Abstract
Morphometric and DNA investigation results of Sarcocystis wobeseri sp. nov. from the barnacle goose (Branta leucopsis) and Sarcocystis sp. (cyst type IV) from the mallard duck (Anas platyrhynchos) are presented. No significant morphometric differences between the investigated Sarcocystis species were found. ITS-1, 18S rRNA, and 28S rRNA gene sequences of these species showed 100% identity. The conclusion is drawn that it is one and the same Sarcocystis species in different intermediate hosts.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Protists of the genus Sarcocystis are parasites with an obligatory prey-predator two-host life cycle. Herbivorous and omnivorous animals (prey) usually serve as intermediate hosts in the muscles of which asexual stages (sarcocysts) form. A sexual cycle with formation of oocysts/sporocysts occurs in the small intestine of carnivorous (predator) hosts. Birds can serve both as intermediate or definitive hosts for many species of Sarcocystis. Representatives of the order Anseriformes are intermediate hosts of these parasites.
Riley (1869) first found cysts of Sarcocystis in ducks. Stiles (1893) recorded macrocysts of these parasites in the shoveller (Anas clypeata) and named them S. rileyi. The striped skunk (Mephitis mephitis) was shown to be the definitive host for this species in North America (Cawthorn et al. 1981), although it is still unknown in Europe. Thus far, S. rileyi has been the only well investigated and named Sarcocystis species in ducks. Later, cysts of Sarcocystis were found in muscles of many species of ducks. Ducks also are intermediate hosts of other Sarcocystis species. Drouin and Mahrt (1980) found as many as five types of cysts in ducks, two of which had a smooth cyst wall of different thickness, and three types of microcysts had radial spines of different length and proximity on the outer surface of the cyst wall. It was established that the same hosts could be infected with more than one type of sarcocysts. Kutkienė and Sruoga (2004) present similar data. They discovered four types of microcysts in ducks: two of them had a smooth or wavy cyst wall that is difficult to distinguish by light microscopy and other two had protrusions of different length and shape on the wall surface (Kutkienė et al. 2008).
Geese have been investigated less extensively than ducks; however, sarcocysts have also been found in them. Wobeser et al. (1981) were the first to discover two types of sarcocysts in wild geese. The first type had finger-like protrusions on a wall surface within which fibrils were evident. This type was found in the lesser snow geese (Anser caerulescens caerulescens) and in the Ross geese (A. rossii). The other type was found in lesser snow geese and Canada geese (Branta canadensis) and had an undulating cyst wall with many small invaginations. Two types of sarcocysts (Kutkienė and Sruoga 2004) have also been found in the geese of Europe (Lithuania). One of them had teat- or finger-like protrusions on the wall surface and was found in the white-fronted geese (A. albifrons), in the lesser white fronted geese (A. erythropus), in the bean geese (A. fabalis) and in the gray-lag geese (A. anser). It has been established experimentally that one of the definitive hosts of this type of cysts was the Arctic fox (Alopex lagopus; Kutkienė et al. 2006). By light microscopy, cysts of another type had a relatively smooth cyst wall and were found in the white-fronted geese and in the gray-lag geese. Sarcocysts in gray-lag goose also were found in Kazakhstan (Pinayeva et al. 1998). Sarcocystis spp. found in all species of the geese examined thus far has been unnamed.
The identification and classification of Sarcocystis species have traditionally been based mainly on ultrastructure of the cysts and knowledge of the life cycle. Traditional morphological criteria could not always solve diagnostic problems of Sarcocystis, especially when similar morphological features of sarcocysts are found in taxonomically related species of hosts (Yang et al. 2001a). In recent years, DNA markers, mainly rRNA genes, have appeared to be able to overcome such complexity. Besides, the DNA analysis combined with the morphological one was successfully used to describe the new Sarcocystis species (Dubey et al. 2001; Dahlgren and Gjerde 2008a; Dahlgren and Gjerde 2008b; Kutkienė et al. 2009).
In the present paper, cysts morphology and DNA analysis results of Sarcocystis wobeseri sp. nov. from the barnacle goose (Branta leucopsis) and Sarcocystis sp. (cyst type IV) from the mallard duck (Anas platyrhynchos) are presented.
Material and methods
In the period of 2008 and 2009, 76 mallards (A. platyrhynchos) hunted in various districts of Lithuania or received from the National Food and Veterinary Risk Assessment Institute and three barnacle geese that were found dead after a fire on the island of Rusnė were investigated.
Light microscopy
To detect Sarcocystis cysts, samples of neck muscles of each bird were examined. For this purpose 28 oath-size pieces of muscles were cut-off, stained with water (1:500) methylene blue solution, lightened with 1.5% acetic acid solution, and pressed in a glass compressor. Sarcocystis infection intensity was evaluated having counted the cysts in 28 cross-sections of the muscles. The morphometric analysis of cyst walls and cystozoites was carried out in fresh preparations after the cysts had been isolated from muscle fibers by two preparation needles. The investigations were carried out using a computerized image analysis system. Cysts were differentiated on the basis of the wall thickness, the length and shape of protrusions and the length and shape of cystozoites.
Transmission electron microscopy
For transmission electron microscopy (TEM), samples of neck muscles containing cysts from one barnacle goose and one mallard were fixed in Karnovsky's fixative, postfixed in 1% osmium tetroxide, dehydrated, and embedded in Epon. Ultrathin sections were stained with 2% uranyl acetate and lead citrate and examined under the JEOL JEM-100B TEM. For TEM, only mature cysts were used.
DNA analysis
Few cysts of Sarcocystis sp. (cyst type I) from one barnacle goose (B. leucopsis) and Sarcocystis sp. (cysts type IV) from one mallard (A. platyrhynchos) isolated from the neck muscles were placed separately in 1.5 ml Eppendorf tubes containing 75% ethanol. Genomic DNA was extracted from sarcocysts using Qiagen DNeasy tissue kit. Taxonomic status of the investigated Sarcocystis spp. was evaluated using highly variable first internal transcribed spacer (ITS-1). Phylogenetic analysis was performed using entire 18S rRNA gene and partial 28S rRNA gene sequences. Primers for ITS-1, 18S rDNA and 28S rDNA were designed from the conserved regions detected by the alignment of published sequences of the Sarcocystidae species using Primer3 program (Rozen and Skaletsky 2000; Table 1). Primers for the 28S rRNA gene containing variable D2 and D3 domains were picked. Phylogenetic trees of the family Sarcocystidae obtained from D2 and D3 domains support the major grouping found for the full-length 28S rRNA gene analyses (Mugridge et al. 2000). Polymerase chain reactions (PCR) were carried out in the final 25-μl volume consisting of 5 μl 10× PCR buffer, 2.5 μl dNTP (2 mM), 0.2 μM each primer, 0.75 μl Taq polymerase, 2.5 μl MgCl2 and 0.2 μg template DNA. Amplification was performed using “hot start” of 95°C for 10 min, followed by 35 cycles of 94°C for 45 s, 56°C (primer pairs 1, 2 and 3), 50°C (fourth primer pair) 53°C (fifth primer pair), 59°C (sixth primer pair) 58°C (seventh primer pair) for 45 s, 72°C for 2 min, and the final extension at 72°C for 10 min. Annealing temperatures and extension time varied depending on the amount of the amplified product and trying to avoid amplification of non-specific fractions. Amplification products were analyzed using 1.7% agarose gel and were purified using the Cyclo-Pure gel extraction kit (Amresco, USA). PCR products were sequenced directly with an ABI Prism 377 automatic DNA sequencer using the same primers as for the PCR reactions. Sequences identity values were determined on the European Molecular Biology Open Software Suite (http://www.ebi.ac.uk/emboss/align/) using the default options. Sequences were aligned using the ClustalW algorithm. The beginning and the end of some sequences were truncated to have all sequences beginning with and ending in the same nucleotide positions. The alignment was then checked manually in order to correct the ambiguously placed nucleotides. Sequenced isolates of the same species were primarily used separately in the phylogenetic analysis. All isolates of the same species were grouped together in the phylogenetic tree; then, they were merged into a consensus sequence using the IUPAC codes for those nucleotide positions with more than one possible character state in the consensus sequence. The phylogenetic trees were constructed using the Bayesian method by MrBayes program, version 3. 1. 2 (Ronquist and Huelsenbeck 2003). The phylogenetic relationships were assessed with the most complex available model, GTR + I + G evolutionary model, which allows all six possible substitutions to vary with a proportion of invariable sites and a gamma-shaped distribution of rates across the sites. The final trees were drawn using TreeView version 1.6.6 (Page 1996). Four species of different genera belonging to the subfamily Toxoplasmatinae and having the shortest evolutionary distance to the subfamily Sarcocystinae were chosen as outgroup of phylogenetic trees (Besnoitia besnoiti, Hammondia hammondi, Neospora caninum, and Toxoplasma gondii). The initial phylogenetic analysis revealed that all Sarcocystis species from birds were placed in one group of the phylogenetic tree. Those species that were grouped together with Sarcocystis parasitizing in birds served as the ingroup. Sequences of 18 representatives of the family Sarcocystidae were included in further phylogenetic analyses (Table 2). S. dispersa, S. mucosa, and Sarcocystis sp. from the domestic pigeon were excluded from further analyses because of a lack of sequences of 18S rRNA or 28S rRNA genes.
Results
Having examined three barnacle geese, Sarcocystis cysts were found in one individual. Infection intensity was average and amounted to 35 cysts in 28 cross-sections of the neck muscles. One type of sarcocysts (temporarily called cyst type I) was found in this bird and proposed as a new species of Sarcocystis.
Sarcocystis wobeseri sp. nov
Description
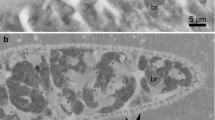
Sarcocysts were ribbon-shaped, very long (the largest fragment found in fresh preparations measured about 5.2 mm) but relatively thin (up to 100 μm). They were divided into chambers by septa filled with cystozoites. By light microscopy, the cyst wall seemed smooth and amounted to 1.0 μm (Fig. 1a). Banana-shaped cystozoites were small and measured 6.4-7.9 μm in length (n = 10; Fig. 1b). Ultrastructurally, the cyst wall amounted to 1.1 μm and was wavy (Fig. 1c). In some places, waves were high and resembled protrusions; however, in other places, the wall was almost smooth (Fig. 1d). The parasitophorous vacuolar membrane had many small invaginations (Fig. 1e). The ground substance layer continued into the interior of the cyst as septa. According to the classification of Dubey et al. (1989), these sarcocysts had type-1 cyst wall.
a-e Structure of the Sarcocystis wobeseri sp. nov. from the neck muscle of the barnacle goose (Branta leucopsis). a, b Light micrographs (computerized image analysis system). Fresh preparations. a Fragment of the cyst; note smooth and thin cyst wall (arrow). b Cystozoites. c-e Electron micrographs of the cyst wall. c Fragment of the cyst wall with waves (arrows). d Fragment of the cyst wall without waves (arrows). e High magnification of the cyst wall; note minute invaginations of the parasitophorous vacuolar membrane (arrows). g Ground substance
Sarcocystis sp. (cyst type IV) from the mallard
Cysts of Sarcocystis were detected in 11 (14.5%) mallards out of the 76 examined ones. Two types of cysts, which we temporarily called cysts type II and type IV, were found. Cyst type II was found during our earlier investigations into ducks and morphological, as well as DNA investigation results were presented in the publication Kutkienė et al 2008.
Sarcocysts type IV was ribbon-shaped, very long, and thick; the largest fragment found measured 8.0 × 0.2 mm. They were divided into large chambers by septa. By light microscopy, the cyst wall seemed wavy and reached up to 1.0 μm (Fig. 2a). Using a computerized image analysis system, it was defined that the cyst surface resembled a honeycomb (Fig. 2c). Banana-shaped cystozoites were small and measured 6.7-8.1 μm in length (n = 20; Fig. 2b). Ultrastructurally, the cyst wall amounted to 1.6 μm and was wavy (Fig. 2d). In some places, the waves were quite high and resembled protrusions; however, in other places, the wall was almost smooth (Fig. 2e). The parasitophorous vacuolar membrane had many small invaginations (Fig. 2f). The ground substance layer continued into the interior of the cyst as septa. These sarcocysts had type-1 cyst wall (Dubey et al. 1989).
a-f Structure of the Sarcocystis sp. (cyst type IV) from the neck muscle of the mallard (Anas platyrhynchos). a-c Light micrographs (computerized image analysis system). Fresh preparations. a Fragment of the cyst with wavy cyst wall (arrows). b Cystozoites. c Fragment of the cyst wall surface, which resembles a honeycomb. d-f Electron micrographs of the cyst wall. d Fragment of the cyst wall with waves, which resemble protrusions (arrows). e Fragment of the cyst wall without waves (arrows). f High magnification of the cyst wall; note minute invaginations of the parasitophorous vacuolar membrane (arrows). g Ground substance
DNA results
ITS-1, 18S rRNA, and 28S rRNA gene sequences of Sarcocystis sp. (cyst type I) from the barnacle goose (we describe as S. wobeseri sp. nov.) and Sarcocystis sp. (cysts type IV) from the mallard were deposited in GenBank (GQ922885–GQ922888, GU475111, GU475112). ITS-1 sequences (844 bp long) gained from S. wobeseri (GU475111) and Sarcocystis sp. (cyst type IV) (GU475112) from the mallard were identical to each other. These sequences had low similarity within ITS-1 comparing with other Sarcocystis species. The highest identity within ITS-1 was found in Sarcocystis sp. from the domestic pigeon (Columbia livia dom.), S. canis, Sarcocystis sp. from wolverine (Gulo gulo) and S. felis species with following values: 93%, 75%, 70%, and 66%, respectively. These two identical sequences were especially different from other Sarcocystis species including S. cruzi, S. dasypi, S. falcatula, S. lindsayi, S. neurona, S. rangiferi, and S. tarandi. A comparison of sequences of 18S rRNA and 28S rRNA genes revealed a 100% sequences identity between S. wobeseri, Sarcocystis sp. (cyst type IV) from the mallard and also Sarcocystis sp. (cyst type I) from the white-fronted goose. According to 18S rRNA and 28S rRNA gene sequences, these three Sarcocystis isolates have the closest sequence identity values to S. cornixi (99.6%) and to Sarcocystis sp. from the domestic pigeon, respectively (99.2%).
In the phylogenetic analysis, three datasets were generated; the first of the 18S rDNA sequences, the second of the 28S rDNA sequences, and the third of the 18S rDNA plus 2S rDNA sequences. The concatenated alignment contained 2,060 aligned nucleotide positions, with 1,598 and 462 aligned nucleotide positions belonging to the 18S rRNA and 28S rRNA genes, respectively. Unfortunately, only ∼500 bp long sequences of the 28S rRNA gene for S. gallotiae, S. lacertae, S. rodentifelis, and Sarcocystis sp. from the shrew have been deposited in GenBank. Obtained phylogenetic trees present similar topologies (Fig. 3). According to phylogenetic analysis, S. wobeseri from barnacle goose is genetically identical to Sarcocystis sp. (cyst type IV) from the mallard and Sarcocystis sp. (cyst type I) from the white-fronted goose and the most genetically related to S. cornixi, Frenkelia glareoli, F. microti, and Sarcocystis sp. from the shrew. In the general, phylogenetic tree S. lacerate and S. gallotiae; S. muris, and S. rodentifelis; Sarcocystis sp. (cyst type III) from the white-fronted goose and Sarcocystis sp. (cyst type II) from the mallard are grouped together with 1.00 support value. The lowest resolution power was observed in 18S rDNA phylogram, where phylogenetic relationships inside S. wobeseri “group” were not robust.
a Phylogenetic tree for the subfamily Sarcocystinae based on 18S rRNA gene sequences. The tree was reconstructed using the Bayesian inference, rooted on B. besnoiti, H. hammondi, N. caninum, and T. gondii and scaled according to the branch length. The numbers in the figure show posterior probability support values. b Phylogenetic tree for the subfamily Sarcocystinae based on 28S rRNA gene sequences. The tree was reconstructed using the Bayesian inference, rooted on B. besnoiti, H. hammondi, N. caninum, and T. gondii and scaled according to the branch length. The numbers in the figure show posterior probability support values. c Phylogenetic tree for the subfamily Sarcocystinae based on the concatenated dataset of 18S rRNA and 28S rRNA gene sequences. The tree was reconstructed using the Bayesian inference, rooted on B. besnoiti, H. hammondi, N. caninum, and T. gondii and scaled according to the branch length. The numbers in the figure show posterior probability support values
Taxonomic summary
Type intermediate host: Barnacle goose (B. leucopsis)
Definitive host: unknown
Locality: Šilutė district (near the Baltic Sea), western Lithuania
GenBank accession numbers: GenBank number as Sarcocystis sp. cyst type I ex B. leucopsis GQ922885, GQ922887, GU475111 for 18S rRNA, partial sequences of 28S rRNA genes and ITS-1, respectively
Specimens deposited: TEM material is deposited at the Institute of Ecology, Vilnius, Lithuania.
Etymology: the species has been named in honor of the famous Canadian scientist of veterinary pathology Gary Wobeser who (together with his co-workers) was the first to have determined this type of cyst wall in Sarcocystis of geese.
Discussion
The present work is a continuation of our earlier investigations into the birds order Anseriformes. In the period of 1997 and 2009, a total of 684 birds of this order were investigated and Sarcocystis cysts were found in 222 (32.5%) individuals (Kutkienė and Sruoga 2004 and unpublished data). By light microscopy, four types of sarcocysts have been determined, which were temporarily called type I, II, III, and IV, and S. rileyi was found. It was stated that cyst type II and III could be easily distinguished by light microscopy not only by the morphology of the cyst wall but also by a very specific shape of cystozoites. Meantime differentiation of cyst type I and IV by light microscopy was really complicated (Kutkienė and Sruoga 2004). Since the same types of cysts were found in different species of birds order Anseriformes, the hypothesis was put forward that the same species of Sarcocystis could parasitize different species of birds order Anseriformes. Later investigations into the ultrastructure of the cyst walls of some bird species and DNA analysis were undertaken (Kutkienė et al 2006; Kutkienė et al. 2008).
There are two opinions as to the host specificity of the species in the problem of sarcosporidiosis. According to the supporters of the first opinion (which is a prevailing one), the majority of species are specific to the intermediate host. In the opinion of other authors, the same Sarcocystis species can parasitize in taxonomically related species of hosts. The latter attitude is based on the morphological similarities of sarcocysts found in different species of hosts, as well as on the same possible definitive hosts (Wesemeier and Sedlaczek 1995; Odening 1998).
Box et al. (1984) experimentally showed that S. falcatula is a species of the passeriform, psitaciform, and columbiform birds. On the basis of these investigations, the idea was put forward that the Sarcocystis species parasitizing birds can be less host-specific than those parasitizing mammals.
Applying the DNA investigation methods in Sarcocystis diagnostics made the solution of the problem of the species specificity in intermediate hosts easier; however, thus far, the structure of the sarcosysts wall and morphology of cystozoites have remained one of the main criteria for diagnosing the Sarcocystis species. This is of especially great value in characterizing species in the same intermediate host (Mehlhorn and Heydorn 1978)
Cysts types I and IV under investigation in the present work are found by light microscopy in the majority of different species of birds order Anserifomes (Kutkienė and Sruoga (2004) and unpublished data). However, as the earlier data and those presented in this article showed, the results of TEM can be at variance with the results of light microscopy. For example, Sarcocystis cyst walls of the common goldeneye (Bucephala clangula; Kutkienė et al. 2008) and the barnacle goose, which seemed to be smooth by light microscopy (according to our classification cyst type I), were clearly wavy by TEM. The case might be that only one cyst type, which has a thin wall with a characteristic wavy surface and many invaginations of the primary cyst wall (according to the classification of Dubey and Odening (2001)—cyst wall type-1) is found in the birds order Anseriformes. Besides, the shape and size of cystozoites of the cysts of types I and IV are the same.
Our investigations into Sarcocystis cysts of the barnacle goose and the mallard carried out by light microscopy showed some morphometric differences. The Sarcocystis wall of the mallard seemed wavy, whereas that of barnacle goose was smooth. The supposition that waviness is dependent on the quality of preparations cannot be rejected. These differences could not be of importance in the species diagnostics.
When comparing the results of the ultrastructure of the barnacle goose and the mallard, it can be stated that no differences of significance to diagnostics of the species were found. Differences of wall thickness found in Sarcocystis species examined can be in dependence on cyst age. Sarcocyst walls from both species of birds correspond to cyst wall type-1 (Dubey and Odening 2001). Cyst wall ultrastructure of other anseriforms, i.e., of S. rileyi (Dubey et al. 2003), Sarcocystis sp. (cyst type II) from the mallard (Kutkienė et al. 2008), Sarcocystis sp. (cyst type III) from the white-fronted goose (Kutkienė et al. 2006) and sarcocysts (with finger-like protrusions on the cyst wall surface) from lesser snow and Ross geese (Wobeser et al. 1981) differed greatly from morphology of Sarcocystis isolates presented in this paper.
As has already been noted earlier, the wall type-1 (Dubey and Odening 2001) has been determined for sarcocysts from the lesser snow and Canada geese (Wobeser et al. 1981) and from the goldeneye (Kutkienė et al. 2008). An undulating cyst wall was seen in the white-rumped swift and the night heron from South Africa (Kaiser and Markus 1983). Thus, our findings and data from literature confirm once again the idea that the cyst wall of the same morphology is characteristic of sarcocysts parasitizing related and taxonomically distant species of animals including those of birds. Speaking about the possibilities of the same Sarcocystis species to parasitize taxonomically related species of animals (e.g., birds order Anseriformes) the morphological similarity of sarcocysts is only a primary feature enabling this supposition to be made. The results of the DNA investigation play an especially significant role in the solution of this problem. The 18S rRNA gene investigation results showed that several morphologically similar Sarcocystis species from the cattle and the water buffalo were the same species in both intermediate hosts (Yang et al. 2001a; Yang et al 2001b). Molecular studies confirm that S. neurona also use several species as intermediate hosts (Mullaney et al. 2005; Mansfield et al. 2008; Miller et al. 2009). Likewise, in our case, S. wobeseri from the barnacle goose and Sarcocystis sp. (cyst type IV) from the mallard had the same nucleotide composition in 4,143 bp long sequenced fragment of ITS-1, 18S rRNA, and 28S rRNA genes. Thus, genetic investigations show that S. wobeseri from the barnacle goose and Sarcocystis sp. (cyst type IV) from the mallard are the same species. According to DNA results S. wobeseri is related but distinct to S. cornixi, F. glareoli, F. microti, and Sarcocystis sp. from the shrew and Sarcocystis sp. from the domestic pigeon. 18S rRNA and 28S rRNA gene sequences of S. wobeseri and Sarcocystis sp. (cyst type I) from the white-fronted goose showed 100% identity. We predict that Sarcocystis sp. (cyst type I) from the white-fronted goose also could be intermediate host for S. wobeseri. To confirm these hypotheses, ITS-1 sequences and cyst wall ultrastructure results of Sarcocystis sp. (cyst type I) from the white-fronted goose should be obtained and compared.
The 18S rRNA gene sequences were successfully used for the phylogenetic analysis within the family Sarcocystidae (Jeffries et al. 1997; Tenter and Johnson 1997; Doležel et al. 1999; Jenkins et al. 1999; Holmdahl et al. 1999; Šlapeta et al. 2001; Šlapeta et al. 2002; Šlapeta et al. 2003; Morrison et al. 2004; Elsheikha et al. 2005; Dahlgren et al. 2008; Dahlgren and Gjerde 2008a; Dahlgren and Gjerde 2008b). In this taxon, several phylogenetic well-supported groups could be distinguished: the first group is made of representatives of the Toxoplasmatinae subfamily, the second group unites species studied in this work as an ingroup, the third group contains Sarcocystis species with a snake-rodent life cycle, and the fourth group comprises Sarcocystis species that uses an even-toed ungulate as their intermediate host and a canine, feline, or unknown predator as their definitive host. Species in the second group have very short branch length as compared with other Sarcocystidae groups, and phylogenetic relationships inside this group are not robust. Evolutionary relationships of some species inside the fourth group are also arguable. In the future perspective, when Sarcocystidae 18S rRNA gene database increases, new difficulties will arise in making a reliable phylogenetic inference. We support the previously stated opinion, that the 18S rRNA marker alone is insufficient to resolve all evolutionary relationships inside this family (Morrison et al 2004). In this study, reliable evolutionary relationships were revealed using entire 18S rRNA gene and partial 28S rRNA gene sequences. The concatenated 18S rRNA and 28S rRNA genes analysis have been successfully used in several studies within Sarcocystidae (Šlapeta et al. 2003; Kutkienė et al. 2009). The ITS-1 region is useful for closely related Sarcocystis species (Marsh et al. 1999; Dubey et al. 2006). However, due to huge sequences variability ITS-1 is a worthless determining evolutionary relationship of whole Sarcocystis genus. In order to reconstruct evolutionary relationships between closely related species of genus Sarcocystis, it could be valuable to use ITS-1 combining with 18S rRNA and/or 28S rRNA genes.
The comparative evaluation of the morphology of the cyst walls and the DNA investigation results enable us to state that S. wobeseri sp. nov. from the barnacle goose and Sarcocystis sp. (cyst type IV) from the mallard are one and the same species.
References
Box ED, Meier JL, Smith JH (1984) Description of Sarcocystis falcatula Stiles, 1893, a parasite of birds and opossums. J Protozool 31:521–524
Cawthorn RJ, Rainnie D, Wobeser G (1981) Experimental transmission of Sarcocystis sp. (Protozoa: Sarcocystidae) between the shoveler (Anas clypeata) duck and striped skunk (Mephitis mephitis). J Wildl Dis 17:389–394
Dahlgren SS, Gjerde B (2008a) Sarcocystis in moose (Alces alces): molecular identification and phylogeny of six Sarcocystis species in moose, and a morphological description of three new species. Parasitol Res 103:93–110
Dahlgren SS, Gjerde B (2008b) Sarcocystis in Norwegian roe deer (Capreolus capreolus) molecular and morphological identification of Sarcocystis oviformis n. sp. and Sarcocystis gracilis and their phylogenetic relationships with other Sarcocystis species. Parasitol Res 104:993–1003
Dahlgren SS, Gouveia-Oliveira R, Gjerde B (2008) Phylogenetic relationships between Sarcocystis species from reindeer and other Sarcocystidae deduced from ssu rRNA gene sequences. Vet Parasitol 151:27–35
Doležel D, Koudela B, Jirků M, Hypša V, Oborník M, Votýpka J, Modrý D, Šlapeta JR, Lukeš J (1999) Phylogenetic analysis of Sarcocystis spp. of mammals and reptiles supports the coevolution of Sarcocystis spp. with their final hosts. Int J Parasitol 29:795–798
Drouin TE, Mahrt JL (1980) The morphology of cysts of Sarcocystis infecting birds in western Canada. Can J Zool 58:1477–1482
Dubey JP, Speer CA, Fayer R (1989) Sarcocystosis of animals and man. CRC Press, Boca Raton, pp 1–215
Dubey JP, Odening K (2001) Toxoplasmosis and related infections. In: Samuel WM, Pybus MJ, Kocan AA (eds) Parasitic diseases of wild mammals. Iowa State University Press, Ames, pp 478–519
Dubey JP, Rosenthal BM, Speer CA (2001) Sarcocystis lindsayi n. sp. (Protozoa: Sarcocystidae) from the South American opossum, Didelphis albiventris from Brazil. J Eukaryot Microbiol 48:595–603
Dubey JP, Cawthorn RJ, Speer CA, Wobeser GA (2003) Redescription of the sarcocysts of Sarcocystis rileyi (Apicomplexa: Sarcocystidae). J Eukaryot Microbiol 50:476–482
Dubey JP, Rosenthal BM, Morales JA, Alfaro A (2006) Morphologic and genetic characterization of Sarcocystis sp. from the African grey parrot, Psittacus erithacus, from Costa Rica. Acta Parasitol 51:161–168
Elsheikha HM, Lacher DW, Mansfield LS (2005) Phylogenetic relationships of Sarcocystis neurona of horses and opossums to other cyst-forming coccidia deduced from SSU rRNA gene sequences. Parasitol Res 97:345–357
Jeffries AC, Schnitzler B, Heydorn AO, Johnson AM, Tenter AM (1997) Identification of synapomorphic characters in the genus Sarcocystis based on 18S rDNA sequences comparison. J Eukaryot Microbiol 44:388–392
Jenkins MC, Ellis JT, Liddell S, Ryce C, Munday BL, Morrison DA, Dubey JP (1999) The relationship of Hammondia hammondi and Sarcocystis mucosa to other heteroxenous cyst forming coccidia as inferred by phylogenetic analysis of the 18S SSU ribosomal DNA sequence. Parasitology 119:135–142
Holmdahl OJ, Morrision DA, Ellis JT, Huong LT (1999) Evolution of ruminant Sarcocystis (Sporozoa) parasites based on small subunit rDNA sequences. Mol Phylogenet Evol 11:27–37
Kaiser IA, Markus MB (1983) Species of Sarcocystis in wild South African birds. Proc Electron Microscop Soc S Afr 13:103–104
Kutkienė L, Sruoga A (2004) Sarcocystis spp. in birds of the order Anseriformes. Parasitol Res 92:171–172
Kutkienė L, Sruoga A, Butkauskas D (2006) Sarcocystis sp. from white-fronted goose (Anser albiforns): cyst morphology and life cycle studies. Parasitol Res 99:562–565
Kutkienė L, Sruoga A, Butkauskas D (2008) Sarcocystis sp. from the goldeneye (Bucephala clangula) and the mallard (Anas platyrhynchos): cyst morphology and ribosomal DNA analysis. Parasitol Res 102:691–696
Kutkienė L, Prakas P, Sruoga A, Butkauskas D (2009) Sarcocystis in the birds family Corvidae with description of Sarcocystis cornixi sp. nov. from the hooded crow (Corvus cornix). Parasitol Res 104:329–336
Mansfield LS, Mehler S, Nelson K, Elsheikha HM, Murphy AJ, Knust B, Tanhauser SM, Gearhart PM, Rossano MG, Bowman DD, Schott HC, Patterson JS (2008) Brown-headed cowbirds (Molothrus ater) harbor Sarcocystis neurona and act as intermediate hosts. Vet Parasitol 153:24–43
Marsh AE, Barr BC, Tell L, Bowmann DD, Conrad PA, Ketcherside C, Green T (1999) Comparison of the internal transcribed spacer, ITS-1, from Sarcocystis falcatula isolates and Sarcocystis neurona. J Parasitol 85:750–757
Mehlhorn H, Heydorn AO (1978) The Sarcosporidia (Protozoa, Sporozoa): life cycle and fine structure. Adv Parasitol 16:43–91
Miller MA, Barr BC, Nordhausen R, James ER, Magargal SL, Murray M, Conrad PA, Toy-Choutka S, Jessup DA, Grigg (2009) Ultrastructural and molecular confirmation of the development of Sarcocystis neurona tissue cysts in the nervous system of southern sea otters (Enhydra lutris nereis). Int J Parasitol 39:1362–1372
Morrison DA, Bornstein S, Thebo P, Wernery U, Kinne J, Mattsson JG (2004) The current status of the small subunit rRNA phylogeny of the coccidia (Sporozoa). Int J Parasitol 34:501–514
Mugridge NB, Morrison DA, Jäkel T, Heckeroth AR, Tenter AM, Johnson AM (2000) Effects of sequence alignment and structural domains of ribosomal DNA on phylogeny reconstruction for the protozoan family Sarcocystidae. Mol Biol Evol 17:1842–1853
Mullaney T, Murphy AJ, Kiupel M, Bell JA, Rossano MG, Mansfield LS (2005) Evidence to support horses as natural intermediate hosts for Sarcocystis neurona. Vet Parasitol 133:27–36
Odening K (1998) The present state of species-systematics in Sarcocystis Lankester, 1882 (Protista, Sporozoa, Coccidia). Syst Parasitol 41:209–233
Page RDM (1996) Treeview: an application to display phylogenetic trees on personal computers. Comput Appl Biosci 12:357–358
Pinayeva LM, Pak CM, Kokhno LI (1998) Sarcocystis of the wild birds of Kazakhstan. Parasitol Int 47:143
Riley CV (1869) A measly wild duck. Am Entomol 1:89
Ronquist F, Huelsenbeck JP (2003) MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19:1572–1574
Rozen S, Skaletsky HJ (2000) Primer 3 on the WWW for the general users and for biologist progarammers. In: Krawetz S, Misener S (eds) Bioinformatics methods and protocols: methods in molecular biology. Humana Press, Totowa, pp 365–386
Stiles CW (1893) On the presence of sarcosporidia in birds. USDA Bur Anim Indus Bull 3:79–89
Šlapeta JR, Modrý D, Votýpka J, Jirků M, Koudela B, Lukeš J (2001) Multiple origin of the dihomoxenous life cycle in Sarcosporidia. Int J Parasitol 31:413–417
Šlapeta JR, Kyselová I, Richardson AO, Modrý D, Lukeš J (2002) Phylogeny and sequence variability of the Sarcocystis singaporensis Zaman and Colley, (1975) 1976 ssrDNA. Parasitol Res 88:810–815
Šlapeta JR, Modrý D, Votýpka J, Jirků M, Lukeš J, Koudela B (2003) Evolutionary relationships among cyst-forming coccidia Sarcocystis spp. (Alveolata: Apicomplexa: Coccidea) in endemic African tree vipers and perspective for evolution of heteroxenous life cycle. Mol Phylogenet Evol 27:464–475
Tenter AM, Johnson AM (1997) Phylogeny of the tissue cyst-forming coccidia. Adv Parasitol 39:69–139
Wesemeier H-H, Sedlaczek J (1995) One known Sarcocystis species and two found for the first time in red deer and wapiti (Cervus elaphus) in Europe. Appl Parasitol 36:245–251
Wobeser G, Leighton FA, Cawthorn RJ (1981) Occurrence of Sarcocystis Lankester, 1882, in wild geese in Saskatchewan. Can J Zool 59:1621–1624
Yang ZQ, Zuo YX, Yao YG, Chen XW, Yang GC, Zhang YP (2001a) Analysis of the 18S rRNA genes of Sarcocystis species suggests that the morphologically similar organisms from cattle and water buffalo should be considered the same species. Mol Biochem Parasitol 115:283–288
Yang ZQ, Zuo YX, Ding B, Chen XW, Luo J, Zhang YP (2001b) Identification of Sarcocystis hominis-like (Protozoa: Sarcocystidae) cyst in water buffalo (Bubalus bubalis) based on 18S rRNA gene sequences. J Parasitol 87:934–937
Acknowledgments
The authors are grateful to Dr. Jan Votýpka of the Department of Parasitology, Faculty of Science, Charles University, Prague, Czech Republic for the valuable comments on the draft of the paper. We thank Mrs. I. Žalakevičienė from the Institute of Experimental and Clinical Medicine for her help in carrying out investigations of electron microscopy.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Kutkienė, L., Prakas, P., Sruoga, A. et al. The mallard duck (Anas platyrhynchos) as intermediate host for Sarcocystis wobeseri sp. nov. from the barnacle goose (Branta leucopsis). Parasitol Res 107, 879–888 (2010). https://doi.org/10.1007/s00436-010-1945-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00436-010-1945-4