Abstract
Epigenetic regulation of the activity of defense genes during onset of diseases or resistance against diseases in plants is an active area of research. In the present study, a pair of wheat NILs for leaf rust resistance gene Lr28 (R) in the background of an Indian cultivar HD2329 (S) was used for a study of DNA methylation mediated regulation of gene expression. Leaf samples were collected at 0 h before (S0 and R0) and 96 h after inoculation (S96 and R96). The DNA samples were subjected to BS-Seq and sequencing data were used for identification of differentially methylated/demethylated regions/genes (DMRs and DMGs). Following four pairs of comparisons were used for this purpose: S0 vs S96; S0 vs R0; R0 vs R96; S96 vs R96. Major role of CHH methylation relative to that of CG and CHG methylation was observed. Some important observations include the following: (i) abundance of CHH methylation among DMRs; (ii) predominance of DMRs in intergenic region, relative to other genomic regions (promoters, exons, introns, TSS and TTS); (iii) abundance of transposable elements (TEs) in DMRs with CHH context; (iv) demethylation mediated high expression of genes during susceptible reaction (S0 vs S96) and methylation mediated low expression of genes during resistant reaction (R0 vs R96 and S96 vs R96); (v) major genes under regulation encode proteins, which differ from those encoded by genes regulated during susceptible reaction and (vi) ~ 500 DMGs carried differential binding sites for H3K4/K27me3 marks suggesting joint involvement of DNA and H3 methylation. Thus, CHH methylation either alone or in combination with histone methylation plays a major role in regulating the expression of genes involved in wheat-leaf rust interaction.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
In recent years, major advances have been made toward understanding the molecular mechanism of plant immunity. As a result, two layers of plant immunity, including pathogen-associated molecular pattern (PAMP)-triggered immunity (PTI) and effector-triggered immunity (ETI) are widely known (Jones and Dangl 2006; Maekawa et al. 2011). The activation of both PTI and ETI causes massive global transcriptional reprogramming, which is controlled by various regulatory mechanisms, including epigenetic regulation (Pandey and Somssich 2009; Rushton et al. 2010), which is poorly understood.
Among mechanisms of epigenetic regulation, DNA methylation is a major component (Lippman et al. 2004; Jablonka and Raz 2009; Zhao et al. 2017), which has already been examined for the following pathosystems: Arabidopsis thaliana and bacterium Pseudomonas syringae pv. tomato (Pst) DC3000 (Dowen et al. 2012), Oryza sativa and fungus Magnaporthe oryzae (Deng et al. 2017), Aegilops tauschii and fungus Blumeria graminis f. sp. tritici (Geng et al. 2019), and Citrullus lanatus and Cucumber Green Mottle Mosaic Virus (Sun et al. 2018).
The resistance against leaf rust in wheat is due to one or more R genes, which are described as Lr genes; ~ 80 Lr genes are already known and the list seems to be ever-growing. In the present study, we focused on the major gene Lr28 that has been deployed for development of a number of resistant cultivars in India. To understand the molecular basis of genetic and epigenetic regulation of Lr28-mediated leaf rust resistance, earlier, we utilized a pair of NILs differing for Lr28 in studies involving analysis of transcriptome, non-coding RNAs, DNA methylation (MeDIP-seq) and histone modifications, etc. (Sharma et al. 2018, 2019; Jain et al. 2020; Saripalli et al. 2020a,b). These earlier studies demonstrated that resistance due to Lr28 involves a complex network with an epigenetic component involving DNA methylation (Saripalli et al 2020a), histone acetylation/methylation and modifications and remodeling of chromatin states (Sharma et al. 2019; Saripalli et al 2020b). Interactions with non-coding RNAs including miRNAs and lncRNAs were also examined (Jain et al 2020). The early studies also demonstrated that the DNA methylation has a significant role in Lr28 mediated leaf rust resistance involving epigenetic regulation of the expression of hundreds of downstream genes (Saripalli et al. 2020a). In our earlier studies using the same material, DNA methylation was examined using MSAP and MeDIP approaches, which have a rather poor resolution and do not allow study of DNA methylation at the level of individual nucleotides precluding the study of cytosine methylation of the two symmetric (CG, CHG) and one asymmetric (CHH) contexts. The present study was thus conducted to examine and understand the role of methylation of the above three cytosine contexts using genome-wide bisulphite sequencing (BS-Seq). A comparison of the results of methylation in three contexts demonstrated that although overall genome-wide methylation with CHH context was rather very low (~ 2%), its involvement in differential methylation between NILs for Lr28 was relatively very high. Chromatin states associated with DNA methylation were also examined, and the results were interpreted utilizing two recent studies on chromatin states in wheat (Li et al. 2019; Ran et al. 2021).
Materials and methods
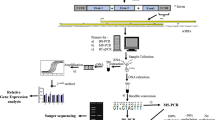
Plant material, treatments and BS-Seq
Plant material utilized in the present study included a pair of NILs that were also used in a number of our earlier studies mentioned above. The methods used for growing this pair of NILs and collecting leaf samples for the present study are also described in our earlier studies (Sharma et al. 2018; Jain et al. 2020; Saripalli et al. 2020a, b). Leaf samples for the present study were collected from both NILs at 0 h before inoculation (hbi) and 96 h after inoculation (hai) to get the following four samples: (i) S0 (susceptible cv. HD2329 at 0 hbi), (ii) S96 (susceptible cv. HD2329 at 96 hai), (iii) R0 (resistant NIL HD2329 + Lr28 at 0 hbi), and (iv) R96 (resistant NIL HD2329 + Lr28 at 96 hai). These four samples were collected in two biological replicates resulting into eight samples. These eight samples were outsourced to Nucleome Informatics Pvt Ltd. for genome-wide bisulfite sequencing (BS-Seq). Standard procedures were followed for bisulphite treatment of DNA samples, followed by sequencing.
Preparation and sequencing of BS-Seq libraries
For the preparation of libraries for whole genome bisulfite sequencing (WGBS), the following protocol was followed: (i) DNA was isolated from each of the eight samples mentioned above and was subjected to bisulfite treatment that was followed by library preparation using EZ DNA methylation gold kit, Zymo Research, USA. This kit integrates DNA denaturation and bisulfite conversion processes into single-step by exploiting temperature denaturation. For this purpose, 200 to ~ 400 bp long DNA fragments were obtained from sonication and adapters were ligated to end-repaired DNA samples. (ii) After desalting, size selection and PCR amplification, the library quality was examined for each sample. (iii) Libraries with satisfactory quality score were sequenced on HiSeq2500 sequencing platform. (iv) The reads with only adaptor sequences and those with low-quality (analysed through QC Toolkit) were removed, resulting in good quality WGBS paired end (PE) reads for further analysis.
BS-Seq data and identification of DMRs/DMGs
Analysis of the WGBS data involved the following steps: (i) The paired-end reads were aligned with wheat reference genome (IWGSC v2.0) utilizing bowtie aligner, multicore and nucleotide coverage parameters using Bismark (Krueger and Andrews 2011). (ii) The aligned reads were subjected to deduplication using Bismark. (iii) Reproducibility was checked using Pearson’s correlation coefficients in the aligned reads between the libraries by deepTools (Ramírez et al. 2014). (iv) Methylations for CG, CHG and CHH contexts were examined using Bismark Extractor; context-wise coverage files were prepared using Perl-script. (v) Differentially methylated regions (DMRs) were extracted using bsseq package (Hansen et al. 2012) of Bioconductor, using the following four comparisons: S0 vs S96; S0 vs R0; S96 vs R96; R0 vs R96. The status of DMRs (e.g., demethylated (hypomethylation) vs methylated (hypermethylation)) was examined in second treatment relative to first treatment in each pair; the cut off was computed using 0.005 and 0.995 quantile range of the t-statistic, at the maximum gap of 100 bases (with at least 5 cytosine sites and at least 20% difference in methylation) for each of the three contexts. (vi) Circular layout of histograms for mean difference in methylation among DMRs was prepared using ClicO to display chromosome-wise distribution of DMRs. (vii) HOMER (Heinz et al. 2010) was used to localize DMRs into five genomic regions including promoters (up to 2 Kb upstream of transcription start site), exons, introns, transcription termination sites (TTS) and intergenic regions. (viii) Differentially methylated genes (DMGs) associated with DMRs were identified using IWGSC RefSeq v2.0 annotation data with the help of a Perl-script. The raw reads generated using BS-Seq analysis for all the eight samples are deposited in Sequence Read Archive (SRA), NCBI, under the accession number PRJNA745483 of Bioproject Database.
Functional annotation/classification and gene ontology analysis of DMGs
Functions of all the DMGs were extracted from IWGSC RefSeq v2.0 annotation data using Perl-script, and DMGs (based on encoded proteins) were classified into the following 13 categories: (i) resistance related; (ii) transcription factors (TFs); (iii) transcription regulators (TRs); (iv) oxidative stress; (v) other stress-related; (vi) transporters; (vii) photosynthesis-related; (viii) chromatin-related; (ix) metabolism-related; (x) kinases; (xi) growth hormone response; (xii) senescence-related, and (xiii) miscellaneous. Gene ontology (GO) terms were extracted by Blast2GO (Conesa and Götz 2008) and plotted by WEGO for all the DMGs for each treatment comparison and were grouped into the following three well-known categories: molecular, biological and cellular processes.
Methylated transposable elements (TE) and their associated genes
Transposable elements (TEs) associated with DMRs (mTEs) were extracted using a Perl-script utilizing IWGSC RefSeq v2.0 TE data. Following three classes of TEs were identified: repeat region, nested repeat and repeat fragment. Transposable element associated genes (TAGs) associated with DMRs were also extracted using a Perl-script; these genes included only those genes, which were also annotated as TAGs in IWGSC RefSeq v2.0 annotation data.
Relationship between DNA methylation and gene expression
Differentially methylated genes (DMGs) were compared with differentially expressed genes (DEGs) earlier identified using RNA-Seq data that was deposited by us in Sequence Read Archive (SRA), NCBI, under the accession number PRJNA588134 of Bioproject Database. The RNA-Seq reads were generated for the same set of samples that were used in the present study. An average of ~ 27 million reads were generated per sample (~ 7.8 GB) of hexaploid wheat genome. These data were also used in our earlier studies involving MeDIP-seq analysis (Saripalli et al. 2020a) and ChIP-seq analysis (Saripalli et al. 2020b).
Relationship between DNA methylation and histone modification
Association of DMRs/DMGs with H3K4me3 and H3K27me3 marks were examined using our ChIP-seq data (Saripalli et al. 2020b). For this analysis, an overlap of at least one base was considered between DMRs and differential binding sites (DBS) for H3K4/K27me3. ChIP-seq raw reads are available at Sequence Read Archive (SRA), NCBI and can be accessed through Bioproject Database under the accession number PRJNA588134.
DNA methylation and chromatin state analysis
Chromatin states due to association of DNA methylation with histone modifications were examined using ChromHMM (Ernst and Kellis 2012). Following steps were involved: (i) files for chromosome size, COORDS and ANCHORFILES were prepared utilizing IWGSC RefSeq v2.0 annotation file using Perl-scripts; (ii) separate genome-wise Cellmarkfiletable for DNA methylation and histone modifications were prepared using controls for two histone marks- H3K4me3 and H3K27me3 and DMRs; (iii) DNA methylation contexts (CG, CHG and CHH) were obtained through binarization performed on bed files by BinarizeBed; (iv) binarization was also performed on BAM files by BinarizeBam for histone modifications; (iv) binary files for DNA methylation and histone modifications were merged chromosome-wise, and (v) the model was learned using LearnModel for 18 chromatin states, which are biologically interpretable.
Co-expression network analysis of DMGs and DEGs
To understand the relationship between DMGs and DEGs, co-expression network analysis was conducted. For this purpose, expression data from 24 RNA-Seq experiments (05 from CS_methylome, 04 from SRP022869 and 15 from ERP009837) were extracted from IWGSC Refseq v2.0 rnaseq mapping data with gene-wise expression values in transcripts per kilobase million (TPM) for only two-leaf and three-leaf stages at 14 and 17 days old seedling without any treatment for all high confidence (HC) genes. Following steps were used to prepare co-expression network: (i) Data from HC genes were normalized by DESeq Bioconductor package; (ii) correlation coefficients were calculated among all genes using a Perl script; (iii) adjacency matrix was prepared using 0.90 cut off value of correlation coefficients using a Perl script; and (iv) later, only DEGs co-expressed with DMGs were extracted from all co-expressed HC genes using a Perl script.
Validation by quantitative RT-PCR (qRT-PCR)
Selection of genes
A set of 26 representative DMGs was used for validation using qRT-PCR; these 26 genes were distributed in all the four treatment pairs as follows: 8 genes from S0 vs S96, 3 genes from S0 vs R0, 8 genes from S96 vs R96 and remaining 7 genes from R0 vs R96 (Table 1). Primers for the above genes were designed using Primer Express (ver. 3.0), and synthesized by Applied Biosystems, USA (Supplementary Table1). All the 26 genes were known to exhibit differential expression in our earlier RNA-Seq study (Sharma et al. 2018); five of these 26 genes were also identified in our earlier ChIP-seq data for differential histone methylation (Saripalli et al. 2020b) involving the same four treatment pairs.
RNA isolation, cDNA synthesis and qRT-PCR
RNA isolation, cDNA synthesis, and qRT-PCR for the selected genes were performed, using the following steps: (i) isolation of total RNA using TRIzol reagent (Ambion, USA); (ii) treatment of extracted RNA with RNase-free DNase I (Invitrogen, USA) for 15 min to degrade any residual genomic DNA; (iii) synthesis of first-strand cDNA from RNA using RevertAid First Strand cDNA Synthesis Kit (Thermo Scientific); (iv) qRT-PCR in 96 well plates with Step OnePlus Real Time PCR system using SYBR Green (both from Applied Biosystem, USA). All reactions were performed using two biological and three technical replicates. Specificity of the primers was assessed and verified through melt curve analysis (Supplementary Figure S1). Target gene expression (fold-change between two treatments) was calculated using 2−∆∆CT method using actin gene for normalization (Livak and Schmittgen 2001). The results were expressed as bar diagrams. The significance of fold-changes was estimated through t test at P < 0.05 and P < 0.01.
Results
BS-Seq: alignment, correlations and extent of methylation
Bisulphite sequencing (BS-Seq) data for 8 libraries contained ~ 2,163 million PE reads (average reads per library = ~ 270 million, ~ 80 GB), which corresponded to sequencing depth of ~ 5X of hexaploid wheat genome. The above PE reads were aligned with wheat reference genome with an average mapping efficiency of 67.07%. An average of ~ 155 million aligned reads per library were recovered in each of the four treatments after de-duplication; average methylation values for CG, CHG and CHH contexts were 90.84%, 63.99% and 2.11%, respectively (Supplementary Table S2). Average rate of bisulfite conversion was ~ 29.75% (Supplementary Table S3). The correlation for aligned reads for pairs of treatments, each involving two replications ranged from 0.78 to 1.00 (Supplementary Figure S2).
DMRs and their chromosomal distribution
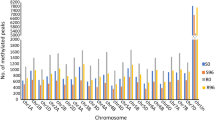
The DMRs were distributed along the length of each of the 21 chromosomes (see circular layout in Fig. 1a). Among three contexts, the number of DMRs was maximum for CHH context (62,853; 51.24%) followed by CHG (36,759; 29.97%) and CG context (23,054; 18.79%). This pattern of relative number of DMRs in different methylation contexts is just the opposite of actual DNA methylation, so that despite overall very low genome-wide methylation for CHH context (2.11%) the differential DNA methylation was high (51.24%). In contrast, the high overall genome-wide DNA methylation for CG and CHG contexts was associated with relatively low differential methylation (see Supplementary Table S2 and Fig. 1b).
a Chromosome-wise distribution of DMRs in the form of histograms, where circles with four colors depict DMRs in four pairs of comparisons and three circles of the same color represent three methylation contexts: red circles = S96 vs S0, cyan circles = R0 vs S0, green circles = R96 vs S96, blue circles = R96 vs R0; in a set of three circles of each color, the outer circle = CG context, middle circle = CHG context and inner circle = CHH context. b Distribution of context-wise DMRs in four pairs of comparisons c Context-wise distribution of demethylated/methylated DMRs in five genomic regions d Relative proportions of DMRs in five genomic regions in four treatment comparisons for each of the three different methylation contexts (CG, CHG and CHH)
While comparing DMRs in four pairs of comparisons, DMRs in S0 vs S96 and S0 vs R0 were almost equal to those in R0 vs R96 and S96 vs R96 in three contexts. However, the DMRs for CHH in R0 vs R96 and S96 vs R96 were almost equal to DMRs for the CHH context in the other two pairs of treatments, but the demethylated DMRs for CG and CHG contexts were markedly low. As shown in Fig. 1b, demethylated DMRs seem to be significantly higher than methylated DMRs in S96 and R0 (both relative to S0) for all three contexts except in CG context of R0 relative to S0. In R96 relative to S96 and R0, methylated DMRs were higher than demethylated DMRs in all the three contexts (for a list of DMRs, see Supplementary Tables S4–S15).
Distribution of DMRs in genomic regions
As shown above, the total numbers of DMRs in four comparisons were largely similar (Fig. 1b; Supplementary Tables S4–S15). However, the pattern of relative proportion of DMRs with three different contexts differed in five different genomic regions, namely (i) intergenic region (62–83%), promoter (7–16%), (ii) TTS (4–11%), (iii) introns (3–9%) and (iv) exons (1–3%) (Fig. 1c). The proportion of methylated and demethylated DMRs in different genomic regions also differed context-wise in different comparisons (Fig. 1d). The intergenic regions are the most prominent genomic region with both demethylated and methylated regions. However, the highest demethylated intergenic regions are available in S0 vs S96 and S0 vs R0, the situation is reverse in S96 vs R96 and R0 vs R96 (Supplementary Table S16). This pattern holds good for all the three methylation contexts, although the differential methylation level is generally minimum in CG and maximum in CHH, when compared among the three contexts within the same treatment comparison. The pattern of distribution of methylation in intergenic region in terms of distance from the nearest gene shows that > 50% of intergenic DMRs were found within a range of ≤ 50 Kb flanking region of genes in case of CG and CHG contexts, which is even higher (~ 80%) in case of CHH. In case of CHH context, > 50% methylation in intergenic region was found within ≤ 10 Kb flanking region of genes.
Methylation context and chromatin state
The chromatin states were examined using three methylation contexts (CG, CHG and CHH) and two histone marks (H3K4me3, H3K27me3). As many as 18 chromatin states were identified, which provided sufficient resolution to understand biologically meaningful patterns, across four different treatments. Following five genomic regions, each with one or more chromatin states were identified: enhancers, intergenic regions, genic regions, promoter regions, repetitive regions (Table 2); methylation levels in each of 20 different permutations and combinations involving four treatments, two histone marks and three methylation contexts are depicted in Fig. 2.
a A heat map of emission probabilities, where each row represents a different chromatin state, and each column represents a different epigenetic mark in four individual treatments (S0, S96, R0, R96); the darker regions correspond to more frequent occurrence of the methylation mark in the corresponding chromatin state; b a heat map of emission probabilities displaying enrichment for six different external genomic annotations based on RefSeq and 18 chromatin states; c a heat map of transition parameters, where rows and columns both represent 18 chromatin states
Differentially methylated genes (DMGs)
The frequencies of DMGs (including both demethylated and methylated) with three different methylation contexts differed (Fig. 3a). The DMGs with CHH context were available in maximum number (20,055), followed by DMGs with CHG (5624) and CG (3189). DMGs that were common in all the three contexts were the least frequent, the frequencies of DMGs that were common in two contexts were relatively few, and the frequencies of unique DMGs were highest for each of the three contexts (Fig. 3b). The DMGs, which figured in more than one comparison were also examined (Fig. 3c). A sum of 2331 DMGs were common in two treatment pairs involving all the three contexts, whereas 1245 DMGs were common in three or four treatment pairs (Fig. 3c). However, some of the unique DMGs have common domains and thus may have similar function. More details of data for demethylated/methylated DMGs for four pairs of treatments are provided in Supplementary Tables S17 to S28. Demethylated DMGs in S96 were relatively more abundant in S96 and R0 (both relative to S0) in all three contexts (except CG context of R0 relative to S0); reverse was the case in R96 relative to S96 and R0.
The details of data on frequencies of methylated (↑) and demethylated (↓) DMGs, were compared among four pairs. Frequencies of similar patterns in two of the four pairs of treatments for three methylation contexts (CG, CHG, CHH) are summarized in Supplementary Figure S2 (a, b, c). The DMGs that are common in more than one pair ranged from 24 to 77 DMGs in CG context, from 90 to 145 in CHG context, and from 221 to 641 in CHH context. Only 46, 98 and 1101 DMGs in CG, CHG and CHH contexts, respectively were common in more than two treatment comparisons, which could not be included in Supplementary Figure S3 (a, b, c). These figures are proportionate to the actual number of unique methylated DMGs in individual comparisons (being maximum in CHH and minimum in CG context), as also mentioned elsewhere. In each comparison, frequencies of both methylated (↑) and demethylated (↓) DMGs were utilized for comparison. For CG context, the maximum number of common DMGs were available between S0 vs S96, S96 vs R96 and R0 vs R96. This data will be further discussed later in this manuscript.
Gene ontology (GO) and functional annotation of DMGs
In each of the three contexts (CG, CHG and CHH), the DMGs were broadly classified into following three well-known categories: (i) cellular component, (ii) molecular function, and (iii) biological process (Supplementary Figs. S4). Biological processes had the maximum terms, the most abundant class among these being the genes involved in ‘cell part’. The DMGs in three contexts did not show any major difference.
Functional annotation of DMGs showed that many of the DMGs had common domains, thus having functional similarity. Based on their domains, DMGs were classified into 13 categories (Fig. 4a). It is apparent from Fig. 4a that highest frequency of DMGs belonged to miscellaneous category (last column), followed by metabolism (category 9), kinases (category 10), transporters (category 6) and oxidative stress (category 4). In this data also, frequencies of DMGs were maximum in CHH context and minimum in CG, with intermediate frequencies in CHG.
a Frequencies of DMGs (shown in y-axis) in 13 different categories (shown in z-axis) established on the basis of functional domains in the proteins, identified in CG, CHG and CHH contexts (shown in x-axis), each in four different comparisons b Frequencies of DMGs and intergenic DMRs (each with three methylation contexts) associated with H3K4/K27me3 marks in four treatment comparisons c Demethylated/methylated TE-associated DMGs (TE-DMGs) in four treatment comparison and three methylation contexts
DNA methylation and gene expression
The data on DNA methylation were also compared with the data on gene expression that were available with us from our earlier studies (Sharma et al. 2018). The results are presented in Tables 3 and 4 (details of genes are available in Supplementary Tables S29–S40). Following are some of the salient features of these results: (i) Methylation in promoter regions influenced the expression of maximum number of DMGs. (ii) The assumption that demethylation induces expression and methylation leads to reduced expression does not always hold good. (iii) More DMGs exhibited improved expression, relative to those, which exhibited reduced expression. (iv) Maximum number of DMGs exhibited methylation in CHH context. (v) Association of high expression with demethylation was more frequent than association of reduced expression with methylation. (vi) Methylation/demethylation within 1 Mb region had more pronounced effect on expression than methylation farther away.
Association of methylated regions with H3K4/K27me3 marks
DMRs/DMGs were also compared with differential binding sites (DBSs) for the two histone marks H3K4/K27me3 (available from our earlier study; Saripalli et al. 2020b). The details of DMGs used for comparison are available in Fig. 4b and Supplementary Tables S41 to S48. Although in our earlier study, both histone marks were found to be equally frequent, association mainly involved H3K4me3 with DMGs and were most abundant in CHH context. Taken both histone marks together, the frequency of DMGs associated with histone marks was predominatly in the genic regions (491 = 97.6%) relative to intergenic regions (only 12; 2.4%); these associations were not observed in the treatment pair S96 vs R96.
Expression of DMGs associated with H3K4/K27me3 marks
Differentially methylated genes with histone marks (H3K4me3 and H3K27me3) were further examined for their expression levels. Among DMGs associated with histone marks, only 30 DMGs could be identified as DEGs using transcriptome data. Ten of these 30 genes showed coordinated expression with both, DNA methylation and histone binding marks (Table 5). Some unique DMGs with histone marks were also identified from across all the three methylation contexts.
Transposable elements (TEs) associated with DMRs
Transposable elements associated with DMRs were classified into following three categories (as provided in IWGSC data): (i) TEs with repeat region; (ii) TEs with nested repeat, and (iii) TEs with repeat fragment. TEs within DMRs were also extracted using RepeatMasker. It was found that most methylated TEs were simple and low complexity repeats; the other classes of TEs occurred in low frequency (Supplementary Table S49). The frequencies of demethylated and methylated TEs of three types for each of the three methylation contexts are summarized in Table 6 (for each of the four different comparisons). In all the four comparisons, the TEs associated with CHH DMRs had the highest frequencies, in each of the three categories of TEs. Frequencies of TEs associated with DMRs were highest in S0 vs S96 and lowest in S0 vs R0. The relative frequencies of TEs associated with demethylated and methylated DMRs also differed, the former being highest in S0 vs S96, and the latter being highest in S96 vs R96 and R0 vs R96.
TE-associated DMGs (TAGs)
TE-associated genes (TAGs) occurred across all the three methylation contexts and all the four treatment comparisons (Table 7 and Fig. 4c; for details of genes see Supplementary Tables S50-S61). The pattern of demethylated TAGs did not differ from the general pattern of methylation in DMGs (see Fig. 4c and Table 7).
Co-expressed methylated transcription factor genes (mTFGs) and DEGs
A large number of co-expressed mTFGs and DEGs were identified using co-expression analysis (Supplementary Table S62). Maximum number of co-expressed mTFGs and DEGs were identified in S96 vs R96, with minimum in S0 vs S96. Further, the co-expressed genes were highest for CHH context in each of the the four treatment pairs. The coexpressed DEGs belonged to a number of classes-protein kinases, oxidoreductases, B3 domain containing genes, histone deacetylases, pentatricopeptide repeat containing genes, etc. The list of mTFGs with maximum number of coexpressed DEGs from each of the context in all treatment pairs are provided in Supplementary Table S63.
Validation of DMRs using qRT-PCR analysis
The results of qRT-PCR are shown in Fig. 5; 19 of 26 genes showed upregulation (11 with demethylation and 8 with methylation) and 7 showed down regulation (4 with methylation and 3 with demethylation). Thus only 15 genes gave expected association of gene expression with methylation (demethylation associated with upregulation and methylation associated with downregulation). The distribution of these 15 genes was as follows: (i) 4 genes in S0 vs S96; (ii) 2 genes in S0 vs R0; (iii) 4 genes in S96 vs R96 and (iv) 5 genes in R0 vs R96; the remaining 11 genes showed just the opposite association of expression with methylation. The expression changes of 13 genes also matched with expression changes observed using RNA-Seq analysis. Most conspicuous results include the genes encoding MLO-like protein (#3 in Fig. 5) showing 6.4-fold downregulation and protein kinase (#5 in Fig. 5) showing 25.3-fold downregulation, both in S0 vs S96.
Differential expression of 26 differentially methylated genes or DMGs (protein encoded by these DMGs are numbered 1 to 26; see details below) distributed in all the four treatment pairs including S0 vs S96, S0 vs R0, S96 vs R96 and R0 vs R96. In each treatment, the expression in second treatment is compared to that of the first. The genes indicated with red colored bars are the genes showing the expected association of downregulation with methylation and upregulation with demethylation. The genes indicated in purple-colored bars show the opposite association with DNA methylation. *Significant expression at P < 0.05; **significant at P < 0.01. 1: Actin cross-linking protein, putative, 2: F-box protein, 3: MLO-like protein, 4: 3-ketoacyl-CoA synthase, 5: Protein kinase, 6: Leucine-rich repeat-containing protein 59, 7: Pentatricopeptide repeat-containing protein, 8: NBS-LRR-like resistance protein, 9: F-Box, 10: UDP-glycosyltransferase, 11: Alpha/beta hydrolase, 12: Disease resistance protein (TIR-NBS-LRR class), 13: basic helix-loop-helix (bHLH), 14: Calcium-dependent lipid-binding (CaLB domain) putative, 15: F-box protein, 16: NADH-quinone oxidoreductase subunit H 1, 17: Cytochrome P450 family protein, putative, 18: Ankyrin repeat-containing protein, 19: Calcium-transporting ATPase, 20: Purple acid phosphatase, 21: Cytochrome b559 subunit alpha, 22: Cytokinin oxidase/dehydrogenase, 23: Copper ion-binding protein, 24: NAD(P)H-quinone oxidoreductase subunit 6, chloroplastic, 25: Peptidase M50B-like protein, 26: transmembrane protein, putative
Discussion
The present study is one of the few studies involving the use of BS-Seq for analysis of genome-wide DNA methylation in bread wheat. The study was intended to analyse the relative role of DNA methylation in three different contexts (CG, CHG and CHH) in Lr28-mediated resistance against leaf rust in bread wheat. There are at least two earlier studies involving genome-wide DNA methylation through BS-Seq, which involved bread wheat, its D sub-genome progenitor Ae. tauschii and tetraploid wheat (Gardiner et al. 2015; Yuan et al. 2020). Another important relevant study on DNA methylation based on BS-Seq was undertaken in Ae. tauschii-powdery mildew (Blumeria graminis) pathosystem (Geng et al. 2019); this study demonstarted that DNA methylation with CHH context represents an important regulatory layer in the complex mechanism of plant immunity, which could be exploited to improve disease resistance in common wheat. The results of our study are in agreement with the results of this earlier study (see later for details).
The results of the present study broadly suggest that only a very small fraction of genome-wide methylation is involved in differential methylation within and between susceptible and resistant NILs. Similar results were obtained in several earlier studies. Another important feature in the results of the present study is that although CHH methylation represents a mere ~ 2% of total genome-wide methylation, the differentially methylated regions (DMRs) involving CHH context accounted for > 50% of genome-wide DMRs. These DMRs/DMGs included both enhanced methylation and demethylation leading to repression/induction of the activity of a large number of genes. The DMRs involved in demethylation were almost double (10,689) the frequency of DMRs involved in enhanced methylation or hypermethylation (4928) (Fig. 1b), suggesting that changes in DNA methylation more often caused activation rather than repression of downstream genes (assuming that demethylation leads to activation of genes, although this assumption is not always true).
The methylation pattern in plants is generally high in CG (90%) followed by CHG and CHH (Matzke and Mosher 2014). Apparently, in the present study, the CG methylation is more conserved in susceptible and resistant lines leading to less frequent appearance of DMRs with CG whereas, the reverse pattern in CHH probably might be due to greater activation of differential sRNAs following RdDM pathways in S and R lines upon pathogen infection, leading to CHH methylation. The dynamic regulation of gene activity predominantly by CHH methylation as observed during our study has also been reported in at least two earlier studies, one involving Arabidopsis-Pseudomonas syringae pathosystem (Dowen et al. 2012) and the other involving Ae tauschii—powdery mildew (Blumeria graminis) pathosystem, mentioned above (Geng et al. 2019).
It is also apparent from the results of the present study that the overall frequency of DMRs with reduced methylation increased with the passage of time after inoculation in S line; reverse was the case in R line (Fig. 1b). This suggests that many more genes are activated and only fewer genes are repressed in S line, the situation being just the opposite in R line. This is in agreement with the findings that adaptation to biotic stress in plants involves DNA methylation leading to repression of genes (Peng and Zhang, 2009).
Genomic distribution of DMRs
The genome-wide identification of DMRs in the present study mainly involved the following activities: (i) identification of DMRs followed by identification of DMGs within these DMRs; (ii) study of the methylation level (enhanced methylation or hypermethylation vs reduced methylation or demethylation) in DMRs and DMGs; (iii) the effect of methylation status on activity of individual specific genes and their relationship with suscpetibility and resistance against leaf rust; and (iv) association of DMRs and histone modifications in five different classes of genomic regions with 18 different chromatin states (Table 2). As mentioned above, perhaps an important observation is the predominance of methylation in asymmetric CHH context relative to symmetric CG and CHG contexts in DMRs and DMGs (Fig. 1c). Methylation in promoters and TTS is supposed to have negative correlation (methylation downregulated gene expression) with gene expression while that in exons, introns and intergenic regions is supposed to have positive correlation with methylation causing increased activity of genes (Curradi et al. 2002; Mette et al. 2000).
The variation in the frequencies of DMRs and DMGs in different genomic regions is though conspicous, their relevance to regulation of gene expression during host pathogen-interaction is not apparent. This variation may actually reflect the relative proportion of these genomic regions in the whole genome. For instance, a very high frequency of DMRs/DMGs in the intergenic regions may actually be due to high proportion of genome occupied by the intergenic regions, as also emphasized by Li et al. (2019). Similarly, very low frequency of DMRs/DMGs in exons may also be attributed to a very small fraction of the genome occupied by these exons. We believe that the significance of this disparity in ditribution of DMRs/DMGs, if there is any, may be the subject of further investigation.
A study of the distribution of 18 chromatin states in five genomic regions and their association with not only DMRs/DMGs, but also with two histone marks (H3K4/K27me3) and relative expression of different classes of genes is another important part of the present study. However, the subject of chromatin states and their relevance to regulation of gene is a recent area of research, which is though important, but not immediately apparent. For instance, one may like to ask the question whether or not these chromatin states depend on DNA methylation and histone marks, and if they do what is the mechanism involved. One may also like to ask the question that how each chromatin state in turn regulates specific genes involved in host–pathogen interaction. We did not find answer to these questions in the published literature, suggesting that these subjects also represent areas of future studies involving more sophisticated techiques.
DMGs identified in R and S NILs
The resistant (R) and susceptible (S) NILs used in the present study were genetically very similar with only 1% polymorphism when examined using 35 K wheat SNP array (our unpublished data). Thus, the NILs constitute suitable material for a comparative study of differential DNA methylation in response to pathogen inoculation (Fig. 3a). In S line, due to infection at 96 hai, the number of DMGs (CG + CHH + CHG = 4373; Fig. 3a) activated due to demethylation are almost one and half-fold the number of genes that are repressed due to methylation (CG + CHH + CHG = 2607; Fig. 3a) after 96 hai. On the contrary, in R line at 96 hai, repression due to methylation of large number of genes (4258 in R0 vs R96 and 4781 in S96 vs R96) seems to be conspicuous from the results of the two comparisons (Fig. 3a). This suggested that susceptibility is relatively controlled by activation of genes whereas resistance is governed by repression of large number of genes (~ 60% genes are activated and ~ 40 repressed in S line). Therefore, it can also be speculated that the genes involved in resistance due to Lr28 are often negatively regulated. This may involve genes like TaLSD1 (encoding for zinc finger protein) and TaNAC35, which have been demonstrated to have a role in negative regulation of stripe rust resistance and Lr14 mediated leaf rust resistance, respectively (Guo et al. 2013; Zhang et al. 2021).
Some of the genes, which undergo demethylation mediated upregulation in S line include those encoding for the following proteins: CC-NBS-LRR, Zinc finger proteins, receptor kinase, WRKY TF, cytochrome P450, calcium transporting ATPase, MYB TF, 3-ketoacyl coA-synthase, ABC transporter etc. Similarly, the genes in R line which showed high methylation mediated downregulation included those encoding for the following: ankyrin repeats, ring-U box superfamily protein, lipid transfer protein, alpha/beta hydrolases, F-box protein, WD40 protein, glutathione S transferase, methyltransferase, bHLH TF, etc. (Supplementary Tables S29–S40). These genes may be involved in signal transduction pathways (e.g. salicyclic and jasmonic acid pathways) following infection.
The genes in R line which show demethylation mediated upregulation may also be important. Some genes in R line which undergo demethylation mediated upregulation include those encoding for the following: CRT binding factor, glycosyltransferase, jasmonate ZIM protein, MADS box TF, NAC TF, bZIP TF, peroxidases, etc. (Supplementary Tables S29–S40).
As mentioned earlier, it is well known that methylation does not always repress gene expression; enhanced methylation in gene body may also lead to higher expression of genes (Dowen et al. 2012). Therefore, the genes showing methylation mediated upregulation may also be important. Such genes in S line showing methylation mediated upregulation due to leaf rust infection (S0 vs S96) include those encoding for NBS-LRR, homeobox leucine zipper protein, glutamate receptor protein, etc. Similar genes in R line (R0 vs R96) involved in providing resistance during leaf rust infection encode for NADH ubiqinone oxidoreductase, transmembrane protein, E3 ubiquitine protein ligase, MADS box TF, peroxidases, etc. The roles of all the above DMGs in S and R lines during susceptibility and resistance are listed in Table 8.
Relationship of DNA methylation with gene expression, histone modification and chromatin state
It is apparent from the results shown in Tables 2 and 5 that the expression level (upregulation vs downregulation) of DMGs depends on several factors including the following: (i) enhanced methylation vs demethylation, (ii) methylation context (CG, CHG, CHH), (iii) binding affinity of two histone marks (H3K4me3 causing induction and H3K27 causing repression); (iv) location of DMGs in one of the five genomic regions (enhancers, intergenic regions, genic regions, promoters and repetitive regions) and (v) chromatin state (18 different chromatin states are known). There are other factors (e.g, ncRNA like miRNA, lncRNA including long intergenic ncRNA or lincRNAs), which may be involved, but have not been included in the present study.
Another relevant area, which is receiving current attention, is the study of the mechanism of DNA methylation and the role of genetic variation in this mechanism (methylation QTLs or mQTLs; Hawe et al. 2022). This suggests that epigenetic regulation of the expression of DMGs, which may be responsible for susceptibility/resistance against leaf rust (like any other trait) is really complex and it is really difficult to partition the role of each of the several factors listed above. It is also difficult to find whether these factors are interdependent or if there is any causal relationship between any two or more of these factors. One may also like to analyse the role of each of three classes of DMGs, representing three different contexts, CG, CHG and CHH and several histone marks including H3K4me3 and H3K27me3 used in the present study. The three different methylation contexts and/or two histone marks perhaps control the activity of DMGs and other genes, whose activity is controlled by these DMGs, through 18 different chromatin states, which in turn are distributed in five different genomic regions, namely enhancers, intergenic regions, genic regions, promoters and repetitive regions. More than half of the DMGs exhibited expected relationship with expression patterns, suggesting that DNA methylation possibly plays a significant role in controlling the expression of genes involved in wheat-leaf rust interaction. Such expected relationships were also observed in earlier studies in responses to biotic stresses involving Ae. tauschii-B. graminis (Geng et al. 2019) and rice-M. oryzeae (Cui et al. 2020) pathosystems. However, this relationship between DNA methylation and gene expression may not be direct and causal, since association of DNA methylhistone marks is also involved. A comparison of results of the present study with the results of our earlier study on the role of H3K4/K27me3 marks using ChIP-seq (Saripalli et al. 2020b) allowed identification of 491 (463 unique) genes which were perhaps regulated by both DNA methylation and binding of histone marks (435 DMGs enriched with H3K4me3 and 28 DMGs enriched with H3K27me3); these results have also been related with chromatin states.
In our own study involving the same experimental material (Saripalli et al. 2020b), ~ 19% of the genes associated with histone methylation due to H3K4me3 (12%) and H3K27me3 (7%) marks were found to exhibit expected expression changes, when compared with DEGs in RNA-seq data. Some of these genes identified in the above study (Saripalli et al. 2020b) were also identified in the present study and the results are discussed later. Similarly, in another study using the same material (Jain et al. 2020), expression of some genes was found to be regulated through ncRNA (miRNA and lncRNA). The results of comparison of DNA methylation have also been related with chromatin state, although the mechanism of action of chromatin state in regulation of expression of specific genes is largely unknown. DNA methylation in enhancer region is perhaps directly involved in regulation of large number of DE genes as observed in our earlier transcriptome study involving wheat-leaf rust interaction (Sharma et al. 2018). This could also be assessed from the results of co-expression analysis in the present study where a large number of methylated TFs were found co-expressed with the DEGs, although the frequency of methylation in promoter region was lower in comparison to frequency of methylated TFs. In case of CHH context, < 50% of methylation in intergenic region was found within < 10 Kb flanking region of the genes, which possibly suggests that methylation in enhancer and/or silencer regions also plays a role in gene regulation during wheat-leaf rust interaction. The results also clearly reveal a role of other epigenetic or genetic mechanisms in the regulation of leaf rust resistance in wheat.
Ten DMGs with H3K4me3 also showed expected negative relationship between H3K4me3 mark and demethylation mostly in S line with CG and CHG methylation contexts; these results do not differ from those reported earlier for Arabidopsis (Zhang et al. 2009; Mathieu et al. 2005). In the remaining four DMGs with H3K27me3 mark, no synergistic relationship with DNA methylation was observed. It is also apparent from the results of the present study that 10 DMGs out of 30 DMGs with H3K4/K27me3 marks showed expected relationship between DNA methylation and gene expression irrespective of the presence of H3K4/K27me3 histone marks.
The results of chromatin state analysis suggested that expression of DMGs encoding the following proteins were also involved (directly/indirectly) in regulation of gene expression through modification of chromatin states: (i) histone-lysine N-methyltransferase, (ii) S-adenosyl-L-methionine-dependent methyltransferases superfamily protein (SAM), (iii) lysine-specific demethylase, (iv) Set1/Ash2 histone methyltransferase complex subunit ASH2, (v) H3 lysine-9 specific, (vi) methyltransferase family protein, (vii) histone acetyltransferase of the CBP family 12, (viii) polycomb group protein and (ix) PHD finger protein. Among the above nine genes whose expression is influenced by chromatin state, four genes listed at numbers (i), (v), (vi) and (ix) were differentially expressed in RNA-Seq data as well. The function of these genes must be important for wheat-leaf rust interaction. Presence of multiple DMGs encoding histone-specific proteins listed at numbers (v) and (vi) suggests that H3K9 methylation mark might play an important role in wheat-leaf rust interaction. Differentially expressed genes (DMGs) encoding basic helix-loop-helix (bHLH) DNA-binding superfamily protein is possibly in agreement with the role of enhancer region and help in DNA folding to assist cis-regulation of genes involving 1 Mb upstream and downstream enhancer regions.
Summary and conclusions
In the present study, context-specific DNA methylation (CG, CHG and CHH) during wheat-leaf rust interaction was examined for the first time using high-resolution BS-Seq approach. A pair of NILs for the gene Lr28 for resistance against leaf rust in the background of the wheat cv. HD2329 was used for this purpose. The major conclusion is that demethylation mediated activation of many genes and enhanced methylation mediated silencing of fewer genes occurs during susceptible reaction (S0 vs S96). The situation is just the reverse in resistance reaction (R0 vs R96 and S96 vs R96) with enhanced methylation mediated repression of many genes and demethylation mediated activation of fewer genes. Genome-wide methylation was maximum for CG context; however, differential methylation mainly involved CHH context. Comparison of DMGs with earlier expression data revealed that more than half of DMGs showed expected relationship with gene expression (i.e. methylation leads to repression and demethylation leads to high activation). Some important DMGs which undergo activation in S line include CC-NBS-LRR, Zinc finger proteins, receptor kinase, WRKY TF, etc. Similarly, the genes in R line which showed methylation mediated downregulation included those encoding for ankyrin repeats, ring-U box superfamily protein, lipid transfer protein, alpha/beta hydrolases, F-box protein, WD40 protein, glutathione S transferase, methyltransferase, bHLH TF, etc. Comparison of the results of BS-Seq with earlier histone methylation data revealed 491 DMGs which were perhaps regulated by differential binding of histone methylation marks (H3K4me3 and H3K27me3); 30 genes also showed differential expression in RNA-Seq data. The results of the present study proved our earlier hypothesis that mechanism of leaf rust resistance in wheat follows a complex mechanism involving one or more epigenetic changes influencing the expression of different downstream genes due to Lr28. Some of the genes identified during the present study may be important candidates for further validation using RNAi or overexpression techniques.
Data availability
The raw reads generated using BS-Seq analysis for all the eight samples which include two biological replicates of each of the resistant (R) and susceptible (S) NILs at 0 hbi and 96 hai are deposited in Sequence Read Archive (SRA), National Center for Biotechnology Information (NCBI), and can be accessed through bioproject accession number PRJNA745483. The raw reads for RNA-Seq and ChIP-Seq analysis are deposited under accession number PRJNA588134 of Bioproject Database.
Code availability
Not applicable.
References
Ahmed SM, Liu P, Xue Q, Ji C, Qi T, Guo J, Guo J, Kang Z (2017) TaDIR1–2, a wheat ortholog of lipid transfer protein AtDIR1 contributes to negative regulation of wheat resistance against Puccinia striiformis f. sp. tritici. Front Plant Sci 11:521. https://doi.org/10.3389/fpls.2017.00521
Casassola A, Brammer SP, Chaves MS, Martinelli JA, Stefanato F, Boyd LA (2015) Changes in gene expression profiles as they relate to adult plant leaf rust resistance in wheat cv. Toropi. Physiol Mol Plant Pathol 89:49–54. https://doi.org/10.1016/j.pmpp.2014.12.004
Changenet V, Macadré C, Boutet-Mercey S et al (2021) Overexpression of a cytochrome P450 monooxygenase involved in orobanchol biosynthesis increases susceptibility to fusarium head blight. Front Plant Sci 12:662025. https://doi.org/10.3389/fpls.2021.662025
Conesa A, Götz S (2008) Blast2GO: a comprehensive suite for functional analysis in plant genomics. Int J Plant Genomics 2008:619832. https://doi.org/10.1155/2008/619832
Cui N, Chen X, Shi Y, Chi M, Hu J, Lai K, Wang Z, Wang H (2020) Changes in the epigenome and transcriptome of rice in response to Magnaporthe oryzae infection. Crop J 9:843–853. https://doi.org/10.1016/j.cj.2020.10.002
Curradi M, Izzo A, Badaracco G, Landsberger N (2002) Molecular mechanisms of gene silencing mediated by DNA methylation. Mol Cell Biol 22:3157–3173. https://doi.org/10.1128/MCB.22.9.3157-3173.2002
Deng Y, Zhai K, Xie Z et al (2017) Epigenetic regulation of antagonistic receptors confers rice blast resistance with yield balance. Science 355:962–965. https://doi.org/10.1126/science.aai8898
Dowen RH, Pelizzola M, Schmitz RJ et al (2012) Widespread dynamic DNA methylation in response to biotic stress. Proc Natl Acad Sci USA 109:E2183–E2191. https://doi.org/10.1073/pnas.1209329109
Ernst J, Kellis M (2012) ChromHMM: automating chromatin-state discovery and characterization. Nat Methods 28:215–216. https://doi.org/10.1038/nmeth.1906
Gardiner LJ, Quinton-Tulloch M, Olohan L et al (2015) A genome-wide survey of DNA methylation in hexaploid wheat. Genome Biol 16:273. https://doi.org/10.1186/s13059-015-0838-3
Geng S, Kong X, Song G et al (2019) DNA methylation dynamics during the interaction of wheat progenitor Aegilops tauschii with the obligate biotrophic fungus Blumeria graminis f. sp. tritici. New Phytol 221:1023–1035. https://doi.org/10.1111/nph.15432
Guo L, Li M, Wang W, Wang L, Hao G, Guo C, Chen L (2012) Over-expression in the nucleotide-binding site-leucine rich repeat gene DEPG1 increases susceptibility to bacterial leaf streak disease in transgenic rice plants. Mol Biol Rep 39:3491–3504. https://doi.org/10.1007/s11033-011-1122-6
Guo J, Bai P, Yang Q, Liu F, Wang X, Huang L, Kang Z (2013) Wheat zinc finger protein TaLSD1, a negative regulator of programmed cell death, is involved in wheat resistance against stripe rust fungus. Plant Physiol Biochem 71:164–172. https://doi.org/10.1016/j.plaphy.2013.07.009
Hansen KD, Langmead B, Irizarry RA (2012) BSmooth: from whole genome bisulfite sequencing reads to differentially methylated regions. Genome Biol 13:R83. https://doi.org/10.1186/gb-2012-13-10-r83
Hawe JS, Wilson R, Schmid KT et al (2022) Genetic variation influencing DNA methylation provides insights into molecular mechanisms regulating genomic function. Nature Genet 54:18–29
He Y, Wu L, Liu X et al (2020) TaUGT6, a novel UDP-glycosyltransferase gene enhances the resistance to FHB and DON accumulation in wheat. Front Plant Sci 11:574775. https://doi.org/10.3389/fpls.2020.574775
Heinz S, Benner C, Spann N, Bertolino E et al (2010) Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell 38:576–589. https://doi.org/10.1016/j.molcel.2010.05.004
Jablonka E, Raz G (2009) Transgenerational epigenetic inheritance: prevalence, mechanisms, and implications for the study of heredity and evolution. Quart Rev Biol 84:131–176. https://doi.org/10.1086/598822
Jain N, Sinha N, Krishna H et al (2020) A study of miRNAs and lncRNAs during Lr28-mediated resistance against leaf rust in wheat (Triticum aestivum L.). Physiol Mol Plant Pathol 112:101552. https://doi.org/10.1016/j.pmpp.2020.101552
Jones JD, Dangl JL (2006) The plant immune system. Nature 444:323–329. https://doi.org/10.1038/nature05286
Krueger F, Andrews SR (2011) Bismark: a flexible aligner and methylation caller for Bisulfite-Seq applications. Bioinformatics 27:1571–1572. https://doi.org/10.1093/bioinformatics/btr167
Li Z, Wang M, Lin K et al (2019) The bread wheat epigenomic map reveals distinct chromatin architectural and evolutionary features of functional genetic elements. Genome Biol 20:139. https://doi.org/10.1186/s13059-019-1746-8
Lippman Z, Gendrel AV, Black M et al (2004) Role of transposable elements in heterochromatin and epigenetic control. Nature 430:471–476. https://doi.org/10.1038/nature02651
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2–DDC. Methods 25:402–408. https://doi.org/10.1006/meth.2001.1262
Maekawa T, Kufer TA, Schulze-Lefert P (2011) NLR functions in plant and animal immune systems: so far and yet so close. Nat Immunol 12:817–826. https://doi.org/10.1038/ni.2083
Mathieu O, Probst AV, Paszkowski J (2005) Distinct regulation of histone H3 methylation at lysines 27 and 9 by CpG methylation in Arabidopsis. EMBO J 24:2783–2791. https://doi.org/10.1038/sj.emboj.7600743
Matzke MA, Mosher RA (2014) RNA-directed DNA methylation: an epigenetic pathway of increasing complexity. Nat Rev Genet 15:394–408. https://doi.org/10.1038/nrg3683
Mette MF, Aufsatz W, Winden J, Matzke MA, Matzke AJM (2000) Transcriptional silencing and promoter methylation triggered by double-stranded RNA. EMBO J 19:5194–5201. https://doi.org/10.1093/emboj/19.19.5194
Pandey SP, Somssich IE (2009) The role of WRKY transcription factors in plant immunity. Plant Physiol 150:1648–1655. https://doi.org/10.1104/pp.109.138990
Peng H, Zhang J (2009) Plant genomic DNA methylation in response to stresses: potential applications and challenges in plant breeding. Prog Nat Sci 19:1037–1045. https://doi.org/10.1016/j.pnsc.2008.10.014
Ramírez F, Dündar F, Diehl S, Grüning BA, Manke T (2014) deepTools: a flexible platform for exploring deep-sequencing data. Nucleic Acids Res 42:W187–W191. https://doi.org/10.1093/nar/gku365
Ran X, Tang T, Wang M et al (2021) CSCS: a chromatin state interface for Chinese Spring bread wheat. aBIOTECH. https://doi.org/10.1007/s42994-021-00048-z
Rushton PJ, Somssic IE, Ringler P, Shen QJ (2010) WRKY transcription factors. Trends Plant Sci 15:247–258. https://doi.org/10.1016/j.tplants.2010.02.006
Saripalli G, Sharma C, Gautam T et al (2020a) Complex relationship of DNA methylation with gene expression during wheat-leaf rust interaction in a pair of NILs differing for Lr28. Mol Bio Rep 47:1339–1360. https://doi.org/10.1007/s11033-019-05236-1
Saripalli G, Singh K, Gautam T, Santosh K, Saurabh R, Prasad P, Jain N, Sharma PK, Balyan HS, Gupta PK (2020b) Genome-wide analysis of H3K4me3 and H3K27me3 modifications due to Lr28 for leaf rust resistance in bread wheat (Triticum aestivum). Plant Mol Biol 104:113–136. https://doi.org/10.1007/s11103-020-01029-4
Sharma C, Saripalli G, Kumar S et al (2018) A study of transcriptome in leaf rust infected bread wheat involving seedling resistance gene Lr28. Funct Plant Biol 45:1046–1064. https://doi.org/10.1071/FP17326
Sharma C, Kumar S, Saripalli G et al (2019) H3K4/K9 acetylation and Lr28-mediated expression of six leaf rust responsive genes in wheat (Triticum aestivum). Mol Genet Genom 294:227–241. https://doi.org/10.1007/s00438-018-1500-z
Sun L, Miao X, Cui J, Deng J, Wang X, Wang Y et al (2018) Genome-wide high-resolution mapping of DNA methylation identifies epigenetic variation across different salt stress in maize (Zea mays L.). Euphytica 214:25. https://doi.org/10.1007/s10681-017-2076-0
Wang F, Ling R, Feng J, Qiu D, Cheng W, Xu S (2015) Wheat bHLH transcription factor gene, TabHLH060, enhances susceptibility of transgenic Arabidopsis thaliana to Pseudomonas syringae. Physiol Mol Plant Pathol 90:123–130. https://doi.org/10.1016/j.pmpp.2015.04.007
Yuan J, Jiao W, Liu Y et al (2020) Dynamic and reversible DNA methylation changes induced by genome separation and merger of polyploid wheat. BMC Biol 18:171. https://doi.org/10.1186/s12915-020-00909-x
Zhang X, Bernatavichute YV, Cokus S et al (2009) Genome-wide analysis of mono-, di- and trimethylation of histone H3 lysine 4 in Arabidopsis thaliana. Genome Biol 10:R62. https://doi.org/10.1186/gb-2009-10-6-r62
Zhang N, Yuan S, Zhao C, Park RF, Wen X, Yang W, Zhang N, Liu D (2021) TaNAC35 acts as a negative regulator for leaf rust resistance in a compatible interaction between common wheat and Puccinia triticina. Mol Genet Genom 296:279–287. https://doi.org/10.1007/s00438-020-01746-x
Zhao G, Zou C, Li K et al (2017) The Aegilops tauschii genome reveals multiple impacts of transposons. Nat Plants 3:946–955. https://doi.org/10.1038/s41477-017-0067-8
Zhu Y, Li Y, Fei F et al (2015) E3 ubiquitin ligase gene CMPG1-V from Haynaldia villosa L. contributes to powdery mildew resistance in common wheat (Triticum aestivum L.). Plant J 84:154–168. https://doi.org/10.1111/tpj.12966
Acknowledgements
The work was carried out, when GS and TG held SRF positions under a research project funded under National Agricultural Science Fund (NASF) -ICAR program of Government of India. KS held the position of RA in a project funded by Department of Biotechnology, Govt. of India, New Delhi. HSB was awarded Senior Scientist and Hony. Scientist positions from Indian National Science Academy (INSA). NASF-ICAR (File No. NASF/ABP-6006/2016-17), New Delhi, India provided funds for carrying out this study. Bioinformatics Infrastructure Facility (BIF) laboratory was used for carrying out most of the bioinformatics analyses. Head, Department of Genetics and Plant Breeding, Chaudhary Charan Singh University, Meerut, provided the necessary infrastructure. Thanks are also due to Ritu Batra for critical reading of the manuscript.
Funding
This work was supported by National Agricultural Science Fund-Indian Council of Agricultural Research (NASF-ICAR). Name of the project: “Epigenetic regulation of host–pathogen genetics of leaf rust resistance in wheat” NASF’s Project No.: NASF-ICAR File No. NASF/ABP-6006/2016-17.
Author information
Authors and Affiliations
Contributions
PKG and HSB conceived the experiment. KS conducted the bioinformatics analysis and wrote the first draft of the manuscript jointly with GS. GS was involved in data interpretation with other authors and also conducted qRT-PCR analysis jointly with TG. NJ raised and provided the seedlings for NILs. PP provided the inoculum for leaf rust pathotype 77-5. PKG and HSB finalized the manuscript with the help of KS and GS. All authors read the final version of the manuscript and approved it.
Corresponding author
Ethics declarations
Conflict of interest
All the authors declare that they have no conflict of interest.
Ethical approval
Not applicable.
Consent to participate
Not applicable.
Consent for publication
Not applicable.
Additional information
Communicated by Bing Yang.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Singh, K., Saripalli, G., Gautam, T. et al. BS-Seq reveals major role of differential CHH methylation during leaf rust resistance in wheat (Triticum aestivum L.). Mol Genet Genomics 297, 731–749 (2022). https://doi.org/10.1007/s00438-022-01879-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00438-022-01879-1