on Pisolithus spp. diversity?
Classically, the soil core sampling methodology is used in studies about ECM fungi diversity (Hui et al. 2017): small amounts of soil (around 50 cm3) are sorted and fine roots and ECM root tips are sampled. The only way to sample enough ECM root tips to realize a diversity study in deep horizons was to explore large volumes of soil (for each depth, we have separated fine roots and ECM root tips from 45 up to 1125 dm3 of soil, depending on the horizons). This time-consuming sampling has limited us in our ability to make several pits, and this does not allow us to conclude on the impact of the reduction of rain on the diversity of Pisolithus.
However, despite the limitations of our sampling, our observations seem to indicate an impact of water availability on the diversity of Pisolithus. On the NMDS result (Fig. 3), a very clear separation between the two pits was observed. All depths considered, a total of 10 replicates were sampled on each plot, which enables us to predict an impact of the reduction of rainfall on the diversity of Pisolithus. This result is in agreement with other works in the literature showing the impact of water stress on fungal communities (Azul et al. 2010; Cregger et al. 2012; Ren et al. 2018; Yuste et al. 2011) and more particularly on ECM communities (Richard et al. 2011). The impact of depth on the vertical structure of Pisolithus seems to be much less marked in the W− than in the W+ plot (Fig. 3). This observation is consistent with the distribution of water content in soil as a function of the depth between the two pits (Supplement data, Fig. S1). Indeed, the W− plot is drier but also more homogeneous between the surface and the depth, which could explain a lower impact on the structure of Pisolithus.
Conclusion
Our study fills a gap in our existing knowledge of the ECM ecology in deep tropical soils and challenges the accepted view of ECM symbiosis. Our work definitely allows showing the significant presence of Pisolithus associated with eucalyptus deep roots in independent pits, although our sampling does not allow us to really conclude on the impact of the rainfall reduction. Our results highlight the importance of taking into account the whole soil profile explored by roots, and suggest that the subsoil can be a reservoir of underestimated biodiversity. The rhizosphere processes down to a depth of 4-m have been investigated (Pradier et al. 2017) but the contribution of ECM fungi in these processes still needs to be assessed, and controlled experiments should complete current field observations, notably to characterize fungal traits related to tree nutrition and water uptake (e.g., aquaporin expression; (Nehls and Dietz 2014; Phillips et al. 2016). Further studies in other perennial crops on deep tropical soils (such as rubber trees, palm trees, coffee, cocoa, …) will be required to assess the role played by mycorrhizal fungi in very deep root functioning.
References
Allen MF, Kitajima K (2013) In situ high-frequency observations of mycorrhizas. New Phytol 200:222–228. https://doi.org/10.1111/nph.12363
Azul AM, Sousa JP, Agerer R, Martin MP, Freitas H (2010) Land use practices and ectomycorrhizal fungal communities from oak woodlands dominated by Quercus suber L. considering drought scenarios. Mycorrhiza 20:73–88. https://doi.org/10.1007/s00572-009-0261-2
Bakker MR, Augusto L, Achat DL (2006) Fine root distribution of trees and understory in mature stands of maritime pine (Pinus pinaster) on dry and humid sites. Plant Soil 286:37–51. https://doi.org/10.1007/s11104-006-9024-4
Battie-Laclau P, Laclau JP, Piccolo MC, Arenque BC, Beri C, Mietton L, Muniz MRA, Jordan-Meille L, Buckeridge MS, Nouvellon Y, Ranger J, Bouillet JP (2013) Influence of potassium and sodium nutrition on leaf area components in Eucalyptus grandis trees. Plant Soil 371:19–35. https://doi.org/10.1007/s11104-013-1663-7
Bordron B, Robin A, Oliveira IR, Guillemot J, Laclau JP, Jourdan C, Nouvellon Y, Abreu-Junior CH, Trivelin PCO, Gonçalves JLM, Plassard C, Bouillet JP (2019) Fertilization increases the functional specialization of fine roots in deep soil layers for young Eucalyptus grandis trees. For Ecol Manag 431:6–19. https://doi.org/10.1016/j.foreco.2018.03.018
Bornyasz MA, Graham RC, Allen MF (2005) Ectomycorrhizae in a soil-weathered granitic bedrock regolith: linking matrix resources to plants. Geoderma 126:141–160. https://doi.org/10.1016/j.geoderma.2004.11.023
Christina M, Laclau JP, Goncalves JLM, Jourdan C, Nouvellon Y, Bouillet JP (2011) Almost symmetrical vertical growth rates above and below ground in one of the world's most productive forests. Ecosphere 2(1-10):art27. https://doi.org/10.1890/es10-00158.1
Christina M, le Maire G, Battie-Laclau P, Nouvellon Y, Bouillet JP, Jourdan C, de Moraes Gonçalves JL, Laclau JP (2015) Measured and modeled interactive effects of potassium deficiency and water deficit on gross primary productivity and light-use efficiency in Eucalyptus grandis plantations. Glob Chang Biol 21:2022–2039. https://doi.org/10.1111/gcb.12817
Christina M, Nouvellon Y, Laclau JP, Stape JL, Bouillet JP, Lambais GR, le Maire G (2017) Importance of deep water uptake in tropical eucalypt forest. Funct Ecol 31:509–519. https://doi.org/10.1111/1365-2435.12727
Christina M, le Maire G, Nouvellon Y, Vezy R, Bordon B, Battie-Laclau P, Gonçalves JLM, Delgado-Rojas JS, Bouillet JP, Laclau JP (2018) Simulating the effects of different potassium and water supply regimes on soil water content and water table depth over a rotation of a tropical Eucalyptus grandis plantation. For Ecol Manag 418:4–14 https://. https://doi.org/10.1016/j.foreco.2017.12.048
Churchland C, Grayston SJ (2014) Specificity of plant-microbe interactions in the tree mycorrhizosphere biome and consequences for soil C cycling. Front Microbiol 5(1-20). https://doi.org/10.3389/fmicb.2014.00261
Clemmensen KE, Bahr A, Ovaskainen O, Dahlberg A, Ekblad A, Wallander H, Stenlid J, Finlay RD, Wardle DA, Lindahl BD (2013) Roots and associated fungi drive long-term carbon sequestration in boreal forest. Science 339:1615–1618. https://doi.org/10.1126/science.1231923
Clemmensen KE, Finlay RD, Dahlberg A, Stenlid J, Wardle DA, Lindahl BD (2015) Carbon sequestration is related to mycorrhizal fungal community shifts during long-term succession in boreal forests. New Phytol 205:1525–1536. https://doi.org/10.1111/nph.13208
Cregger MA, Schadt CW, McDowell NG, Pockman WT, Classen AT (2012) Response of the soil microbial community to changes in precipitation in a semiarid ecosystem. Appl Environ Microbiol 78:8587–8594. https://doi.org/10.1128/aem.02050-12
da Silva EV, Bouillet JP, de Moraes Gonçalves JL, Junior CHA, Trivelin PCO, Hinsinger P, Jourdan C, Nouvellon Y, Stape JL, Laclau JP (2011) Functional specialization of Eucalyptus fine roots: contrasting potential uptake rates for nitrogen, potassium and calcium tracers at varying soil depths. Funct Ecol 25:996–1006. https://doi.org/10.1111/j.1365-2435.2011.01867.x
De Beeck MO, Lievens B, Busschaert P, Declerck S, Vangronsveld J, Colpaert JV (2014) Comparison and validation of some ITS primer pairs useful for fungal metabarcoding studies. PLoS One 9(1-11):e97629. https://doi.org/10.1371/journal.pone.0097629
Dickie IA, Koide RT (2014) Deep thoughts on ectomycorrhizal fungal communities. New Phytol 201:1083–1085. https://doi.org/10.1111/nph.12674
Dickie IA, Xu B, Koide RT (2002) Vertical niche differentiation of ectomycorrhizal hyphae in soil as shown by T-RFLP analysis. New Phytol 156:527–535. https://doi.org/10.1046/j.1469-8137.2002.00535.x
Ducousso M, Duponnois R, Thoen D, Prin Y (2012) Diversity of ectomycorrhizal fungi associated with eucalyptus in Africa and Madagascar. Int J For Res 2012:1–10. https://doi.org/10.1155/2012/450715
Edgar RC, Haas BJ, Clemente JC, Quince C, Knight R (2011) UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 27:2194–2200. https://doi.org/10.1093/bioinformatics/btr381
Egerton-Warburton L (2015) Aluminum-tolerant Pisolithus ectomycorrhizas confer increased growth, mineral nutrition, and metal tolerance to eucalyptus in acidic mine spoil. Appl Environ Soil Sci 803821:1–9. https://doi.org/10.1155/2015/803821
Genney DR, Anderson IC, Alexander IJ (2006) Fine-scale distribution of pine ectomycorrhizas and their extramatrical mycelium. New Phytol 170:381–390. https://doi.org/10.1111/j.1469-8137.2006.01669.x
Giachini AJ, Oliveira VL, Castellano MA, Trappe JM (2000) Ectomycorrhizal fungi in Eucalyptus and Pinus plantations in southern Brazil. Mycologia 92:1166–1177. https://doi.org/10.2307/3761484
Gocke MI, Huguet A, Derenne S, Kolb S, Dippold MA, Wiesenberg GLB (2017) Disentangling interactions between microbial communities and roots in deep subsoil. Sci Total Environ 575:135–145. https://doi.org/10.1016/j.scitotenv.2016.09.184
Hogberg MN, Hogberg P (2002) Extramatrical ectomycorrhizal mycelium contributes one-third of microbial biomass and produces, together with associated roots, half the dissolved organic carbon in a forest soil. New Phytol 154:791–795. https://doi.org/10.1046/j.1469-8137.2002.00417.x
Hogberg MN, Hogberg P, Myrold DD (2007) Is microbial community composition in boreal forest soils determined by pH, C-to-N ratio, the trees, or all three? Oecologia 150:590–601. https://doi.org/10.1007/s00442-006-0562-5
Hui N, Liu XX, Kotze DJ, Jumpponen A, Francini G, Setala H (2017) Ectomycorrhizal fungal communities in urban parks are similar to those in natural forests but shaped by vegetation and park age. Appl Environ Microbiol 83(1-12). https://doi.org/10.1128/aem.01797-17
IBÁ (2015) Report. Indústria Brasileira de Árvores – Brazilian tree industry. Brasília, 64 pp. https://www.iba.org/datafiles/publicacoes/pdf/iba-2015.pdf
Joergensen RG, Wichern F (2008) Quantitative assessment of the fungal contribution to microbial tissue in soil. Soil Biol Biochem 40:2977–2991. https://doi.org/10.1016/j.soilbio.2008.08.017
Johnson NC, Gehring CA (2007) Mycorrhizas: symbiotic mediators of rhizosphere and ecosystem processes. In: Cardon ZWJ (ed) The rhizosphere: an ecological perspective. Academic, New York, pp 73–100
Koljalg U et al (2013) Towards a unified paradigm for sequence-based identification of fungi. Mol Ecol 22:5271–5277. https://doi.org/10.1111/mec.12481
Laclau JP, Ranger J, de Moraes Gonçalves JL, Maquère V, Krusche AV, M’Bou AT, Nouvellon Y, Saint-André L, Bouillet JP, de Cassia Piccolo M, Deleporte P (2010) Biogeochemical cycles of nutrients in tropical eucalyptus plantations main features shown by intensive monitoring in Congo and Brazil. For Ecol Manag 259:1771–1785. https://doi.org/10.1016/j.foreco.2009.06.010
Laclau J-P, da Silva EA, Rodrigues Lambais G, Bernoux M, le Maire G, Stape JL, Bouillet JP, Gonçalves JLM, Jourdan C, Nouvellon Y (2013) Dynamics of soil exploration by fine roots down to a depth of 10 m throughout the entire rotation in Eucalyptus grandis plantations. Front Plant Sci 4(1-12). https://doi.org/10.3389/fpls.2013.00243
Lagrange H, Jay-Allgmand C, Lapeyrie F (2001) Rutin, the phenolglycoside from eucalyptus root exudates, stimulates Pisolithus hyphal growth at picomolar concentration. New Phytol 149:349–355. https://doi.org/10.1046/j.1469-8137.2001.00027.x
Lambais GR, Jourdan C, de Cássia Piccolo M, Germon A, Pinheiro RC, Nouvellon Y, Stape JL, Campoe OC, Robin A, Bouillet JP, le Maire G, Laclau JP (2017) Contrasting phenology of Eucalyptus grandis fine roots in upper and very deep soil layers in Brazil. Plant Soil 421:301–318. https://doi.org/10.1007/s11104-017-3460-1
Lehto T, Zwiazek JJ (2011) Ectomycorrhizas and water relations of trees: a review. Mycorrhiza 21:71–90. https://doi.org/10.1007/s00572-010-0348-9
Li CH, Yan K, Tang LS, Jia ZJ, Li Y (2014) Change in deep soil microbial communities due to long-term fertilization. Soil Biol Biochem 75:264–272. https://doi.org/10.1016/j.soilbio.2014.04.023
Lindahl BD, Ihrmark K, Boberg J, Trumbore SE, Hogberg P, Stenlid J, Finlay RD (2007) Spatial separation of litter decomposition and mycorrhizal nitrogen uptake in a boreal forest. New Phytol 173:611–620. https://doi.org/10.1111/j.1469-8137.2006.01936.x
Maghnia FZ, Abbas Y, Mahé F, Kerdouh B, Tournier E, Ouadji M, Tisseyre P, Prin Y, el Ghachtouli N, Bakkali Yakhlef SE, Duponnois R, Sanguin H (2017) Habitat- and soil-related drivers of the root-associated fungal community of Quercus suber in the Northern Moroccan forest. PLoS One 12:e0187758. https://doi.org/10.1371/journal.pone.0187758
Mahé F, Rognes T, Quince C, de Vargas C, Dunthorn M (2014) Swarm: robust and fast clustering method for amplicon-based studies. Peerj 2. https://doi.org/10.7717/peerj.593
Maquere V (2008) Dynamics of mineral elements under a fest-growing eucalyptus plantation in Brazil. In: Implications for soil sustainability. Institut des Sciences et Industries du Vivant et de l’Environnement (Agro Paris Tech), Universidade de São Paulo, Escola Superior de Agricultura Luiz de Queiroz, Piracicaba
Martin M (2011) Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet.journal 17:10–12. https://doi.org/10.14806/ej.17.1.200
McCormack ML, Fernandez CW, Brooks H, Pritchard SG (2017) Production dynamics of Cenococcum geophilum ectomycorrhizas in response to long-term elevated CO2 and N fertilization. Fungal Ecol 26:11–19. https://doi.org/10.1016/j.funeco.2016.11.001
Moyersoen B, Fitter AH, Alexander IJ (1998) Spatial distribution of ectomycorrhizas and arbuscular mycorrhizas in Korup National Park rain forest, Cameroon, in relation to edaphic parameters. New Phytol 139:311–320. https://doi.org/10.1046/j.1469-8137.1998.00190.x
Nehls U, Dietz S (2014) Fungal aquaporins: cellular functions and ecophysiological perspectives. Appl Microbiol Biotechnol 98:8835–8851. https://doi.org/10.1007/s00253-014-6049-0
Pagano M, Lugo M (eds) (2019) Mycorrhizal fungi in South America. Springer, Berlin Fungal Biology Series. 374 p. https://doi.org/10.1007/978-3-030-15228-4
Pereira APD et al (2017) Shifts in the bacterial community composition along deep soil profiles in monospecific and mixed stands of Eucalyptus grandis and Acacia mangium. PLoS One 12:e0180371. https://doi.org/10.1371/journal.pone.0180371
Phillips RP, Ibanez I, D'Orangeville L, Hanson PJ, Ryan MG, McDowell NG (2016) A belowground perspective on the drought sensitivity of forests: towards improved understanding and simulation. For Ecol Manag 380:309–320. https://doi.org/10.1016/j.foreco.2016.08.043
Pradier C, Hinsinger P, Laclau JP, Bouillet JP, Guerrini IA, Gonçalves JLM, Asensio V, Abreu-Junior CH, Jourdan C (2017) Rainfall reduction impacts rhizosphere biogeochemistry in eucalypts grown in a deep Ferralsol in Brazil. Plant Soil 414:339–354. https://doi.org/10.1007/s11104-016-3107-7
Querejeta JI, Egerton-Warburton LM, Allen MF (2007) Hydraulic lift may buffer rhizosphere hyphae against the negative effects of severe soil drying in a California Oak savanna. Soil Biol Biochem 39:409–417. https://doi.org/10.1016/j.soilbio.2006.08.008
R Core Team (2017) R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna Available online at https://www.R-project.org/
Ren CJ, Chen J, Lu X, Doughty R, Zhao F, Zhong Z, Han X, Yang G, Feng Y, Ren G (2018) Responses of soil total microbial biomass and community compositions to rainfall reductions. Soil Biol Biochem 116:4–10. https://doi.org/10.1016/j.soilbio.2017.09.028
Richard F, Roy M, Shahin O, Sthultz C, Duchemin M, Joffre R, Selosse M-A (2011) Ectomycorrhizal communities in a Mediterranean forest ecosystem dominated by Quercus ilex: seasonal dynamics and response to drought in the surface organic horizon. Ann For Sci 68:57–68. https://doi.org/10.1007/s13595-010-0007-5
Rognes T, Flouri T, Nichols B, Quince C, Mahe F (2016) VSEARCH: a versatile open source tool for metagenomics. Peerj 4:e2584. https://doi.org/10.7717/peerj.2584
Rosling A, Landeweert R, Lindahl BD, Larsson KH, Kuyper TW, Taylor AFS, Finlay RD (2003) Vertical distribution of ectomycorrhizal fungal taxa in a podzol soil profile. New Phytol 159:775–783. https://doi.org/10.1046/j.1469-8137.2003.00829.x
Rousk J, Brookes PC, Baath E (2009) Contrasting soil pH effects on fungal and bacterial growth suggest functional redundancy in carbon mineralization. Appl Environ Microbiol 75:1589–1596. https://doi.org/10.1128/aem.02775-08
Scattolin L, Montecchio L, Agerer R (2008) The ectomycorrhizal community structure in high mountain Norway spruce stands. Trees Struct Funct 22:13–22. https://doi.org/10.1007/s00468-007-0164-9
Schmidt PA, Balint M, Greshake B, Bandow C, Rombke J, Schmitt I (2013) Illumina metabarcoding of a soil fungal community. Soil Biol Biochem 65:128-132. https://doi.org/10.1016/j.soilbio.2013.05.014
Silva RF, Lupatini M, Trindade L, Antoniolli ZI, Steffen RB, Andreazza R (2013) Copper resistance of different ectomycorrhizal fungi such as Pisolithus microcarpus, Pisolithus sp., Scleroderma sp. and Suillus sp. Braz J Microbiol 44:619–627. https://doi.org/10.1590/S1517-83822013005000039
Steffen RB, Antoniolli ZI, Steffen GPK, Jacques RJS, dos Santos ML, Godoy HT, Bogusz S (2013) Eucalyptus essential oil as bio-stimulator of the growth of in vitro ectomycorrhizal fungi. Cienc Florest 23:403–414. https://doi.org/10.5902/198050989285
Stone MM, DeForest JL, Plante AF (2014) Changes in extracellular enzyme activity and microbial community structure with soil depth at the Luquillo Critical Zone Observatory. Soil Biol Biochem 75:237–247. https://doi.org/10.1016/j.soilbio.2014.04.017
Sulzbacher MA, Grebenc T, Jacques RJS, Antoniolli ZI (2013) Ectomycorrhizal fungi from southern Brazil - a literature-based review, their origin and potential hosts. Mycosphere 4:61–95. https://doi.org/10.5943/mycosphere/4/1/5
Taylor DL, Bruns TD (1999) Community structure of ectomycorrhizal fungi in a Pinus muricata forest: minimal overlap between the mature forest and resistant propagule communities. Mol Ecol 8:1837–1850. https://doi.org/10.1046/j.1365-294x.1999.00773.x
Tedersoo L, Koljalg U, Hallenberg N, Larsson KH (2003) Fine scale distribution of ectomycorrhizal fungi and roots across substrate layers including coarse woody debris in a mixed forest. New Phytol 159:153–165. https://doi.org/10.1046/j.0028-646x.2003.00792.x
Tournier E et al (2015) Modification of a commercial DNA extraction kit to recover safely and rapidly DNA and RNA simultaneously from soil, for total and active molecular biomass quantification. MethodsX 2:182–191. https://doi.org/10.1016/j.mex.2015.03.007 eCollection 2015
Trocha LK, Bulaj B, Kutczynska P, Mucha J, Rutkowski P, Zadworny M (2017) The interactive impact of root branch order and soil genetic horizon on root respiration and nitrogen concentration. Tree Physiol 37:1055–1068. https://doi.org/10.1093/treephys/tpx096
Wickham H (2009) Ggplot2: Elegant Graphics for Data Analysis. 2nd Edition, Springer, New York. https://doi.org/10.1007/978-0-387-98141-3
Wickham H (2017) Tidyverse: Easily Install and Load "Tidyverse" Packages. R package version 1.1.1. Available: https://CRAN.R-project.org/package=tidyverse
Yuste JC et al (2011) Drought-resistant fungi control soil organic matter decomposition and its response to temperature. Glob Chang Biol 17:1475–1486. https://doi.org/10.1111/j.1365-2486.2010.02300.x
Zheng L, Zhao X, Zhu GB, Yang W, Xia C, Xu T (2017) Occurrence and abundance of ammonia-oxidizing archaea and bacteria from the surface to below the water table, in deep soil, and their contributions to nitrification. Microbiologyopen 6:8. https://doi.org/10.1002/mbo3.488
Acknowledgments
We would like to thank the staff of the Itatinga Experimental Station (ESALQ-USP), as well as all the students present at the station during the sampling, and Eder Araujo da Silva and Floragro for their technical support. The site belongs to the SOERE F-ORE-T network, which is supported annually by ECOFOR, AllEnvi and the French national research infrastructure ANAEE (http://www.anaeefrance.fr/fr/). This work was performed in collaboration with the GeT core facility, Toulouse, France (http://get.genotoul.fr/), and was supported by France Génomique National infrastructure, funded as part of “Investissement d’avenir” program managed by Agence Nationale pour la Recherche (contract ANR-10-INBS-09).
Funding
This research was funded by the Centre de Coopération Internationale en Recherche Agronomique pour le Développement (CIRAD) and the project FAPESP 2016/18944-3 “Climate change and energy efficiency in agriculture: a focus on water stress, organic management and soil biology”
Author information
Authors and Affiliations
Contributions
A.R., J.P.L., J.P.B., F.D.A., E.C., C.P., J.L.M.G, P.H., C.J., designed the study; A.R., C.P., G.R.L., A.P.A.P., A.G., M.C.S. A.L.P., P.T., P.H., performed research; F.M., H.S, A.R., A.G., M.S., analyzed data; A.R. wrote the paper with contributions of C.P. and of all authors; all the authors helped to interpret the results of the study, reviewed the manuscript and contributed substantially to the revisions.
Corresponding author
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Table S1.
Physical and chemical properties of the soil in the W+ and W- pits. The data comes from Pradier et al. (2017). The soil samples were collected at the same time, from the same pits, as the fine roots and ECM root tips analyzed in this study. Different letters indicate significant differences between depths. P-values in bold with an asterisk indicate significant effects (Tukey test, P < 0.05). pH: pH measured in CaCl2, %H: humidity percentage, %C and %N: total C and N contents, P: P extracted with an anion exchange resin, K: K extracted with a cation exchange resin. (DOCX 46 kb)
Figure S1.
Soil water content (v/v%) in the W+ and W- plots down to 4 meters. The 4 months around the sampling period are outlined. SWC was interpolated by a contour line interpolation using marching square algorithm (R software version 3.2.5 and the plotly package version 4.5.6). (PDF 146 kb)
File S1.
All codes and representative OTU sequences in HTML format. The purpose of this document is to provide the reader with details about the bioinformatic methods used to prepare this paper. The code snippets and shell commands presented here were executed on a Debian GNU/Linux 8, and might have to be adapted to your particular system. (HTML 206 kb)
Rights and permissions
About this article
Cite this article
Robin, A., Pradier, C., Sanguin, H. et al. How deep can ectomycorrhizas go? A case study on Pisolithus down to 4 meters in a Brazilian eucalypt plantation. Mycorrhiza 29, 637–648 (2019). https://doi.org/10.1007/s00572-019-00917-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00572-019-00917-y