Abstract
In the present study, we describe Sarcocystis entzerothi n. sp. from the European roe deer (Capreolus capreolus) based on the microscopical and DNA analysis. By light microscopy (LM), cysts of S. entzerothi were spindle-shaped with pointed tips, 950–1900 × 70–150 μm in size and had 5–6 μm long finger-like cyst wall protrusions. Cyst wall of S. entzerothi by transmission electron microscopy (TEM) was type 10a-like; villar protrusions were up to 1.2 μm wide, densely packed, lying about 0.1 μm between each other, had profuse microgranules and microfilaments, parasitophorous vacuolar membrane had many minute invaginations, and the ground substance layer measured up to 0.4 μm. This species is morphologically similar to Sarcocystis silva, previously found in the roe deer and the moose (Alces alces). By LM, cysts of S. silva were cigar-shaped with blunted tips, measured 1000–1500 × 130–184 μm, and had 7–8 μm long finger-like cyst wall protrusions. Under TEM, S. silva had no clear differences from S. entzerothi in their cyst wall ultrastructure. Having examined six roe deer hunted in Lithuania, cysts of S. entzerothi and S. silva were identified in four and two animals, respectively. These two Sarcocystis species could be morphologically differentiated according to the shape of the cysts and the length of protrusions. The species examined showed 95.6–96.1 % and 85.6–86.9 % sequence identity within 18S ribosomal DNA (rDNA) and cox1, respectively, and therefore they could be clearly distinguished by means of molecular methods. It should be noted that in the 18S rDNA phylogenetic tree, S. entzerothi from the roe deer was placed together with one sequence of Sarcocystis sp. from the Lithuanian red deer (Cervus elaphus) demonstrating the same species. Based on 18S rDNA and cox1 sequences, S. entzerothi was more closely related to Sarcocystis species transmitted via felids than canids.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Protozoan parasites of the genus Sarcocystis (Apicomplexa: Sarcocystidae) have an obligatory two-host prey-predator life cycle. Oocysts/sporocysts develop in a small intestine of a definitive host, while sarcocysts are mainly formed in striated muscles of an intermediate host. Generally, herbivores and omnivores serve as intermediate hosts, while carnivores act as definitive hosts of these parasites (Dubey et al. 2015).
Up till now, the European roe deer (Capreolus capreolus) is known to be an intermediate host of four Sarcocystis species, Sarcocystis gracilis, Sarcocystis capreolicanis, Sarcocystis oviformis, and Sarcocystis silva (Rátz 1909; Erber et al. 1978; Dahlgren and Gjerde 2009; Gjerde 2012). The validity of these species was confirmed by molecular methods using 18S ribosomal DNA (rDNA) and cox1 (mitochondrial gene encoding subunit I of cytochrome c oxidase) sequences analysis (Dahlgren and Gjerde 2009; Gjerde 2012, 2013; Kolenda et al. 2014). The first three species were observed exclusively in the roe deer, while S. silva was identified in the roe deer and the moose (Alces alces) (Dahlgren and Gjerde 2008; Gjerde 2012). The morphology of two Sarcocystis species from the roe deer, S. gracilis and S. capreolicanis, which are transmitted via canids (Blažek et al. 1978; Erber et al. 1978), were extensively examined by light microscopy (LM), transmission electron microscopy (TEM), and scanning electron microscopy (SEM) (Bergmann and Kinder 1976; Erber et al. 1978; Entzeroth 1982, 1985; Sedlaczek and Wesemeier 1995; Kutkienė 2001; Spickschen and Pohlmeyer 2002; Gjerde 2012). Morphological characteristics of S. oviformis cysts have been studied by LM and SEM so far (Dahlgren and Gjerde 2009). The fourth species, S. silva, characterized by thick-walled cysts with finger-like protrusions, was described on the basis of the results of LM and DNA analysis (Gjerde 2012). Before the description of S. silva, some authors assigned cysts of this particular type found in the roe deer to Sarcocystis hofmanni (López et al. 2003), S. cf. hofmanni (Sedlaczek and Wesemeier 1995; Kutkienė 2001), or S. hofmanni-like (Prakas 2011) due to their resemblance to S. hofmanni cysts described in the European badger (Meles meles) (Odening et al. 1994).
In a previous study, Prakas (2011) molecularly examined 11 cysts with finger-like protrusions, 10 of them were isolated from the Lithuanian roe deer and one was isolated from the Lithuanian red deer (Cervus elaphus). Based on the 18S rDNA sequence comparison, cysts belonged to two species, S. silva (three identical sequences assigned to JN256132) and Sarcocystis sp. (JN256133–JN256137 from the roe deer and JN256125 from the red deer). Despite relatively high (3.8–4.0 %) 18S rDNA sequences variation between S. silva and Sarcocystis sp. extracted from the roe deer, clear taxonomically important morphological differences of these likely two species were not established by LM. Therefore, the aim of the present study was to determine whether cysts with finger-like protrusions found in the roe deer represented one or more Sarcocystis species based on their morphological (LM and TEM) and molecular (18S rDNA and cox1) characterization.
Material and methods
Samples
In 2015, diaphragm muscles of six roe deer hunted in Central Lithuania were examined for the Sarcocystis cysts. The muscle samples were kept frozen (−20 °C) until microscopic examination of the cysts.
Morphological analysis
The morphological analysis of sarcocysts and bradyzoites was performed in fresh-squashed preparations. By LM, the cysts were differentiated according to the size and shape of sarcocysts, the structure of the cyst wall and bradyzoites after the cysts had been isolated from muscle fibers with the help of two fine needles. Two morphologically different cysts, both with finger-like protrusions, were processed for TEM and examined in the previously described way (Prakas et al. 2016).
Molecular analysis
For molecular analysis, six cysts with finger-like protrusions on their surface were isolated from the muscle tissues, placed in individual 1.5 ml tubes containing 20 μl of 96 % ethanol and kept at −20 °C. Genomic DNA was extracted from individual sarcocysts using QIAamp® DNA Micro Kit (Qiagen, Hilden, Germany) according to the manufacturer’s recommendations. The resulting DNA samples were used for PCR amplification of 18S rDNA and cox1 sequences. SarAF/SarBR and SarCF/SarDR primer pairs were used for nearly complete 18S rDNA sequence amplification (Kutkienė et al. 2010), while partial cox1 sequences were amplified with the help of SF1 forward primer in combination with one of the following reverse primers SR5 (Gjerde 2013) or in the present study newly designed SR12H (5′-AAATACCTTGGTGCCCGTAG-3′), depending on Sarcocystis species. Each PCR reaction mixture contained 12.5 μl DreamTaq PCR Master Mix (2×) (Thermo Fisher Scientific Baltics, Vilnius, Lithuania), 0.05 μg template DNA, 1 μM of each primer, and nuclease-free water to make the final 25-μl volume. PCR cycling conditions, PCR products evaluation and sequencing were carried out as described previously (Prakas et al. 2016). The resulting sequences were imported into the MEGA7 (Kumar et al. 2016), edited manually if necessary and merged into a single fragment of the analyzed genes.
The 18S rDNA and cox1 sequences obtained in this study were compared with those of various Sarcocystis spp. using the Nucleotide BLAST program megablast option (http://blast.ncbi.nlm.nih.gov/) in order to find remarkably similar DNA sequences and to compute the sequence identity values. Previously published sequences of selected Sarcocystis spp. using ruminants as intermediate hosts were retrieved from GenBank for the phylogenetic analyses. The multiple 18S rDNA and cox1 sequence alignments were obtained using MUSCLE algorithm (Edgar 2004) implemented in the MEGA7 software. TOPALi v2.5 software (Milne et al. 2004) was used to select a nucleotide substitution model with the best fit to the aligned sequence dataset and to construct the phylogenetic trees under the Bayesian inference.
Results
Sarcocystis spp. found in Lithuanian roe deer
Under LM, five Sarcocystis species were found in the roe deer examined. Three of them that had previously been described, and morphologically well distinguished species (S. gracilis, S. capreolicanis, and S. oviformis) were not analyzed by molecular methods. Detailed examinations were carried out with the particular Sarcocystis species having cysts with finger-like protrusions. According to the molecular results, these sarcocysts were identified as belonging to one of two distinct species, Sarcocystis entzerothi n. sp. or S. silva. Cysts of S. entzerothi were detected in four of six roe deer examined (isolates CcLttvz1, CcLttvz2, CcLttvz4, and CcLttvz6), whereas cysts of S. silva were found in two animals (isolates CcLttva2 and CcLttva3). One roe deer harbored both species (isolates CcLttvz2 and CcLttva2).
Description of S. entzerothi n. sp.
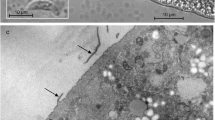
By LM, cysts of S. entzerothi were microscopic, spindle-shaped with pointed tips, and measured 1138.8 × 107.5 μm (950–1900 × 70–150; n = 6) (Fig. 1a). The cyst wall had 5–6 μm long finger-like protrusions (Fig. 1b). The contents of the cysts were divided by septa into large chambers filled with banana-shaped 10.7 × 3.3 μm (9.29–12.69 × 2.6–4.23; n = 18) bradyzoites (Fig. 1c). Under TEM, the cyst wall of S. entzerothi had thick, up to 4.5 μm long, finger-like villar protrusions. The width of the protrusions varied from 0.9 to 1.2 μm at the bases (Fig. 1d). The ground substance layer was thin, 0.4–0.5 μm, and seemed electron pale. The cyst wall protrusions of S. entzerothi had profuse microgranules and microfilaments; the protrusions were tightly packed, lying about 0.1 μm between each other (Fig. 1e). The surface of the protrusions appeared to be wavy, since the parasitophorous vacuolar membrane had many minute invaginations. The cyst wall was type 10a-like (Dubey et al. 2015).
Morphology of sarcocysts of Sarcocystis entzerothi n. sp. from the diaphragm muscles of the roe deer. a–c Light micrographs. Fresh preparations. a Fragment of the cyst. Note pointed tip of the cyst (arrow). b Image showing finger-like cyst wall protrusions (arrows). c Banana-shaped bradyzoites. d, e TEM micrographs. d Portion of the cyst wall showing finger-like protrusions (arrows). The image indicates atypical appearance of the cyst wall, whereas protrusions were squeezed by host myofibrils and consequently bent sideways. e Higher magnification of cyst wall protrusions at the proximal end; note the presence of numerous microfilaments and microgranules (arrowheads). Ground substance (g)
Initially, the amplification of cox1 fragment of S. entzerothi was unsuccessful using previously published reverse primers suitable for Sarcocystis spp. from cervids (Gjerde 2013, 2014b); therefore, a new reverse primer SR12H was designed. Thus, cox1 sequences of S. entzerothi were amplified with SF1/SR12H primer pair. 18S rDNA (1821 bp long) and cox1 (907 bp long) sequences of S. entzerothi were deposited in GenBank with accession numbers KX643334–KX643337 and KX643340–KX643343, respectively. Four S. entzerothi isolates examined were identical in the cox1 and composed two 18S rDNA haplotypes differing in two substitutions. Comparing 18S rDNA sequences of S. entzerothi with Sarcocystis sp. from the Lithuanian red deer (JN256125) and the Lithuanian roe deer (JN256133–7), 99.7–100 % sequence identity indicating the same species was determined. It should be pointed out that S. entzerothi clearly genetically differed from S. silva, indicating 95.6–96.1 % and 85.6–86.9 % sequence identity in 18S rDNA and cox1, respectively. 18S rDNA sequences of S. entzerothi shared the highest identity values, similar to those calculated when S. entzerothi and S. silva were compared, with sequences of Sarcocystis truncata (95.7–96.2 %), Sarcocystis elongata (95.5–95.9 %), and Sarcocystis tarandi (95.5–95.9 %), whereas, S. entzerothi had the highest sequence identity within cox1 to Sarcocystis bovifelis (86.6–86.9 %), S. truncata (86.1–86.6 %), Sarcocystis bovini (86.1–86.4 %), and S. silva.
Taxonomic summary of S. entzerothi n. sp.
Type intermediate host The roe deer (Capreolus capreolus).
Definitive host Unknown.
Locality Central Lithuania.
Sarcocyst morphology
By LM, sarcocysts were spindle-shaped with pointed tips, 950–1900 × 70–150 μm in size, having 5–6 μm long finger-like cyst wall protrusions. Banana-shaped bradyzoites measured 10.7 × 3.3 μm. Under TEM, protrusions were tightly packed, with 0.1 μm distances between them, up to 4.5 μm long, 0.9–1.2 μm wide, filled with microfilaments and microgranules. The parasitophorous vacuolar membrane had many minute invaginations. The ground substance layer measured about 0.4–0.5 μm. The cyst wall was type 10a-like.
Specimens deposited
TEM material deposited at the National Centre of Pathology, Vilnius, Lithuania. Sequences deposited in NCBI GenBank with accession numbers KX643334–KX643337 (18S rDNA) and KX643340–KX643343 (cox1).
Etymology
Species was named after Prof. Rolf Entzeroth who contributed significantly to knowledge of Sarcocystis in cervids.
Morphological and molecular characteristics of S. silva
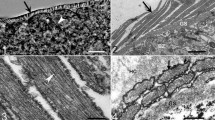
By LM, cysts of S. silva were cigar-shaped with blunted tips, 1200 × 154.6 μm (1000–1500 × 130–184; n = 3) in size (Fig. 2a). Finger-like cyst wall protrusions were 7–8-μm long (Fig. 2b). Bradyzoites were banana-shaped, 11.06 × 3.7 μm (10.08–12.25 × 3.2–4.91; n = 9) in size (Fig. 2c). Under TEM, protrusions of S. silva were up to 7 μm long, 0.9–1.5 μm wide, tightly packed, lying about 0.1 μm apart, and tapered at the distal ends (Fig. 2d). The cyst wall protrusions had numerous microgranules and microfilaments. The parasitophorous vacuolar membrane had many minute invaginations (Fig. 2e). The ground substance layer was thin, 0.4–0.7 μm. The ground substance and base of the protrusions seemed darker, probably due to a huge number of microgranules. The cyst wall was type 10a-like (Dubey et al. 2015).
Morphology of sarcocysts of Sarcocystis silva from the diaphragm muscles of the roe deer. a–c Light micrographs. Fresh preparations. a Fragment of the cyst. Note blunted tip of the cyst (arrow). b Image showing finger-like cyst wall protrusions (arrows). c Banana-shaped bradyzoites. d, e TEM micrographs. d Fragment of the cyst wall. Arrows pointed at the finger-like protrusions. e Higher magnification of the base of cyst wall protrusions; note invaginations of the parasitophorous vacuolar membrane (arrows), microfilaments, and microgranules (arrowheads). Ground substance (g)
Two newly obtained 1804-bp-long 18S rDNA sequences of S. silva (KX643338–KX643339) were identical with the previously published sequence of S. silva from the Lithuanian roe deer (JN256132), whereas sequence identity of two new 1053-bp-long cox1 haplotypes of S. silva (KX643344–KX643345) accounted for 99.4 %. Comparing sequences of S. silva from the Lithuanian roe deer with other available S. silva sequences from the Norwegian and Polish roe deer and the Norwegian moose, 99.7–100 % and 98.4–99.5 % sequence identity for 18S rDNA and cox1 was determined.
Phylogeny
Phylogenetic relationships of some Sarcocystis species, S. truncata, Sarcocystis rangiferi, S. tarandi, S. elongata, S. bovifelis, S. bovini, Sarcocystis sinensis, Sarcocystis buffalonis, and Sarcocystis hirsuta were not fully defined on the basis of 18S rDNA sequences (Fig. 3). In contrast, all Sarcocystis species analyzed formed monophyletic groups in the phylogram of cox1 sequences (Fig. 4). In the 18S rDNA phylogenetic tree, S. entzerothi sequences interleaved with sequences, accession nos. JN256125, JN256133–7, and together formed one clade. Thus, the 18S rDNA phylogenetic analysis was in congruence with the molecular data demonstrating the isolates of S. entzerothi and Sarcocystis sp. from the roe deer and the red deer as the representatives of the same species. Based on 18S rDNA and cox1 sequences, S. entzerothi was most closely related to Sarcocystis spp. using the representatives of the families Bovidae and Cervidae as an intermediate host. In the phylogenetic analyses, S. entzerothi did not group with Sarcocystis species employing canids as definitive hosts. Based on the 18S rDNA sequence analysis, S. entzerothi was placed with S. tarandi, S. elongata, S. silva, S. truncata, and S. rangiferi. However, S. entzerothi phylogenetic placement in the 18S rDNA sequences analysis is equivocal due to a poor posterior probability support value (58), whereas S. entzerothi was a sister taxon to S. sinensis, S. bovini, and S. bovifelis in the cox1 phylogenetic tree. Definitive hosts of S. truncata, S. rangiferi, S. silva, S. tarandi, S. elongata, and S. bovini are unknown, while S. sinensis and S. bovifelis are transmittable via felids (Dahlgren et al. 2008; Gjerde 2012, 2014b, 2016a, 2016b; Gjerde and Hilali 2016). Therefore, S. entzerothi was more closely related to species transmitted by felids than canids.
The phylogenetic tree of selected Sarcocystis species based on 18S rDNA sequences. The tree was constructed using the Bayesian methods, scaled according to the branch length, and rooted on S. cruzi. The final alignment contained 186 sequences and 1777 aligned nucleotide positions. The HKY + G evolutionary model was set for the phylogenetic analysis. The figures next to branches show the posterior probability support values. GenBank accession numbers or the number of sequences for the corresponding taxon is given behind the Sarcocystis species name
The phylogenetic tree for selected Sarcocystis species based on cox1 sequences. The tree was constructed using the Bayesian methods, scaled according to the branch length, and rooted on S. cruzi. The final alignment contained 308 sequences and 766 aligned nucleotide positions. The K80 + G evolutionary model was set for the phylogenetic analysis. GenBank accession numbers or number of sequences for the corresponding taxon is given behind the Sarcocystis species name
Discussion
Numerous Sarcocystis species have been described in cervids (Gjerde 2014a, 2014b; Calero-Bernal et al. 2015; Prakas et al. 2016). Morphological criteria are mostly insufficient to differentiate Sarcocystis species found in cervids, since morphologically similar Sarcocystis spp. cysts are found in the same or closely related hosts (Dubey 1980; Colwell and Mahrt 1981; Speer and Dubey 1982; Dubey and Lozier 1983; Dubey et al. 1983; Dubey and Speer 1985, 1986; Gjerde 1986; Atkinson et al. 1993; Sedlaczek and Wesemeier 1995; Wesemeier and Sedlaczek 1995a, 1995b). DNA analysis has been shown to be a powerful tool for resolving taxonomic uncertainties of Sarcocystis spp. infecting the representatives of the family Cervidae (Dahlgren and Gjerde 2008, 2009, 2010; Gjerde 2012, 2014a, 2014b; Calero-Bernal et al. 2015; Prakas et al. 2016; Reissig et al. 2016). Therefore, it is necessary to combine morphological and molecular data in order to identify cryptic Sarcocystis species or avoid species oversplitting. Historically, 18S rDNA was the first molecular locus for the genetic characterization of Sarcocystis species with ruminants as intermediate hosts (Tenter et al. 1992; Jeffries et al. 1997; Holmdahl et al. 1999; Morrison et al. 2004; Dahlgren and Gjerde 2007). However, 18S rDNA is an inappropriate choice discriminating closely related Sarcocystis species employing cervids as intermediate hosts. Specifically, S. tarandi and S. elongata were not clearly separated by 18S rDNA as S. rangiferi from S. truncata and Sarcocystis hjorti from Sarcocystis pilosa, and these species were distinguished on the basis of the cox1 sequence analysis (Gjerde 2014b; Prakas et al. 2016).
In the present study, we described S. entzerothi n. sp. based on LM, TEM, 18S rDNA, and cox1 analysis. The newly characterized species is morphologically similar to S. silva, previously found in the roe deer (Gjerde 2012) and the moose (Dahlgren and Gjerde 2008). Gjerde (2012) described S. silva cysts in the roe deer as having tightly packed, upright, finger-like protrusions about 8 μm long and 1.5 μm wide. Having examined six roe deer hunted in Lithuania, we identified S. entzerothi cysts in four of them and S. silva in two animals. Cysts of S. silva were morphologically consistent with those of S. silva from the Norwegian roe deer (Gjerde 2012). In the present work, two species examined shared 95.6–96.1 % and 85.6–86.9 % sequence identity within 18S rDNA and cox1, respectively. Therefore, S. entzerothi could be clearly differentiated from S. silva using DNA analysis. Sarcocysts of both species discussed were similar in size, having densely packed finger-like protrusions on their surface (Figs. 1 and 2). However, S. entzerothi and S. silva were distinguishable by the shape of the cysts and by the length of villar protrusions. Cysts of S. entzerothi were spindle-shaped with pointed tips, whereas cysts of S. silva were cigar-shaped with blunt ends. It should be pointed out that the tips of the cysts of S. entzerothi sometimes seemed rounded in muscle fibers. Cyst wall protrusions of S. entzerothi were 5–6 μm long, while those of S. silva measured 7–8 μm. Nevertheless, the lengths of the protrusions may depend on the cyst state during fixation and on the age of the cyst. In conclusion, morphological separation between S. entzerothi and S. silva sometimes might be difficult; therefore DNA analysis is necessary for a definitive discrimination of these two species.
Cysts having finger-like cyst wall protrusions were reported in the roe deer in several publications before the descriptions of S. silva and S. entzerothi. The cysts of this type were morphologically characterized by LM in histological sections or fresh preparations and using TEM. Bergmann and Kinder (1976) were the first to notice the thick-walled cysts with finger-like protrusions in the roe deer using LM and TEM analyses. Subsequently, Schramlová and Blažek (1978) studied the thick-walled cyst under TEM and characterized them as 4.49–7.49 μm in thickness, forming palisade-like protrusions. Erber et al. (1978) examined fresh muscle preparations of the roe deer under LM and distinguished three cyst types. Type 3 cysts had rigid 5–6 μm long and about 0.5-μm-thick finger-like protrusions. Afterwards, Entzeroth (1982) examined most exhaustively the wall ultrastructure of the cysts found in the roe deer and described six cyst wall types. The first three types were distinguished by finger-like protrusions varying in length and width. The author assumed that cyst walls of types 1–3 might belong to the same species at different stages of growth. Sedlaczek and Wesemeier (1995) found cysts with finger-like protrusions in the esophagus, heart, diaphragm, and skeletal musculature (thigh, loin, thorax, ribs) of 42 roe deer out of the 66 examined. Under LM, cysts had 7.8–8.6 μm long and 1.5–1.6 μm wide finger-like protrusions, whereas under TEM, the protrusions were 6.3–7.2 μm long and 1.2–1.7 μm wide. Kutkienė (2001) found cysts with tightly packed finger-like protrusions measuring up to 8.3 μm in length. The cysts showing a similar villar protrusions were also detected by Spickschen and Pohlmeyer (2002) and López et al. (2003). According to the contradictory morphological descriptions of the cysts with finger-like protrusions in the muscles of the roe deer, we could not state accurately, whether S. entzerothi, S. silva, or both of them were found in the above-mentioned. studies as no DNA analysis has been performed in those studies.
Earlier, one cyst with finger-like protrusions isolated from the Lithuanian red deer was characterized in 18S rDNA and the obtained sequence was deposited in GenBank with accession number JN256125 (Prakas 2011). Based on the results of the present study, this sequence belongs to S. entzerothi. Therefore, the case might be that S. entzerothi is not strictly host-specific and could employ both roe deer and red deer as intermediate hosts. However, molecularly based identification of S. entzerothi in one red deer is insufficient evidence suggesting that this Sarcocystis species uses red deer as another intermediate host. Furthermore, S. entzerothi cysts were not identified in numerous red deer examined in comprehensive studies in Norway (Dahlgren and Gjerde 2010; Gjerde 2014b). Therefore, it is necessary to carry out thorough investigations of red deer for the presence of S. entzerothi.
Based on 18S rDNA sequences, phylogenetic placement of S. entzerothi was unreliable (Fig. 3). By contrast, grouping of S. entzerothi together with S. sinensis, S. bovini, and S. bovifelis in the cox1 phylogenetic tree was well-supported (Fig. 4). Two of these species, S. bovifelis and S. sinensis, use representatives of the family Bovidae as intermediate hosts and felids as definitive hosts (Gjerde 2016b; Gjerde and Hilali 2016). Thus, phylogenetic results suggest that definitive hosts of S. entzerothi might be felids. However, in previous experiments, domestic cats, wildcats (Felis silvestris), jungle cats (Felis chaus), bobcats (Lynx rufus), and the tiger (Panthera tigris) did not shed Sarcocystis sporocysts after ingesting roe deer meat infected with Sarcocystis spp. cysts (Entzeroth et al. 1978; Erber et al. 1978; Entzeroth 1981). Consequently, either felids do not act as intermediate hosts of S. entzerothi or the muscles of the roe deer used in these experiments did not contain S. entzerothi cysts.
In the present study, five Sarcocystis species, S. entzerothi, S. silva, S. gracilis, S. capreolicanis, and S. oviformis, in the diaphragm of the roe deer were found. According to the previous morphological investigations, the sixth Sarcocystis species might exist in this host. As observed under LM, the cysts of Sarcocystis sp. appeared to have a thin smooth wall (Kutkienė 2001), whereas the TEM analysis showed cysts having a highly folded primary cyst wall that formed hair-like protrusions running in parallel with the cyst surface (Schramlová and Blažek 1978; Entzeroth 1982; Santini et al. 1997). The cyst wall structure similar to that of Sarcocystis sp. from the roe deer was established for Sarcocystis cervicanis (Hernández-Rodríguez et al. 1981) and Sarcocystis sp. (Entzeroth et al. 1983) from the red deer, Sarcocystis wapiti from the wapiti (Cervus canadensis) (Speer and Dubey 1982), Sarcocystis grueneri from the reindeer (Rangifer tarandus) (Gjerde 1985), Sarcocystis sp. from the moose (Colwell and Mahrt 1981), and Sarcocystis sp. from the fallow deer (Dama dama) (Entzeroth et al. 1985; Poli et al. 1988). Comparative molecular studies are needed to determine the actual number of Sarcocystis species representing a cyst of this type found in different cervids.
References
Atkinson CT, Wright SD, Telford SR, Mclaughlin GS, Forrester DJ, Roelke ME, Mccown JW (1993) Morphology, prevalence, and distribution of Sarcocystis spp. in white-tailed deer (Odocoileus virginianus) from Florida. J Wildl Dis 29:73–84. doi:10.7589/0090-3558-29.1.73
Bergmann V, Kinder E (1976) Elektronenmikroskopische Untersuchungen zur Wandstruktur von Sarkozysten in der Skelettmuskulatur von Wildschwein und Reh. Monatsh Vet Med 31:785–788
Blažek K, Schramlová J, Ippen R (1978) Dog as definitive host of sarcosporidia infecting roe deer. Folia Parasitol 25:95–96
Calero-Bernal R, Verma SK, Cerqueira-Cézar CK, Schafer LM, Van Wilpe E, Dubey JP (2015) Sarcocystis mehlhorni, n. sp. (Apicomplexa: Sarcocystidae) from the black-tailed deer (Odocoileus hemionus columbianus). Parasitol Res 114:4397–4403. doi:10.1007/s00436-015-4679-5
Colwell DD, Mahrt JL (1981) Ultrastructure of the cyst wall and merozoites of Sarcocystis from moose (Alces alces) in Alberta, Canada. Z Parasitenkd 65:317–329. doi:10.1007/bf00926727
Dahlgren SS, Gjerde B (2007) Genetic characterisation of six Sarcocystis species from reindeer (Rangifer tarandus tarandus) in Norway based on the small subunit rRNA gene. Vet Parasitol 146:204–213. doi:10.1016/j.vetpar.2007.02.023
Dahlgren SS, Gjerde B (2008) Sarcocystis in moose (Alces alces): molecular identification and phylogeny of six Sarcocystis species in moose, and a morphological description of three new species. Parasitol Res 103:93–110. doi:10.1007/s00436-008-0936-1
Dahlgren SS, Gjerde B (2009) Sarcocystis in Norwegian roe deer (Capreolus capreolus): molecular and morphological identification of Sarcocystis oviformis n. sp. and Sarcocystis gracilis and their phylogenetic relationship with other Sarcocystis species. Parasitol Res 104:993–1003. doi:10.1007/s00436-008-1281-0
Dahlgren SS, Gjerde B (2010) Molecular characterization of five Sarcocystis species in red deer (Cervus elaphus), including Sarcocystis hjorti n. sp., reveals that these species are not intermediate host specific. Parasitology 137:815–840. doi:10.1017/S0031182009991569
Dahlgren SS, Gouveia-Oliveira R, Gjerde B (2008) Phylogenetic relationships between Sarcocystis species from reindeer and other Sarcocystidae deduced from ssu rRNA gene sequences. Vet Parasitol 151:27–35. doi:10.1016/j.vetpar.2007.09.029
Dubey JP (1980) Sarcocystis species in moose (Alces alces), bison (Bison bison), and pronghorn (Antilocapra americana) in Montana. Am J Vet Res 41:2063–2065
Dubey JP, Lozier SC (1983) Sarcocystis infection in the white-tailed deer (Odocoileus virginianus) in Montana: intensity and description of Sarcocystis odoi n. sp. Am J Vet Res 44:1738–1743
Dubey JP, Speer CA (1985) Prevalence and ultrastructure of three types of Sarcocystis in mule deer, Odocoileus hemionus (Rafinesque), in Montana. J Wildl Dis 21:219–228. doi:10.7589/0090-3558-21.3.219
Dubey JP, Speer CA (1986) Sarcocystis infections in mule deer (Odocoileus hemionus) in Montana and the description of three new species. Am J Vet Res 47:1052–1055
Dubey JP, Jolley WR, Thorne ET (1983) Sarcocystis sybillensis sp. nov. from the North American elk (Cervus elaphus). Can J Zool 61:737–742. doi:10.1139/z83-098
Dubey JP, Calero-Bernal R, Rosenthal BM, Speer CA, Fayer R (2015) Sarcocystosis of animals and humans, 2nd edn. CRC Press, Boca Raton
Edgar RC (2004) MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32:1792–1797. doi:10.1093/nar/gkh340
Entzeroth R (1981) Untersuchungen an Sarkosporidien (Mieschersche Schläuche) des einheimischen Rehwildes (Capreolus capreolus L). Z Jagdwiss 27:247–257. doi:10.1007/BF02243674
Entzeroth R (1982) A comparative light and electron microscope study of the cysts of Sarcocystis species of roe deer (Capreolus capreolus). Z Parasitenkd 66:281–292. doi:10.1007/bf00925345
Entzeroth R (1985) Light-, scanning-, and transmission electron microscope study of the cyst wall of Sarcocystis gracilis Rátz, 1909 (Sporozoa, Coccidia) from the roe deer (Capreolus capreolus L.). Arch Protistenkd 129:183–186. doi:10.1016/s0003-9365(85)80020-8
Entzeroth R, Scholtyseck E, Greuel E (1978) The roe deer intermediate host of different Coccidia. Naturwissenschaften 65:395. doi:10.1007/bf00439714
Entzeroth R, Nemeséri L, Scholtyseck E (1983) Prevalence and ultrastructure of Sarcocystis sp. from the red deer (Cervus elaphus L.) in Hungary. Parasitol Hung 16:47–52
Entzeroth R, Chobotar B, Scholtyseck E, Neméseri L (1985) Light and electron microscope study of Sarcocystis sp. from the fallow deer (Cervus dama). Z Parasitenkd 71:33–39. doi:10.1007/bf00932916
Erber M, Boch J, Barth D (1978) Drei Sarkosporidienarten des Rehwildes. Berl Münch Tierärztl Wsch 91:482–486
Gjerde B (1985) Ultrastructure of the cysts of Sarcocystis grueneri from cardiac muscle of reindeer (Rangifer tarandus tarandus). Z Parasitenkd 71:189–198. doi:10.1007/BF00926269
Gjerde B (1986) Scanning electron microscopy of the sarcocysts of six species of Sarcocystis from reindeer (Rangifer tarandus tarandus). Acta Pathol Microbiol Immunol Scand [B] 94:309–317. doi:10.1111/j.1699-0463.1986.tb03058.x
Gjerde B (2012) Morphological and molecular characterization and phylogenetic placement of Sarcocystis capreolicanis and Sarcocystis silva n. sp. from roe deer (Capreolus capreolus) in Norway. Parasitol Res 110:1225–1237. doi:10.1007/s00436-011-2619-6
Gjerde B (2013) Phylogenetic relationships among Sarcocystis species in cervids, cattle and sheep inferred from the mitochondrial cytochrome c oxidase subunit I gene. Int J Parasitol 43:579–591. doi:10.1016/j.ijpara.2013.02.004
Gjerde B (2014a) Morphological and molecular characteristics of four Sarcocystis spp. in Canadian moose (Alces alces), including Sarcocystis taeniata n. sp. Parasitol Res 113:1591–1604. doi:10.1007/s00436-014-3806-z
Gjerde B (2014b) Sarcocystis species in red deer revisited: with a redescription of two known species as Sarcocystis elongata n. sp. and Sarcocystis truncata n. sp. based on mitochondrial cox1 sequences. Parasitology 141:441–452. doi:10.1017/S0031182013001819
Gjerde B (2016a) Molecular characterisation of Sarcocystis bovifelis, Sarcocystis bovini n. sp., Sarcocystis hirsuta and Sarcocystis cruzi from cattle (Bos taurus) and Sarcocystis sinensis from water buffaloes (Bubalus bubalis). Parasitol Res 115:1473–1492. doi:10.1007/s00436-015-4881-5
Gjerde B (2016b) The resurrection of a species: Sarcocystis bovifelis Heydorn et al., 1975 is distinct from the current Sarcocystis hirsuta in cattle and morphologically indistinguishable from Sarcocystis sinensis in water buffaloes. Parasitol Res 115:1–21. doi:10.1007/s00436-015-4785-4
Gjerde B, Hilali M (2016) Domestic cats (Felis catus) are definitive hosts for Sarcocystis sinensis from water buffaloes (Bubalus bubalis). J Vet Med Sci. doi:10.1292/jvms.16-0127
Hernández-Rodríguez S, Martínez-Gómez F, Navarrete I, Acosta-García I (1981) Estudio al microscopio optico y electronico del quiste de Sarcocystis cervicanis. Rev Ibérica Parasitol 41:351–361
Holmdahl OJ, Morrison DA, Ellis JT, Huong LT (1999) Evolution of ruminant Sarcocystis (Sporozoa) parasites based on small subunit rDNA sequences. Mol Phylogenet Evol 11:27–37. doi:10.1006/mpev.1998.0556
Jeffries AC, Schnitzler B, Heydorn AO, Johnson AM, Tenter AM (1997) Identification of synapomorphic characters in the genus Sarcocystis based on 18S rDNA sequence comparison. J Eukaryot Microbiol 44:388–392. doi:10.1111/j.1550-7408.1997.tb05713.x
Kolenda R, Ugorski M, Bednarski M (2014) Molecular characterization of Sarcocystis species from Polish roe deer based on ssu rRNA and cox1 sequence analysis. Parasitol Res 113:3029–3039. doi:10.1007/s00436-014-3966-x
Kumar S, Stecher G, Tamura K (2016) MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol 33:1870–1874. doi:10.1093/molbev/msw054
Kutkienė L (2001) The species composition of European roe deer (Capreolus capreolus) Sarcocystis in Lithuania. Acta Zool Lituanica 11:97–101. doi:10.1080/13921657.2001.10512363
Kutkienė L, Prakas P, Sruoga A, Butkauskas D (2010) The mallard duck (Anas platyrhynchos) as intermediate host for Sarcocystis wobeseri sp. nov. from the barnacle goose (Branta leucopsis). Parasitol Res 107:879–888. doi:10.1007/s00436-010-1945-4
López C, Panadero R, Bravo A, Paz A, Sánchez-Andrade R, Díez- Banos P, Morrondo P (2003) Sarcocystis spp. infection in roe deer (Capreolus capreolus) from the north-west of Spain. Z Jagdwiss 49:211–218. doi:10.1007/bf02189739
Milne I, Wright F, Rowe G, Marshall DF, Husmeier D, McGuire G (2004) TOPALi: software for automatic identification of recombinant sequences within DNA multiple alignments. Bioinformatics 20:1806–1807. doi:10.1093/bioinformatics/bth155
Morrison DA, Bornstein S, Thebo P, Wernery U, Kinne J, Mattsson JG (2004) The current status of the small subunit rRNA phylogeny of the coccidian (Sporozoa). Int J Parasitol 34:501–514. doi:10.1016/j.ijpara.2003.11.006
Odening K, Stolte M, Walter G, Bockhardt I (1994) The European badger (Carnivora: Mustelidae) as intermediate host of further three Sarcocystis species (Sporozoa). Parasite 1:23–30. doi:10.1051/parasite/1994011023
Poli A, Mancianti F, Marconcini A, Nigro M, Colagreco R (1988) Prevalence, ultrastructure of the cyst wall and infectivity for the dog and cat of Sarcocystis sp. from fallow deer (Cervus dama). J Wildl Dis 24:97–104. doi:10.7589/0090-3558-24.1.97
Prakas P (2011) Diversity and ecology of Sarcocystis in Lithuanian game fauna. PhD thesis, Vilnius University, Vilnius, Lithuania
Prakas P, Butkauskas D, Rudaitytė E, Kutkienė L, Sruoga A, Pūraitė I (2016) Morphological and molecular characterization of Sarcocystis taeniata and Sarcocystis pilosa n. sp. from the sika deer (Cervus nippon) in Lithuania. Parasitol Res 115:3021–3032. doi:10.1007/s00436-016-5057-7
Rátz S (1909) Die Sarcosporidien und ihre in Ungarn vorkommenden Arten. Allattani Közlemények 8:1–37
Reissig EC, Moré G, Massone A, Uzal FA (2016) Sarcocystosis in wild red deer (Cervus elaphus) in Patagonia. Argentina Parasitol Res 115:1773–1778. doi:10.1007/s00436-016-4915-7
Santini S, Mancianti F, Nigro M, Poli A (1997) Ultrastructure of the cyst wall of Sarcocystis sp. in roe deer. J Wildl Dis 33:853–859. doi:10.7589/0090-3558-33.4.853
Schramlová J, Blažek K (1978) Ultrastruktur der Cystenwand der Sarkosporidien des Rehes (Capreolus capreolus L.). Z Parasitenkd 55:43–48. doi:10.1007/bf00383473
Sedlaczek J, Wesemeier HH (1995) On the diagnostics and nomenclature of Sarcocystis species (Sporozoa) in roe deer (Capreolus capreolus). Appl Parasitol 36:73–82
Speer CA, Dubey JP (1982) Sarcocystis wapiti sp. nov. from the North American wapiti (Cervus elaphus). Can J Zool 60:881–888. doi:10.1139/z82-120
Spickschen C, Pohlmeyer K (2002) Untersuchung zum Vorkommen von Sarkosporidien bei Reh-, Rot- und Muffelwild in zwei unterschiedlichen Naturräumen des Bundeslandes Niedersachsen. Z Jagdwiss 48:35–48. doi:10.1007/bf02285355
Tenter AM, Baverstock PR, Johnson AM (1992) Phylogenetic relationships of Sarcocystis species from sheep, goats, cattle and mice based on ribosomal RNA sequences. Int J Parasitol 22:503–513. doi:10.1016/0020-7519(92)90151-a
Wesemeier HH, Sedlaczek J (1995a) One known Sarcocystis species and one found for the first time in fallow deer (Dama dama). Appl Parasitol 36:299–302
Wesemeier HH, Sedlaczek J (1995b) One known Sarcocystis species and two found for the first time in red deer and wapiti (Cervus elaphus) in Europe. Appl Parasitol 36:245–251
Acknowledgments
This research was supported by the Open Access to research infrastructure of the Nature Research Centre under Lithuanian open access network initiative. The authors are grateful to Ms. S. Amšiejienė from the National Centre of Pathology (Vilnius, Lithuania) for her help in carrying out electron microscopy investigations.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Prakas, P., Rudaitytė, E., Butkauskas, D. et al. Sarcocystis entzerothi n. sp. from the European roe deer (Capreolus capreolus). Parasitol Res 116, 271–279 (2017). https://doi.org/10.1007/s00436-016-5288-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00436-016-5288-7