Abstract
Main conclusion
SmMYB35, a light-responsive R2R3-MYB transcription factor, positively regulates anthocyanin biosynthesis in eggplant by binding to the promoters of SmCHS, SmF3H, SmDFR, and SmANS and enhancing their activities. In addition, SmMYB35 interacts with SmTT8 and SmTTG1 to form a MBW complex, thereby enhancing anthocyanin biosynthesis.
Abstract
Eggplant is a vegetable rich in anthocyanins. SmMYB35, a light-responsive R2R3-MYB transcription factor, was isolated from eggplant and investigated for its biological functions. The results suggested that the expression of SmMYB35 was regulated by SmHY5 through directly binding to G-box in the promoter region, and the overexpression of SmMYB35 could increase the anthocyanin content in the stems and petals of the transgenic eggplants. SmMYB35 could also bind to the promoters of SmCHS, SmF3H, SmDFR, and SmANS and enhance their activities. In addition, SmMYB35 interacted with SmTT8 and SmTTG1 to form a MBW complex which enhanced anthocyanin biosynthesis. Taking together, we firstly verified that SmMYB35 promoted anthocyanin biosynthesis in plants. The results provide new insights into the regulatory effects of SmMYB35 on key anthocyanin biosynthetic genes and advance our understanding of the molecular mechanism of light-induced anthocyanin synthesis in eggplants.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Eggplant (Solanum melongena L.) is consumed and cultivated in many countries. Purple eggplants are more attractive to consumers because of its high anthocyanin content. However, low light conditions often result in poor coloration and reduce its consumer acceptance. Anthocyanins are a type of water-soluble pigments, widely detected in plant stems, leaves, petals and fruit peels (Kayesh et al. 2013). Anthocyanins not only supply plants with vivid colors, but also protect them from various stresses, such as drought, extreme temperature, and high salt conditions (Dixon et al. 1995).
Anthocyanins are synthesized via a branch of the flavonoid biosynthesis pathway (Belwal et al. 2020). Several anthocyanin biosynthetic enzymes have been characterized in different plants, including chalcone synthase (CHS), chalcone isomerase (CHI), flavanone 3-hydroxylase (F3H), flavonoid 3′ 5'-hydroxylase (F3′5'H), dihydroflavonol-reductase (DFR), and anthocyanidin synthase (ANS) (Jiang et al. 2016a; Shoeva et al. 2017). The genes encoding these enzymes are regulated by a group of transcription factors (TFs), including MYB, bHLH and WD40 (Xu et al. 2013). The MYB family is one of the largest groups of transcriptional regulators involving in the regulation of anthocyanin biosynthesis. There are AtMYB75, AtMYB90, AtMYB113, AtMYB114, and AtMYB4 in Arabidopsis thaliana (Hemm et al. 2001; Gonzalez et al. 2008), SlANT1, SlAN2 and SlMYBATV in S. lycopersicum (Kiferle et al. 2015; Cao et al. 2017), StAN1 and StMYB44 in S. tuberosum (Liu et al. 2016, 2019), SmMYB1 and SmMYB86 in S. melongena L (Jiang et al. 2016b; Li et al. 2021), and MdMYB10 and MdMYB6 in Malus × domestica (Espley et al. 2009; Xu et al. 2020). These MYBs usually promote or inhibit structural genes expression to exhibit their regulatory roles in anthocyanin synthesis. BHLH proteins are the second largest class of TFs (Toledo-Ortiz et al. 2003). Numerous bHLH TFs, such as TT8, GL3, and EGL3, interact with R2R3-MYB proteins to regulate the anthocyanin biosynthesis in Arabidopsis (Feller et al. 2011). In eggplant, SmTT8 binds to the promoter of SmCHS and promotes its activation (He et al. 2019). The regulatory capability of SmMYB1 on anthocyanin biosynthetic genes was strengthened when SmTT8 was added (Zhou et al. 2019). So far, a few WD40 proteins and their functionalities in the biosynthesis of anthocyanin, proanthocyanidin and trichome formation have been characterized, including AtTTG1 from Arabidopsis (Shan et al. 2019), PgWD40 from pomegranate (Ben-Simhon et al. 2011), and SmTTG1 from eggplant (Li et al. 2021). The main function of WD40 is to provide a stable platform for MYB and bHLH proteins and promotes their interactions to form a (MYB-bHLH-WD40) MBW complex (Liu et al. 2018a; Tang et al. 2020).
Light is one of the most influential environmental factors not only affecting plant growth and development but also regulating the production of secondary metabolites by regulating the expression of light-response genes. In the light, ELONGATED HYPOCOTYL5 (HY5) acts downstream of photoreceptors to promotes photomorphogenesis (Xu et al. 2018). In addition, SmHY5 directly binds to the promoters of SmCHS and SmDFR, to promotes anthocyanin biosynthesis in eggplant (Jiang et al. 2016b). What's more, HY5 also directly activates the transcription of several MYB genes containing the HY5 potential binding sites in their promoters to regulate physiological processes (Lee et al. 2007; An et al. 2017). Taken together, HY5 promotes anthocyanin accumulation by regulating the expression of MYB genes and anthocyanin biosynthetic genes. Although light-induced anthocyanin accumulation has been documented in several plants, the molecular mechanism underlining HY5-dependent was not fully understood.
Our previous study found that the synthesis of anthocyanin in the peel of ‘Lanshan Hexian’ eggplant was regulated by light (Jiang et al. 2016b; Li et al. 2017). Our previous work also screened for several genes involving in anthocyanin biosynthesis (Li et al. 2017, 2018). SmMYB35 was one of the differentially expressed genes in the transcriptome data at 0, 5, and 12 d with up-down-regulated expression pattern in ‘Lanshan Hexian’. Moreover, SmMYB35 affected the anthocyanin accumulation and expression level of structural genes when co-infiltrated with SmMYB1 and SmTT8 in Nicotiana benthamiana (Li et al. 2017). However, how SmMYB35 may promote anthocyanin synthesis is still unclear. This study was conducted to investigate the potential regulatory roles of SmMYB35 in eggplant.
Materials and methods
Plant materials and growth conditions
The eggplant cultivar ‘Lanshan Hexian’ (purple-black peel), ‘140’ (white peel), from my own lab, and the SmMYB35-OE transgenic eggplant seeds were sown in chambers with a growth condition at 25 °C/22 °C (day/night) and 16 h/8 h photoperiod. The seedlings in the 4-leaf stage were transplanted to the greenhouse of Shanghai Hangyu Seed Base, Shanghai, China.
Arabidopsis thaliana of Columbia-0 (Col-0) and SmMYB35-OE transgenic Arabidopsis thaliana were cultured in 10 cm diameter plastic chambers with a growth condition at 25 °C/22 °C (day/night) and 16 h/8 h photoperiod.
The seedlings of N benthamiana were growth under the same condition as that of Arabidopsis thaliana.
Gene isolation and subcellular localization
The coding sequences of SmMYB35 (Sme2.5_00216.1_g00001.1) were downloaded from the Eggplant Genome DataBase (http://eggplant.kazusa.or.jp/keyword.html). The total RNA was extracted from the peel of photosensitive eggplant cultivar, ‘Lanshan Hexian’ using Plant RNA Extraction Kit (TaKaRa, Otsu, Japan). The cDNA was synthesized using PrimeScript™ RT reagent Kit with gDNA Eraser (TaKaRa). Total DNA was extracted from eggplant leaves by CTAB method to isolate SmMYB35 promoter. The online programmer PlantCARE (http://bioinformatics.psb.ugent.be/webtools/plantcare) was used to analyze the number and location of cis-elements harbored in the promoter region. For subcellular localization experiments, the PHB-SmMYB35-YFP vector was constructed with homologous recombination method. The GV3101 strains harboring PHB-SmMYB35-YFP or PHB-YFP were transformed into 5-week-old N. benthamiana leaves, and the nuclei were stained with DAPI (Sangon Biotech, shanghai, China), as described in my own published paper (Li et al. 2021). All primers of this research were designed by primer 5 and listed in Supplementary Table S1.
Plant transformation and phenotype analysis
The coding sequences of SmMYB35 were ligated to the PHB vector containing yellow fluorescent protein and a 35S CaMV promoter to generate the 35S::SmMYB35-YFP plasmids. The recombinant plasmids were transformed into Agrobacterium tumefaciens strain GV3101 and then transfected into Arabidopsis thaliana by the floral dip method. Transgenic seedlings were verified by resistance screening and confirmed with PCR, and self-crossed for homozygous selection until T3 lines for analysis. The Agrobacterium-mediated transformation was performed using cotyledons of the eggplant cultivar ‘140’, as previously described (Jiang et al. 2016a; Li et al. 2021). The phenotypes of SmMYB35-OE Arabidopsis thaliana, ‘140’ transformed eggplants and their WT were observed at the same growth conditions.
Extraction and quantification of anthocyanins
Anthocyanins of eggplant tissues were extracted and measured as described by Li et al. (2017). At least three biological replicates were collected and three measurements for each replicate were performed.
Quantitative real-time polymerase chain reaction (qRT-PCR)
To discover the expression of SmCHS, SmF3H, SmANS, SmDFR in both the Col-0 and SmMYB35-OE transgenic lines, the expression of SmMYB35, SmCHI, SmCHS, SmF3H, SmF35H, SmANS, SmDFR in both the ‘140’ and SmMYB35-OE transgenic lines were anaylzed. The expression patterns of SmCHI, SmCHS, SmF3H, SmF35H, SmANS, SmDFR in different tissues of ‘140’, ‘140’ transformed plants and their WT, different tissues of ‘Lanshan Hexian’ and ‘140’, including roots, stems, leaves, petals, flesh, peel, and sepals were studied. In bagging experiments, the fastest anthocyanin synthesis rate was at around 5 days as shown in our previous study; the time points of sampling for qRT-PCR and RNA-seq were designed to be at 0, 5, and 12 d. The expression level of SmMYB35 was differentially in the transcriptome data at 0d, 5d, and 12d, as was found in our previous studies (Li et al. 2017). Then eggplant fruit peel was collected at 0 ~ 8 h after bag removal from 8:00 am to 16:00 pm to further explore the expression of SmMYB35 under light condition. Total RNA was treated with DNase to remove traces of DNA. RNA (1 μg) was used for cDNA synthesis with the PrimeScript RT Master Mix Perfect Real-Time Kit (TaKaRa). RT-qPCR was performed using SYBR Premix Ex Taq II Kit (TaKaRa) on Light Cycler 96 (Roche, Basel, Switzerland). The reaction procedure was as follows: 95 °C for 30 s, followed by 40 cycles of 95 °C for 5 s, 60 °C for 30 s, and 72 °C for 30 s. Three technical replicates were run for the RT-qPCR of each sample using the gene-specific primers shown in Table S1. The Actin (GU984779.1) from the eggplant was amplified in parallel as an internal reference gene. The data were analyzed using the 2−ΔΔCT method.
Yeast one-hybrid assay (Y1H)
The yeast one-hybrid experiment was performed as mentioned earlier (Jiang et al. 2016b). The coding sequences (CDSs) of SmHY5 (Sme2.5_03211.1_g00004.1) and SmMYB35 were amplified and ligated to the pB42AD vector. The promoter sequences of anthocyanin structural genes SmCHS, SmF3H, SmDFR, and SmANS as reported previously (Jiang et al. 2016a), and three tandem copies of the G-box-1 and G-box-2 motifs from the SmMYB35 promoter were fused to the pLacZ vector.
Dual-luciferase assay (Dual-LUC)
The dual-luciferase assay experiment as described in my own paper (Li et al. 2021). The CDS of SmHY5, SmMYB35, SmTT8, and SmTTG1 were inserted into a PHB vector as effectors; the empty PHB vector was used as a negative control. The promoter sequences (SmMYB35, SmCHS, SmF3H, SmDFR and SmANS) were cloned and inserted into the cloning sites of the pGreen0800-LUC vector as reporters.
Yeast two-hybrid assay (Y2H)
The yeast two-hybrid experiment was mentioned earlier (Jiang et al. 2016b; Li et al. 2021). The CDS of SmMYB35 and SmTT8 were inserted into the bait vector pGBKT7 (BD) as bait, respectively. SmMYB35 and SmTTG1 were fused to the prey vector pGADT7 (AD) as prey, respectively. Because of the strong autoactivation of full-length SmMYB35, the 60 mmol/L 3-AT solution was used for Y2H screening assays whose procedure was described in the Yeast Protocols Handbook (Takara).
Bimolecular fluorescence complementation (BiFC)
For BiFC assay, the vectors pXY104 and pXY106 were described previously (Jiang et al. 2016b; Li et al. 2021). For the construction of the BiFC vectors, CDS of the SmMYB35 and SmTT8 were cloned into pXY106 to produce SmMYB35-nYFP and, SmTT8-nYFP (the N-terminal fragment of YFP) fusion proteins, respectively. CDS of the SmMYB35 and SmTTG1 were cloned into pXY104, to obtain SmMYB35-cYFP and, SmTTG1-cYFP (the C-terminal fragment of YFP) fusion proteins, respectively. Infiltration and detection were performed the same as subcellular localization experiments described.
Results
SmMYB35 is a nuclear-localized protein and highly expressed in anthocyanin-rich tissues
The subcellular localization of an SmMYB35-YFP fusion protein was determined to discover where the SmMYB35 protein functions within the cell. In contrast to YFP, which was detected in both nucleus and cytoplasm, the SmMYB35-YFP fusion protein was only exclusively observed in the nucleus of N. benthamiana leaf cells (Fig. 1a), which was different to YFP detected in both nucleus and cytoplasm, suggesting that SmMYB35 is a nuclear-localized protein.
Subcellular localization and expression patterns of SmMYB35. a Subcellular localization of the SmMYB35 protein in N. benthamiana leaves epidermal cells. DAPI served as a marker for the nucleus. Scale bars = 22 μm. b, c The expression level of SmMYB35 in different eggplant tissues in ‘Lanshan Hexian’ and ‘140’, respectively. Data are represented by three biological replicates, each of which included three technical replicates. One-way analysis of variance was used to test the statistical significance. which was indicated by different letters (P < 0.05)
The spatial and temporal expression patterns of SmMYB35 were analyzed in different tissues at the same developmental stage of the ‘Lanshan Hexian’ and ‘140’ by qRT-PCR. The expression level of SmMYB35 was high in peel, moderate in stems, and very low in roots of ‘Lanshan Hexian’ (Fig. 1b). The results also showed that SmMYB35 was highly expressed in stems and moderately expressed in petals of '140' (Fig. 1c). SmMYB35 was highly expressed in anthocyanin-rich tissues of both ‘Lanshan Hexian’ and ‘140’.
Overexpression of SmMYB35 increases anthocyanin content and induces the expression of anthocyanin biosynthetic genes in Arabidopsis and eggplant
To study the function of the SmMYB35 in Arabidopsis, the CDS of SmMYB35 driven by the 35S promoter was overexpressed in Arabidopsis (Fig. S1a). The transgenic plants were identified by basta screening to T3 generation. Three independent transgenic lines harboring 35S::SmMYB35 were then randomly selected for further molecular and phenotypic characterization. Compared to wild-type (WT) plants, SmMYB35-OE transgenic plants certified enhanced anthocyanin accumulation in leaves (Fig. S1b). qRT-PCR showed that the expression of key enzyme genes in anthocyanin biosynthesis was increased compared with that of the WT (Fig. S1d). To further verify the function of SmMYB35 in the eggplant cultivar ‘140’ (white peel), the T1 SmMYB35-OE transgenic eggplant was obtained by the cotyledonary explants mediated by Agrobacterium tumefaciens-mediated transformation (Fig. S2a) and was tested by qRT-PCR. Compared to WT, the expression of SmMYB35 and anthocyanin biosynthetic genes were also increased (Fig. S2b). These results suggested that SmMYB35 induced the expression of anthocyanin biosynthetic genes. To further ensure that SmMYB35 acted as an activator in anthocyanin accumulation, the T1 transgenic plant was self-pollinated to obtain homozygous transgenic eggplants. As a result, we observed a higher anthocyanin accumulation in T2 SmMYB35-OE than in WT in stems and petals (Fig. 2a, b). Then, qRT-PCR expression analysis was performed in three randomly selected T2 SmMYB35-OE transgenic eggplants. The transcript levels of SmMYB35 in stems and petals of three transgenic eggplants were considerably higher than in WT (Fig. 2c). The expression of key anthocyanin biosynthetic genes was also drastically increased in three T2 SmMYB35-OE compared to WT (Fig. 2d, e). These results indicated that a significant increase in transcript levels in stems and petals of SmMYB35-OE compared to that of WT. Above all, overexpressing of SmMYB35 in eggplant promoted the expression level of anthocyanin biosynthetic genes and increased the anthocyanin content in stems and petals.
Effects of SmMYB35-OE on eggplant. a Phenotypic difference between WT and SmMYB35-OE. b Quantitative analysis of anthocyanins contents in the stems and petals from WT and SmMYB35-OE. c qRT-PCR analysis of the expression of SmMYB35 in stems and petals of transgenic eggplants. d and e qRT-PCR analysis of the expression of SmCHI, SmCHS, SmF3H, flavonoid 3′5'-hydroxylase (SmF3′5'H), SmDFR, and SmANS in stems and petals of transgenic eggplants, respectively. Data are represented by three biological replicates, each of which included three technical replicates. The error bars indicate the standard errors of the means. One-way analysis of variance was utilized to test the statistical significance, different letters on the bars indicate significant differences (P < 0.05)
SmMYB35 acts as a positive regulator to activate the transcription of SmCHS, SmF3H, SmDFR, and SmANS
To uncover the regulatory mechanism of SmMYB35 on anthocyanin biosynthetic genes (Fig. 3a). We found that SmMYB35 activated the transcription of SmCHS, SmF3H, SmDFR, and SmANS in the yeast one-hybrid assay. As shown in Fig. 3b, SmMYB35 bind to the promoters of anthocyanin structural genes, their combinations exhibited blue color on the defective medium (-TU) with x-Gal. However, negative controls failed to express β-galactosidase. These results suggested that SmMYB35 directly attached to the promoters of SmCHS, SmF3H, SmDFR, and SmANS, respectively.
SmMYB35 acts as a positive regulator to activate the transcription of SmCHS, SmF3H, SmDFR, and SmANS a Schematic representation of the anthocyanin biosynthetic pathway. b SmMYB35 specifically binded to the promoters of SmCHS, SmF3H, SmDFR, and SmANS in one-hybrid system. c Schematic diagram of the reporter and effector constructs used in the Dual-LUC assay. d, e, f and g Transient Dual-LUC assay in N. benthamiana leaves. The promoter activity of SmCHS, SmF3H, SmDFR, and SmANS was measured by a ratio of LUC to REN. Data are represented by three biological replicates. Significance is indicated by different letters (P < 0.05)
To further confirm whether SmMYB35 activates these biosynthetic gene promoters, Dual-LUC was conducted. The promoter of SmCHS, SmF3H, SmDFR, and SmANS were fused to LUC luciferase as a reporter. The SmMYB35 driven by 35S promoter as effector (Fig. 3c). The LUC experiment showed that SmMYB35 could activate the expression of SmCHS, SmF3H, SmDFR, and SmANS, indicating that SmMYB35 was a positive regulator of anthocyanin synthesis (Fig. 3d–g). The above results demonstrated that SmMYB35 stimulated anthocyanin synthesis through binding to promoters of SmCHS, SmF3H, SmDFR, and SmANS, and positively activated the expression of these biosynthetic genes.
SmTT8 and SmTTG1 enhance the regulatory function of SmMYB35 on anthocyanin biosynthetic genes by physical protein–protein interaction
The proteins interacting with SmMYB35 were screened by yeast two-hybrid, to elucidate the molecular basis for its regulatory role in anthocyanin biosynthesis. Selected genes belonging to bHLH and WD40 family, which have been reported in our previous studies on eggplant, were SmTT8 and SmTTG1, specifically. Y2H assays indicated that both SmTT8 and SmTTG1 interacted with SmMYB35 in yeast cells (Fig. 4a). Then BiFC assays were carried out to further confirm these interactions in N. benthamiana leaf cells (Fig. 4b). When SmMYB35-cYFP with SmTT8-nYFP, SmMYB35-cYFP with SmTTG1-nYFP, and SmTT8-cYFP with SmTTG1-nYFP were transiently co-expressed, those YFP fluorescence signals were observed in the nucleus. In contrast, no fluorescence was observed in negative controls. Together, these results demonstrated that two of them interacted with each other in vitro and in vivo.
SmMYB35 interacts with both SmTT8 and SmTTG1, and promotes the transcriptional activation activity of SmCHS, SmF3H, SmDFR, and SmANS. a Y2H assays to detect the interactions between SmMYB35, SmTT8, and SmTTG1 with GAL4 system in yeast cells. b BiFC analysis to detect the pair interactions between SmMYB35, SmTT8, and SmTTG1 in N. benthamiana leaves epidermal cells. Scale bars = 22 μm. c Transient Dual-LUC assay in N. benthamiana leaves. The promoter activity of SmCHS, SmF3H, SmDFR, and SmANS was expressed as a ratio of LUC to REN. Data are represented by three biological replicates. Significance is indicated by different letters (P < 0.05)
To further investigate whether the interactions between SmMYB35, SmTT8, and SmTTG1 affected the transcriptional activation of anthocyanin biosynthetic genes, Dual-LUC assay was conducted. The transcriptional activation of SmCHS, SmF3H, SmDFR, and SmANS was significantly enhanced when any combination of two TFs was injected compared to SmMYB35, SmTT8 or SmTTG1 injection alone, and the transcriptional activation was more significant after the co-expression of the three TFs (Fig. 4c). These results indicated that the interaction between these three TFs could facilitate the SmCHS, SmF3H, SmDFR, and SmANS expression in varied degrees.
SmHY5 is a transcriptional activator of SmMYB35
Our previous bagging experiments demonstrated that the expression level of SmMYB35 was induced by light in eggplant peel. Based on the differential expression of SmMYB35 in transcriptome data at 0d, 5d, and 12d, the expression of SmMYB35 was peaked at 5 d, and down-regulated at 12 d. The qRT-PCR and RNA-seq results of SmMYB35 were highly correlated (r = 0.994, Fig. 5a). To explore the relationship between expression of SmMYB35 and light-exposed time in eggplants, the expression of SmMYB35 at 0, 1, 2, 3, 4, 5, 6, 7, and 8 h was measured. The expression of SmMYB35 peaked at 5 h (the highest light intensity of the day) and gradually turned down after that with weakening light intensity (Fig. 5b). Clearly, the expression of SmMYB35 was triggered when exposed to light.
Expression patterns of SmMYB35 in bagging experiment. a QRT-PCR was performed to validate the RNA-seq results. b The expression patterns of SmMYB35 from nine-time points at 0, 1, 2, 3, 4, 5, 6, 7, and 8 h after bag removal. Data are represented by three biological replicates, each of which included three technical replicates. One-way analysis of variance was used to test the statistical significance which is indicated by different letters (P < 0.05)
To understand the light-response mechanism of SmMYB35, we analyzed the promoter of SmMYB35 and found two G-box motifs (G-box-1 and G-box-2) locating at 795 and 1074 bps, upstream of ATG, respectively, which were the potential binding cis-elements of SmHY5 (Fig. 6a, b). To further investigate whether SmHY5 recognizes the G-boxes harbored in the promoter of SmMYB35, Y1H assays were conducted. The results demonstrated that the pB42AD-SmHY5 fusion protein could strongly activate the expression of LacZ reporter gene through binding to three tandem repeats of the G-box-1 and G-box-2 motifs, indicating that SmHY5 binds to the G-box-1 and G-box-2 motifs in the SmMYB35 promoter, respectively. However, SmHY5 did not bind to the full-length promoter (1766 bps upstream ATG) of SmMYB35 (Fig. 6c). Then, a Dual-LUC assay was performed in N. benthamiana leaves to detect whether SmHY5 activated the full-length promoter of SmMYB35. The promoter of SmMYB35 fused to LUC luciferase acting as a reporter, and REN luciferase driven by 35S promoter acting as an inner reference. The effector constructs contained the SmHY5 driven by 35S promoter (Fig. 6d). The results showed that SmHY5 significantly activated the promoter of SmMYB35 (Fig. 6e). These results indicated that SmHY5 could bind to the promoter of SmMYB35 and stimulate its expression.
SmHY5 intensifies the transcription of SmMYB35 by binding to its promoter. a Cis-acting regulatory elements in SmMYB35 promoter sequences. b Schematic diagrams of the SmMYB35 promoter. The positions of potential HY5 DNA binding sites (G-box-1, G-box-2) are signed by black boxes. c Y1H assays indicate protein-DNA interactions. d Schematic diagram of the reporter and effector constructs used in the Dual-LUC assay. e Transient Dual-LUC assay in N. benthamiana leaves. The promoter activity of SmMYB35 promoter is expressed as a ratio of LUC to REN. Data are represented by three biological replicates. Significance is indicated by different letters (P < 0.05)
Discussion
Fruits with uniform coloration conferred by anthocyanin are more attractive to consumers, for the benefits of anthocyanin to the health of the human body. Anthocyanin biosynthesis is determined by a series of enzymes, which gene expressions are regulated by numerous transcription factor families (Gonzalez et al. 2008; Nuraini et al. 2020). Among which MYB, bHLH and WD40 are important for activating the expression of structural genes, which function in a MBW transcription regulatory complex manner (Li et al. 2020). Usually, environmental factors, such as temperature and light, regulate anthocyanin biosynthesis by modulating the expression of anthocyanin-related genes or protein stabilities of MYB and bHLH (Gu et al. 2019). Light is crucial for the coloration of eggplant, which can drastically induce anthocyanin-related genes expression (Jiang et al. 2016b; Li et al. 2017), demonstrating that the light signal participates in the regulation of anthocyanin synthesis in eggplants.
SmMYB35 is an activator of anthocyanin biosynthesis in eggplant
Many R2R3-MYBs which involved in the regulation of anthocyanin synthesis have been identified at the genetic and molecular levels (Liu et al. 2018b). Based on their well-conserved DNA-binding domains, 73 R2R3-MYB transcriptional factors have been screened in the eggplant database (Wang et al. 2016). SmMYB1 was the earliest discovered and most critical positive-regulator for anthocyanin synthesis (Zhang et al. 2014; Jiang et al. 2016b). The overexpression of SmMYB1 can drive the rich anthocyanin accumulation in all tissues except the fruit flesh (Zhang et al. 2016). In the present study, the tissue expression specificity of SmMYB35 was highly correlative with that of SmMYB1 (Jiang et al. 2016b) and the anthocyanin content reported in different tissues of eggplant (Jiang et al. 2016a), implying that SmMYB35 may play a similar role as SmMYB1 in regulating anthocyanin synthesis. Interestingly, both Y2H and BiFC showed that SmMYB1 and SmMYB35 could interact with each other (Fig. S3), which illustrated our previous study that SmMYB35 increased the anthocyanin accumulation when co-infiltrated with SmMYB1 and SmTT8 in N. benthamiana (Li et al. 2017), suggesting that SmMYB35 acts as an enhancer for SmMYB1 in anthocyanin synthesis in eggplant.
Anthocyanin biosynthesis is generally determined by structural and regulatory genes. Pigment accumulation in plant tissues is mainly controlled by the expression profiles of regulatory genes. SlAN2-like acts as a master regulator that drives rich anthocyanin accumulation both in peel and flesh (Sun et al. 2019). In addition, MdMYB1 and MdMYBA are responsible for red skin coloration (Lin-Wang et al. 2010), while MdMYB10 controls red flesh coloration of apple fruit (Chagné et al. 2007). In the present study, SmMYB35-OE transgenic eggplants displayed a higher anthocyanin accumulation ubiquitously in the stems and petals than those in WT, but not in other tissues (Fig. 4). It is worth noting that most structural genes showed the highest expression levels in the stems and petals in ‘140’ (Fig. S4). Interestingly, both WT and transgenic plants have white fruit peels and no anthocyanin accumulation. According to previous research, the white peel of eggplant resulted from the mutation and low expression level of a major anthocyanin activator SmMYB1 (Zhang et al. 2014). Our investigation declared that the expression of SmMYB1 was low in the peel of WT (Fig. S5), suggesting that overexpression of SmMYB35 may not produce anthocyanin in the peel. The regulatory effect of SmMYB35 on structural genes is weakened, when SmMYB1, a major anthocyanin activator, was absent or lowly expressed in peels. However, other mechanisms cannot be excluded at present. One study also found that VvMYB5a does not participate in the regulation of anthocyanins, whereas VvMYB5b can activate a subset of late biosynthetic genes of the flavonoid pathway (Cavallini et al. 2014). There are also findings that transcription factors activate a precise pathway on post-transcriptional modification. Such as MYB75 phosphorylation by MPK4 is required for full function of MYB75 in Arabidopsis (Li et al. 2016), and apple MPK4 mediates phosphorylation of MYB1 to enhance light-induced anthocyanin accumulation (Yang et al. 2021).
How does SmMYB35 promote anthocyanin accumulation in eggplant
The regulation of anthocyanin biosynthesis in plants has been well documented. There are two mechanisms which MYB activators control anthocyanin biosynthesis by binding to the promoters of the target genes and taking a part in MBW complexes (Koes et al. 2005; Premathilake et al. 2020). Many MYB TFs have also been found to activate the expression of anthocyanin biosynthetic genes in Rosaceae, such as apple, grape, and strawberry (Deluc et al. 2008; Espley et al. 2009; Zhang et al. 2020). MBW complexes could more efficiently activate the promoters of anthocyanin biosynthetic genes than MYB alone (Chagné et al. 2013). Other findings in Arabidopsis thaliana report that MYBs (such as PAP1, TT2, MYB5 and MYB4) and bHLHs (such as GL3, EGL3, MYC1 and TT8) can directly interact with TTG1 to form a MBW complex and have an important transcription role in the biosynthesis of anthocyanin synthesis (Broun 2005). Another study also indicated that AtTT8 exhibited a strong interaction with AtTTG1 in the presence of all the tested R2R3MYBs including AtMYB61 (Zhang et al. 2019). TTG1 often acts as a s caffold for the combinatorial interactions between MYB and bHLH and is essential for regulating the transcriptional activity of MBW complexes (Shan et al. 2019). The interaction among MYB, bHLH and TTG1 is a necessary condition for anthocyanin accumulation. Overall, the regulation of flavonoid synthesis in plants is considered to be the original function of the MBW complex. When a R3-MYB type repressor competes with one of the R2R3-MYB partners in the MBW complex, which blocks the formation of MBW, the expression of anthocyanin biosynthetic genes is attenuated (Nguyen et al. 2015). Hence, there is a dynamic balance in the accumulation of anthocyanin. In the present work, SmMYB35 directly binds to the promoters of SmCHS, SmF3H, SmDFR, and SmANS to stimulate their activities (Fig. 3). In addition, we found that the interactions of SmMYB35 protein with SmTT8 and SmTTG1 formed a SmMYB35-SmTT8-SmTTG1 ternary transcription complex to participate in the regulation of the anthocyanin pathway (Fig. 4). These results elucidated the phenotype of SmMYB35-OE transgenic eggplant, and suggested that the molecular mechanism of MYBs participating in the regulation of anthocyanin biosynthesis is highly conservative among different species.
Light is the most important environmental factors for anthocyanin biosynthesis (Shin et al. 2013). HY5 is a central modulator of light signaling mediating many aspects of light-response by targeting the G-box of the target genes (Lee et al. 2007). Electrophoretic mobility shift assay and ChIP-qPCR assays revealed that SlHY5 could recognize and bind to the G-box and ACGT-containing element contained in the promoters of anthocyanin biosynthesis genes (Liu et al. 2018a, b, c). PyHY5 binding to G-box motifs of the promoters of PyMYB10 enhanced its expression, and then promoted the accumulation of anthocyanin in red pear (Wang et al. 2020), implying that harbor of the G-box in the promoter might be the potential target gene of HY5. Jiang Minming’s study also found that the expression level of SmHY5 displayed no tissue-specificity in ‘Lanshan Hexian’ and increased immediately after bag removal for light irradiation (Jiang et al. 2016b). In the present paper, we found the expression level of SmMYB35 was induced by exposure to light, and was positively correlated with anthocyanin accumulation at 0 d, 5 d, and 12 d after bag removal. Moreover, the expression of SmMYB35 gene was affected by light period and light intensity under natural conditions, and the expression level was highest in the middle of the day (Fig. 5b). Besides, SmHY5 directly binds to the promoter region G-box to induce the transcriptional activation of SmMYB35, thereby mediating light signal modulating the expression of SmMYB35, same as AtMYB75 described in Arabidopsis (Ho et al. 2013). We suggested that anthocyanin biosynthetic genes was regulation via HY5 and via MYB TF.
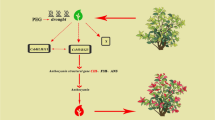
In conclusion, our findings firstly verified that SmMYB35 enhanced anthocyanin biosynthesis in plants and we established a model diagram for analyzing the mechanism of light-responsive transcription factor SmMYB35 involving in regulating anthocyanin biosynthesis in eggplant. In light conditions, nucleus-localized TF SmHY5 binds to the G-box in SmMYB35 promoter and activates the expression of SmMYB35. Then SmMYB35 increases the transcriptional activation of anthocyanin biosynthetic genes by binding to their promoters. Furthermore, the regulatory function was enhanced when SmMYB35 interacts with SmTT8, and SmTTG1 to form a SmMYB35-SmTT8-SmTTG1 transcriptional complex (Fig. 7). This model deepens our understanding of the molecular mechanism of SmMYB35 mediating light modulating anthocyanin biosynthesis in eggplant and provides the basis for innovating anthocyanin-rich eggplant cultivars under low light conditions.
Model of light-induced anthocyanin biosynthesis in eggplant. In light conditions, nucleus-localized TF SmHY5 induced expression of SmMYB35, then positive-regulators such as SmMYB35, SmTT8, and SmTTG1 form a ternary complex induce expression of anthocyanin biosynthetic genes and increased anthocyanin accumulation. Arrows with polyline lines indicate the promoters of SmMYB35 and anthocyanin biosynthetic genes. Straight lines indicate promotion
Author contribution statement
HYC, YL, and LZL conceived and designed the research. LZL SHL, HYG, LDL, and SLS performed experiments. LZL analyzed data and wrote the manuscript. All authors read and approved the manuscript.
Data availability
All data generated or analysed during this study are included in this article [and its supplementary information files].
Abbreviations
- ANS:
-
Anthocyanidin synthase
- CHI:
-
Chalcone isomerasess
- CHS:
-
Chalcone synthase
- DFR:
-
Dihydroflavonol-reductase
- F3H:
-
Flavanone 3-hydroxylase
- HY5:
-
ELONGATED HYPOCOTYL5
- TF:
-
Transcription factor
- YFP:
-
Yellow fluorescent protein
References
An JP, Qu FJ, Yao JF et al (2017) The bZIP transcription factor MdHY5 regulates anthocyanin accumulation and nitrate assimilation in apple. Hortic Res 4:1–9
Belwal T, Singh G, Jeandet P et al (2020) Anthocyanins, multi-functional natural products of industrial relevance. Recent biotechnological advances. Biotechnol Adv 43:107600
Ben-Simhon Z, Judeinstein S, Nadler-Hassar T et al (2011) A pomegranate (Punica granatum L.) WD40-repeat gene is a functional homologue of Arabidopsis TTG1 and is involved in the regulation of anthocyanin biosynthesis during pomegranate fruit development. Planta 234:865–881
Broun P (2005) Transcriptional control of flavonoid biosynthesis: a complex network of conserved regulators involved in multiple aspects of differentiation in Arabidopsis. Current Opin Plant Biol 8:272–279
Cao X, Qiu Z, Wang X et al (2017) A putative R3 MYB repressor is the candidate gene underlying atroviolacium, a locus for anthocyanin pigmentation in tomato fruit. J Exp Bot 68:5745–5758
Cavallini E, Zenoni S, Finezzo L et al (2014) Functional diversification of grapevine MYB5a and MYB5b in the control of flavonoid biosynthesis in a petunia anthocyanin regulatory mutant. Plant Cell Physiol 55:517–534
Chagné D, Carlisle CM, Blond C et al (2007) Mapping a candidate gene (MdMYB10) for red flesh and foliage colour in apple. BMC Genomics 8:1–11
Chagné D, Lin-Wang K, Espley RV et al (2013) An ancient duplication of apple MYB transcription factors is responsible for novel red fruit-flesh phenotypes. Plant Physiol 161:225–239
Deluc L, Bogs J, Walker AR et al (2008) The transcription factor VvMYB5b contributes to the regulation of anthocyanin and proanthocyanidin biosynthesis in developing grape berries. Plant Physiol 147:2041–2053
Dixon RA, Paiva NL (1995) Stress-induced phenylpropanoid metabolism. Plant Cell 7:1085–1097
Espley RV, Brendolise C, Chagné D et al (2009) Multiple repeats of a promoter segment causes transcription factor autoregulation in red apples. Plant Cell 21:168–183
Feller A, MacHemer K, Braun EL, Grotewold E (2011) Evolutionary and comparative analysis of MYB and bHLH plant transcription factors. Plant J 66:94–116
Gonzalez A, Zhao M, Leavitt JM, Lloyd AM (2008) Regulation of the anthocyanin biosynthetic pathway by the TTG1/bHLH/Myb transcriptional complex in Arabidopsis seedlings. Plant J 53:814–827
Gu K-D, Wang C-K, Hu D-G, Hao Y-J (2019) How do anthocyanins paint our horticultural products? Sci Hortic 249:257–262
He YJ, Chen H, Zhou L et al (2019) Comparative transcription analysis of photosensitive and non-photosensitive eggplants to identify genes involved in dark regulated anthocyanin synthesis. BMC Genomics 20:678
Hemm MR, Herrmann KM, Chapple C (2001) AtMYB4: a transcription factor general in the battle against UV. Trends Plant Sci 6:135–136
Ho D, Choi M, Kim K et al (2013) HY5 regulates anthocyanin biosynthesis by inducing the transcriptional activation of the MYB75/PAP1 transcription factor in Arabidopsis. FEBS Lett 587:1543–1547
Jiang MM, Liu Y, Ren L et al (2016a) Molecular cloning and characterization of anthocyanin biosynthesis genes in eggplant (Solanum melongena L.). Acta Physiol Plant 38:1–14
Jiang MM, Ren L, Lian HL et al (2016b) Novel insight into the mechanism underlying light-controlled anthocyanin accumulation in eggplant (Solanum melongena L.). Plant Sci 249:46–58
Kayesh E, Shangguan L, Korir NK et al (2013) Fruit skin color and the role of anthocyanin. Acta Physiol Plant 35:2879–2890
Kiferle C, Fantini E, Bassolino L et al (2015) Tomato R2R3-MYB proteins SlANT1 and SlAN2: Same protein activity, different roles. PLoS ONE 10:0136365
Koes R, Verweij W, Quattrocchio F (2005) Flavonoids : a colorful model for the regulation and evolution of biochemical pathways. Trends Plant Sci 10:236–242
Lee J, He K, Stolc V et al (2007) Analysis of transcription factor HY5 genomic binding sites revealed its hierarchical role in light regulation of development. Plant Cell 19:731–749
Li S, Wang W, Gao J et al (2016) MYB75 phosphorylation by MPK4 is required for light-induced anthocyanin accumulation in Arabidopsis. Plant Cell 28:2866–2883
Li J, Ren L, Gao Z et al (2017) Combined transcriptomic and proteomic analysis constructs a new model for light-induced anthocyanin biosynthesis in eggplant (Solanum melongena L.). Plant Cell Environ 40:3069–3087
Li J, He Y, Zhou L et al (2018) Transcriptome profiling of genes related to light-induced anthocyanin biosynthesis in eggplant (Solanum melongena L.) before purple color becomes evident. BMC Genomics 19:201
Li J, Luan Q, Han J et al (2020) CsMYB60 directly and indirectly activates structural genes to promote the biosynthesis of flavonols and proanthocyanidins in cucumber. Hortic Res 7:1–15
Li LZ, He YJ, Ge HY et al (2021) Functional characterization of SmMYB86, a negative regulator of anthocyanin biosynthesis in eggplant (Solanum melongena L). Plant Sci 302:110696
Lin-Wang K, Bolitho K, Grafton K et al (2010) An R2R3 MYB transcription factor associated with regulation of the anthocyanin biosynthetic pathway in Rosaceae. BMC Plant Biol 10:50
Liu Y, Lin-Wang K, Espley RV et al (2016) Functional diversification of the potato R2R3 MYB anthocyanin activators AN1, MYBA1, and MYB113 and their interaction with basic helix-loop-helix cofactors. J Exp Bot 67:2159–2176
Liu CC, Chi C, Jin LJ et al (2018a) The bZip transcription factor HY5 mediates CRY1a-induced anthocyanin biosynthesis in tomato. Plant Cell Environ 41:1762–1775
Liu Y, Hou H, Jiang X et al (2018b) A WD40 repeat protein from camellia sinensis regulates anthocyanin and proanthocyanidin accumulation through the formation of MYB–bHLH–WD40 ternary complexes. Int J Mol Sci 19:1686
Liu Y, Tikunov Y, Schouten RE et al (2018c) Anthocyanin biosynthesis and degradation mechanisms in Solanaceous vegetables: a review. Front Chem 6:52
Liu Y, Lin-Wang K, Espley RV et al (2019) StMYB44 negatively regulates anthocyanin biosynthesis at high temperatures in tuber flesh of potato. J Exp Bot 70:3809–3824
Nguyen NH, Jeong CY, Kang GH et al (2015) MYBD employed by HY5 increases anthocyanin accumulation via repression of MYBL2 in Arabidopsis. Plant J 84:1192–1205
Nuraini L, Ando Y, Kawai K et al (2020) Anthocyanin regulatory and structural genes associated with violet flower color of Matthiola incana. Planta 251:1–15
Premathilake AT, Ni J, Bai S et al (2020) R2R3-MYB transcription factor PpMYB17 positively regulates flavonoid biosynthesis in pear fruit. Planta 252(4):59
Shan X, Li Y, Yang S et al (2019) A functional homologue of Arabidopsis TTG1 from Freesia interacts with bHLH proteins to regulate anthocyanin and proanthocyanidin biosynthesis in both Freesia hybrida and Arabidopsis thaliana. Plant Physiol Bioch 141:60–72
Shin DH, Choi M, Kim K et al (2013) HY5 regulates anthocyanin biosynthesis by inducing the transcriptional activation of the MYB75/PAP1 transcription factor in Arabidopsis. FEBS Lett 587:1543–1547
Shoeva OY, Glagoleva AY, Khlestkina EK (2017) The factors affecting the evolution of the anthocyanin biosynthesis pathway genes in monocot and dicot plant species. BMC Plant Biol 7:5–14
Sun C, Deng L, Du M et al (2019) A transcriptional network promotes anthocyanin biosynthesis in tomato flesh. Mol Plant 13:42–58
Tang B, Li L, Hu Z et al (2020) Anthocyanin accumulation and transcriptional regulation of anthocyanin biosynthesis in purple pepper. J Agr Food Chem 68:12152–12163
Toledo-Ortiz G, Huq E, Quail PH (2003) The Arabidopsis basic/helix-loop-helix transcription factor family. Plant Cell 15:1749–1770
Wang SJ, Chen Z, Ji T et al (2016) Genome-wide identification and characterization of the R2R3MYB transcription factor superfamily in eggplant (Solanum melongena L.). Agri Gene 2:38–52
Wang Y, Zhang X, Zhao Y et al (2020) Transcription factor PyHY5 binds to the promoters of PyWD40 and PyMYB10 and regulates its expression in red pear 'Yunhongli No. 1’. Plant Physiol Bioch 154:665–674
Xu W, Grain D, Bobet S et al (2014) Complexity and robustness of the flavonoid transcriptional regulatory network revealed by comprehensive analyses of MYB–bHLH–WDR complexes and their targets in Arabidopsis seed. New Phytol 202:132–144
Xu P, Zawora C, Li Y et al (2018) Transcriptome sequencing reveals role of light in promoting anthocyanin accumulation of strawberry fruit. Plant Growth Regul 86:121–132
Xu H, Zou Q, Yang G et al (2020) MdMYB6 regulates anthocyanin formation in apple both through direct inhibition of the biosynthesis pathway and through substrate removal. Hortic Res 7:1–17
Yang T, Ma H, Li Y et al (2021) Apple MPK4 mediates phosphorylation of MYB1 to enhance light-induced anthocyanin accumulation. Plant J 106:1728–1745
Zhang YJ, Hu Z, Chu G et al (2014) Anthocyanin accumulation and molecular analysis of anthocyanin biosynthesis-associated genes in eggplant (Solanum melongena L.). J Agr Food Chem 62:2906–2912
Zhang YJ, Chu G, Hu Z et al (2016) Genetically engineered anthocyanin pathway for high health-promoting pigment production in eggplant. Mol Breeding 36:54
Zhang B, Chopra D, Schrader A et al (2019) Evolutionary comparison of competitive protein-complex formation of MYB, bHLH, and WDR proteins in plants. J Exp Bot 70:3197–3209
Zhang Z, Shi Y, Ma Y et al (2020) The strawberry transcription factor FaRAV1 positively regulates anthocyanin accumulation by activation of FaMYB10 and anthocyanin pathway genes. Plant Biotechnol J 18:2267–2279
Zhou L, He YJ, Li J et al (2019) CBFs function in anthocyanin biosynthesis by interacting with MYB113 in eggplant (Solanum melongena L.). Plant Cell Physiol 61:416–426
Acknowledgements
The project was supported by the National Natural Science Foundation of China and Shanghai Agriculture Applied Technology Development Program, China, (Grant No.201902080008F01106).
Funding
The project was supported by the National Natural Science Foundation of China (31872944) and Shanghai Agriculture Applied Technology Development Program, China (Grant No.201902080008F01106).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare no conflicts of interest.
Additional information
Communicated by Dorothea Bartels.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Li, L., Li, S., Ge, H. et al. A light-responsive transcription factor SmMYB35 enhances anthocyanin biosynthesis in eggplant (Solanum melongena L.). Planta 255, 12 (2022). https://doi.org/10.1007/s00425-021-03698-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00425-021-03698-x