Abstract
Infection with Sarcocystis is common in many species of wild cervids but none is reported from the black-tailed deer (Odocoileus hemionus columbianus). Here, we report Sarcocystis infection in two black-tailed deer from northwest USA for the first time. Sarcocysts were microscopic, up to 556 μm long and mature. The sarcocyst wall was up to 1.39 μm thick and had rectangular 1.17-μm-long villar protrusions, type 17, with thin (230 nm) electron dense ground substance layer. Molecular characterization and phylogenetic analysis indicated that Sarcocystis in the black-tailed deer is related to structurally distinct Sarcocystis species in cervids. A new name, Sarcocystis mehlhorni, is proposed for the Sarcocystis species in black-tailed deer.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Infection with Sarcocystis is common in many species of wild cervids, but none is reported from the black-tailed deer (Odocoileus hemionus columbianus) (Dubey et al. 2015a). Here, we report Sarcocystis infection in two black-tailed deer from northwest USA for the first time.
Material and methods
Tongue and myocardium were collected from two (#14183, #15030) adult black-tailed deer (O. hemionus columbianus) from Pierce County, WA, USA, on 11 December 2014 and 19 April 2015. Animals were lethally removed by US Department of Agriculture, Animal and Plant Health Inspection Service personnel during an authorized direct control project. Samples were immediately collected. Refrigerated tongues and hearts were shipped (overnight delivery) to the Animal Parasitic Diseases Laboratory, USDA, Beltsville, Maryland, the following day, for testing for protozoal infections.
Microscopical examination
A portion of tissues was fixed in buffered formalin and processed for histology using hematoxylin and eosin (HE) stain of 5-μm-thick sections. The number of cysts per section (2 × 0.7 cm) was recorded. Fresh muscles were examined for sarcocyst presence by squeezing; individual cysts were excised and kept frozen until molecular assays. Five cysts detected in the HE sections of deer #14183 were excised and processed for transmission electron microscopy (TEM) as reported by Dubey et al. (2015b).
Molecular analyses
Thirteen morphologically similar individual cysts were excised from tissues in deer #15030 and subjected to DNA isolation using DNeasy Blood and Tissue Kit (Qiagen Inc., Valencia, CA, USA) according to manufacturer’s instructions. DNA quantification and quality were determined by Thermo Scientific NanoDrop Lite Spectrophotometer (Thermo Scientific, Waltham, MA, USA).
The DNA was characterized by PCR amplification and sequencing of two regions of the nuclear ribosomal DNA unit, 18S rRNA and 28S rRNA, and the mitochondrial cytochrome c oxidase subunit 1 (cox1) locus. The complete regions of 18S rRNA and 28S rRNA were amplified using overlapping fragments and primer pairs; ERIB1/S2r, S5f/S4r, S3f/Primer Bsarc, and KL1/LS2R, LS1F/KL3, respectively, as described previously (Gjerde and Josefsen 2015). The partial sequence of cox1 locus was also amplified using primer pair SF1/SR5 (Gjerde 2013, 2014; Gjerde and Josefsen 2015). The amplified PCR products were run on 2.5 % (w/v) agarose gel.
The single PCR amplicons of 18S rRNA, 28S rRNA, and cox1 were excised from the gel and purified using QIAquick Gel Extraction (Qiagen, Inc., Valencia, CA, USA) according to the manufacturer’s recommendations. The purified PCR products were sent to Macrogen Corporation (Rockville, MD, USA) for direct sequencing using the same primer pair used in PCR amplification to obtain both strands reads. The resulting sequences were edited manually if necessary, and analyzed using the software Geneious version 8.0.4 (Biomatters Ltd. Auckland, New Zealand). The sequences obtained were aligned against each other and the published sequences of various Sarcocystis spp. to detect intraspecies and interspecies variation on these DNA regions, respectively.
Phylogenetic trees were estimated to show relationship among reference Sarcocystis species by the neighbor-joining algorithm applied to Tamura-Nei genetic distances, as implemented by Geneious version 8.0.4. Trees were tested by selecting bootstrap method with a value of 1000 bootstrap replicates. Phylogenetic trees based on 18S rRNA and cox1 sequences were constructed using sequences obtained from the black-tailed deer sarcocysts and previously published sequences of various species of Sarcocystis in NCBI GenBank.
Results
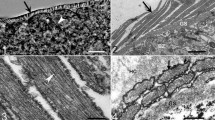
In wet mounts, sarcocysts were fusiform and up to 600 μm (n = 13); released bradyzoites from broken cysts were banana-shaped and 12 μm long (n = 10). Microscopic examination of HE-stained tissue sections revealed 9 cysts in heart and 74 in tongue of deer #14183, and 19 sarcocysts (3 in heart and 16 in tongue) in deer #15030. Only 2 sarcocysts were cut longitudinally and they measured 330 and 556 μm long. In cross-sections, sarcocysts were 36.0–163 μm wide. Under lower magnification, the sarcocyst wall appeared smooth, but at ×1000 illumination, polygonal or elongated villar protrusions were evident (Fig. 1). Bradyzoites were separated in groups by septa.
Light micrographs of Sarcocystis mehlhorni, n. sp. from black-tailed deer. a, b Longitudinally cut cysts. HE stain. c, d Transversally cut cysts. HE stain. a Note the septa and the thin wall. b Appearance of vp resembling keys of a piano. c Thin-walled sarcocyst. d Vp with rectangular appearance and remarkable dark ground substance
One mild focus of myocarditis was observed in deer #15030.
By TEM, the parasitophorous vacuolar membrane was folded into villar protrusions (vp) and lined by a 15–20-nm thick electron dense layer (Fig. 2a, b). Total thickness of primary cyst wall ranged from 29 to 44 nm. The vp were rectangular, type 17 (Dubey et al. 2015a), and measured 1.17 μm (1.50–0.92) long and 1.42 μm (1.98–1.20) wide (n = 66). The ground substance layer (gs) was remarkably thin (184–278 nm; studied in 4 cysts) and electron dense, that continues as septa (88.0–312 nm wide). Up to 6 disc-shaped plaques were present in the vp (Fig. 2c). Structurally, plaques were 200 nm wide, with at least 3 different layers, numerous microtubules connected them to the core of the vp, and several microfilaments can be seen arising from the parasitophorous vacuolar membrane (Fig. 2d). The total thickness of the sarcocyst wall including the vp and ground substance was 1.39 μm (1.70–1.16) thick. Bradyzoites were 10.4 × 2.1 μm (8.3–11.5 × 1.6–2.7; n = 12) in size and with a prominent mitochondrion, nucleus (nu), numerous micronemes (mn), dense granules (dg), amylopectin granules (am) (Fig. 3a), two rhoptries (rh1, rh2; Fig. 3b), and a conoid (co) with 22 subpellicular tubules and 7 internal tubules (Fig. 3c). Metrocytes, seen in the periphery, were 5.6 μm (5.0–6.0; n = 4) in size but are not described in detail because they were not well preserved.
TEM micrographs of Sarcocystis mehlhorni, n. sp. from black-tailed deer. a Sarcocyst with polygonal-shaped villar protrusions (vp) on the wall and enclosed bradyzoites (br). b Details of the cyst wall; note rectangular vp with disc-shaped plaques (ds), vesicles (vs), and thin electron dense ground substance (gs). c Cross/oblique section of vp showing several ds (arrows) from cyst #4. d Higher magnification of two ds; note the presence of microtubules (mt), three visible layers (arrowheads), and microfilaments (arrows) that arise from the parasitophorous vacuolar membrane
TEM micrographs of Sarcocystis mehlhorni, n. sp. from black-tailed deer. a Longitudinally cut bradyzoite, micronemes (mn), nucleus (nu), amylopectin (am), and dense granules (dg). b Conoidal part of a bradyzoite showing two rhoptries (rh1, rh2), conoid (co), and mn. c Detail of the co of a bradyzoite; note 7 internal microtubules (it) and 22 subpellicular microtubules (sm)
The 18S rRNA (in three fragments), 28S rRNA (in two fragments), and cox1 loci were amplified by PCR using DNA obtained from the individual sarcocysts of black-tailed deer. DNA sequencing of PCR amplicons resulted the unambiguous sequences of two nuclear DNA regions: 18S rRNA (1819 bp), 28S rRNA (1598 bp), and the mitochondrial DNA locus, cox1 (1002 bp, 1022 bp). These sequences were submitted to NCBI GenBank with accession numbers KT378042 (18S rRNA), KT378043 (28S rRNA), and KT378044, KT378045 (cox1). Phylogenetic analysis based on both the 18S rRNA and the cox1 sequences obtained from the DNA of individual sarcocysts of Sarcocystis mehlhorni indicated an especially close relationship to another parasite in this genus that employs Canidae as their definitive host, Sarcocystis tarandivulpes (Figs. 4, 5). The 18S rRNA, 28S rRNA, and cox1 sequences shared the highest identity with sequences of S. tarandivulpes (99.0 %, EF056012), Sarcocystis tenella (94.0 %, AF076899), and S. tarandivulpes (92.0 %, KC209718), respectively.
Phylogenetic tree based on 18S rRNA sequences. Input sequences were the 18S rRNA regions of various species retrieved from NCBI GenBank and 1819-bp long sequence obtained from Sarcocystis mehlhorni sarcocysts from black-tailed deer. Accession number of gene sequence was given in parenthesis following the species name. Tree was built by selecting the Tamura-Nei genetic distance model and neighbor-joining tree methods (Geneious version 8.0.4). Tree was tested by selecting bootstrap method with value of 1,000 replicates. Sarcocystis mehlhorni indicated an especially close relationship to other parasites in this genus that employs Canidae as their definitive hosts, S. tarandivulpes
In addition, for 18S rRNA, similarities were found for other Sarcocystis spp. that parasitize Cervidae, to note, Sarcocystis rangi (EF056011) from reindeer (Rangifer tarandus) or Sarcocystis taeniata (KF831278) and Sarcocystis alceslatrans (KF831275) from moose (Alces alces) (Fig. 4). When studying cox1, a genetic relationship was observed with S. taeniata (KF831253), Sarcocystis grueneri (KC209616), and S. alceslatrans (KF831248) (Fig. 5).
Phylogenetic tree based on cox1 sequences. Input sequences were the cox1 regions of various species retrieved from NCBI GenBank and sequences of Sarcocystis mehlhorni sarcocysts from black-tailed deer (1002 and 1022 bp). Accession numbers of gene sequence were given in parenthesis following the species name. Tree was built by selecting the Tamura-Nei genetic distance model and neighbor-joining tree methods (Geneious version 8.0.4). Tree was tested by selecting bootstrap method with value of 1,000 replicates. Sarcocystis mehlhorni clustered together with Sarcocystis species that employs Canidae as its definitive hosts, S. tarandivulpes
Taxonomic summary of S. mehlhorni n. sp. (Figs. 1, 2, 3, 4, and 5)
Diagnosis
Sarcocysts were microscopic, up to 556 μm long and mature. The sarcocyst wall was up to 1.39 μm thick and had rectangular 1.17-μm-long villar protrusions with up to 6 disc-shaped plaques, type 17, with thin (230 nm) electron dense ground substance layer.
Host
Black-tailed deer (O. hemionus columbianus).
Distribution
Washington State (USA), probably other areas in North America.
Definitive host
Unknown, possibly Canidae.
Etymology
Species named after Prof. Heinz Mehlhorn who, with Otto Heydorn and Michel Rommel, contributed immensely to our knowledge on biology and structure of Sarcocystis.
Specimens deposited
Histological sections stained with H and E and Toluidine blue deposited in the United States National Parasite Collection in the Division of Invertebrate Zoology and National Museum of Natural History, Smithsonian Institution, Washington, D.C. (USNM numbers 1283156 to 128158). Sequences were deposited in NCBI GenBank accession number KT378042 (18S rRNA), KT378043 (28S rRNA), and KT378044, KT378045 (cox1).
Discussion
Only one morphological type of sarcocyst was found in the black-tailed deer (BTD) examined and is named S. mehlhorni. Its sarcocysts resembled Sarcocystis odocoileocanis from the white-tailed deer (WTD) from the USA (Table 1). Crum et al. (1981) named S. odocoileocanis for the parasite in WTD from Georgia with canids as the definitive hosts but did not describe its ultrastructure. Entzeroth et al. (1982) first reported the ultrastructure of sarcocysts in WTD from Michigan, USA. The sarcocyst wall was stated to be 15 to 20 μm thick (Entzeroth et al. 1982); however, the authors meant to say 1.5 to 2.0 μm thick (personal communication from B. Chobotar to J. P. Dubey, May 2015). The gs layer was 0.4–1.0 μm thick (Entzeroth et al. 1982); these measurements for the gs in BTD are thinner than in WTD. Otherwise, the descriptions of villar protrusions in WTD and BTD appear similar, including description of S. odocoileocanis from WTD from Florida (Atkinson et al. 1993) and Montana (Dubey and Lozier 1983). One of the prominent feature of the sarcocyst in BTD was the presence of disc-shaped plaques on villar protrusions; these structures were not commented upon but are visible in illustrations provided by Entzeroth et al. (1982), Atkinson et al. (1993), and Dubey and Lozier (1983).
The disc-shaped plaques were first described by Speer and Dubey (1986) in vp of Sarcocystis hemionilatrantis in mule deer from the USA. The ultrastructure of S. odocoileocanis from the BTD broadly resembles the structure of sarcocysts of S. hemionilatrantis in mule deer from the USA and S. tarandivulpes from the reindeer in Scandinavia (Gjerde 1984a, 1985, 1986) (Table 1).
For the morphological-related/similar species in wild ruminants, molecular information is only available for S. tarandivulpes (Dahlgren and Gjerde 2007; Gjerde 2013). When examining the three markers, molecular similarities were present. Further molecular characterization of S. odocoileocanis and S. hemionilatrantis will be of interest. Phylogenetic relationship of Sarcocystis sp. from the black-tailed deer with Canidae-transmitted Sarcocystis species in wild ruminants suggests that the present species is transmitted by these carnivores.
Transmission experiments are necessary to confirm the final identity of Sarcocystis spp. from the black-tailed deer. For example, S. odocoileocanis was infective to cattle and sheep (Crum et al. 1981) but not to goat (Lindsay et al. 1988).
Whether the differences on morphological description of parasites in white-tailed deer and mule deer from the USA and S. tarandivulpes in reindeer from Norway is related to techniques, description cannot be resolved from the literature.
In conclusion, there is uncertainty concerning the identity of S. odocoileocanis-like sarcocysts in cervids. Available morphological and biological data are summarized in Table 1. It is evident that without extensive investigation and transmission experiments (that will not be easily done), the question of species cannot be resolved. To facilitate further studies, we propose a new name, S. mehlhorni.
References
Atkinson CT, Wright SD, Telford SR, Mclaughlin GS, Forrester DJ, Roelke ME, McCown JW (1993) Morphology, prevalence, and distribution of Sarcocystis spp. in white-tailed deer (Odocoileus virginianus) from Florida. J Wildl Dis 29:73–84. doi:10.7589/0090-3558-29.1.73
Crum JM, Fayer R, Prestwood AK (1981) Sarcocystis spp. in white-tailed deer I. Definitive and intermediate host spectrum with a description of Sarcocystis odocoileocanis n. sp. J Wildl Dis 17:567–579. doi:10.7589/0090-3558-17.4.567
Dahlgren SS, Gjerde B (2007) Genetic characterisation of six Sarcocystis species from reindeer (Rangifer tarandus tarandus) in Norway based on the small subunit rRNA gene. Vet Parasitol 146:204–213. doi:10.1016/j.vetpar.2007.02.023
Dahlgren SS, Gjerde B, Skirnisson K, Gudmundsdottir B (2007) Morphological and molecular identification of three species of Sarcocystis in reindeer (Rangifer tarandus tarandus) in Iceland. Vet Parasitol 149:191–198. doi:10.1016/j.vetpar.2007.08.015
Dubey JP, Lozier SC (1983) Sarcocystis infection in the white-tailed deer (Odocoileus virginianus) in Montana: intensity and description of Sarcocystis odoi n. sp. Am J Vet Res 44:1738–1743
Dubey JP, Speer CA (1985) Prevalence and ultrastructure of three types of Sarcocystis in mule deer, Odocoileus hemionus (Rafinesque), in Montana. J Wildl Dis 21:219–228. doi:10.7589/0090-3558-21.3.219
Dubey JP, Speer CA (1986) Sarcocystis infections in mule deer (Odocoileus hemionus) in Montana and the description of three new species. Am J Vet Res 47:1052–1055
Dubey JP, Kistner TP, Callis G (1983) Development of Sarcocystis in mule deer transmitted through dogs and coyotes. Can J Zool 61:2904–2912. doi:10.1139/z83-378
Dubey JP, Calero-Bernal R, Rosenthal BM, Speer CA, Fayer R (2015a) Sarcocystosis of animals and humans, 2nd edn. CRC Press, Boca Raton
Dubey JP, Hilali M, van Wilpe E, Verma SK, Calero-Bernal R, Abdel-Wahab A (2015b) Redescription of Sarcocystis fusiformis sarcocysts from the water buffalo (Bubalus bubalis). Parasitology 142:385–394. doi:10.1017/S003118201400122X
Entzeroth R, Chobotar B, Scholtyseck E (1982) Ultrastructure of Sarcocystis sp. from the muscle of a white-tailed deer (Odocoileus virginianus). Z Parasitenkd 68:33–38
Gjerde B (1984a) A light microscopic comparison of the cysts of four species of Sarcocystis infecting the domestic reindeer (Rangifer tarandus) in Northern Norway. Acta Vet Scand 25:195–204
Gjerde B (1984b) The fox as definitive host for Sarcocystis sp. Gjerde, 1984 from skeletal muscle of reindeer (Rangifer tarandus) with a proposal for Sarcocystis tarandivulpes n. sp. as replacement name. Acta Vet Scand 25:403–410
Gjerde B (1985) Ultrastructure of the cysts of Sarcocystis tarandivulpes from skeletal muscle of reindeer (Rangifer tarandus tarandus). Acta Vet Scand 26:91–104
Gjerde B (1986) Scanning electron microscopy of the sarcocysts of six species of Sarcocystis from reindeer (Rangifer tarandus tarandus). APMIS 94:309–317
Gjerde B (2013) Phylogenetic relationships among Sarcocystis species in cervids, cattle and sheep inferred from the mitochondrial cytochrome c oxidase subunit I gene. Int J Parasitol 43:579–591. doi:10.1016/j.ijpara.2013.02.004
Gjerde B (2014) Morphological and molecular characteristics of four Sarcocystis spp. in Canadian moose (Alces alces), including Sarcocystis taeniata n. sp. Parasitol Res 113:1591–1604. doi:10.1007/s00436-014-3806-z
Gjerde B, Josefsen TD (2015) Molecular characterisation of Sarcocystis lutrae n. sp. and Toxoplasma gondii from the musculature of two Eurasian otters (Lutra lutra) in Norway. Parasitol Res 114:873–886. doi:10.1007/s00436-014-4251-8
Hudkins G, Kistner TP (1977) Sarcocystis hemionilatrantis (sp. n.) life cycle in mule deer and coyotes. J Wildl Dis 13:80–84. doi:10.7589/0090-3558-13.1.80
Lindsay DS, Blagburn BL, Mason WH, Frandsen JC (1988) Prevalence of Sarcocystis odocoileocanis from white-tailed deer in Alabama and its attempted transmission to goats. J Wildl Dis 24:154–156. doi:10.7589/0090-3558-24.1.154
Speer CA, Dubey JP (1986) An unusual structure in the primary cyst wall of Sarcocystis hemionilatrantis. J Protozool 33:130–132. doi:10.1111/j.1550-7408.1986.tb05573.x
Speer CA, Pond DB, Ernst JV (1980) Development of Sarcocystis hemionilatrantis Hudkins and Kistner, 1977 in the small intestine of coyotes. Proc Helminthol Soc Wash 47:106–113
Acknowledgments
The authors thank Mr. Efrain Pérez and Joseph Madary, Joint Pathology Center, Veterinary Services, US Army, Silver Spring, MD, USA, for the excellent technical help with electron microscopy.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Calero-Bernal, R., Verma, S.K., Cerqueira-Cézar, C.K. et al. Sarcocystis mehlhorni, n. sp. (Apicomplexa: Sarcocystidae) from the black-tailed deer (Odocoileus hemionus columbianus). Parasitol Res 114, 4397–4403 (2015). https://doi.org/10.1007/s00436-015-4679-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00436-015-4679-5