Abstract
The woodland strawberry, Fragaria vesca, is a model plant for the diverse Rosaceae family, which contains many valuable fruit and ornamental crops. Light is an important external environment factor which influences all aspects of plant growth and development, including fruit ripening. Transcription regulation of light will provide insights into effect of light for fruit ripening. We treat strawberry fruit with dark or light, and by the RNA-Seq method, compare transcriptional level between dark-treated and light-treated strawberry fruits. Additionally, we detect anthocyanin and sugar accumulation in two-treated conditions of fruits. Moreover, we detect the protein change of FvMYB10, a key regulator of anthocyanin biosynthesis, under dark and light. Light can promote anthocyanin and soluble sugar accumulation in mature fruit, and also increases the expression of aroma-related genes and stabilizes FvMYB10 protein. Our findings reveal that light is essential for anthocyanin and sugar accumulation and regulates FvMYB10 at transcriptional level and post-translation level.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Fruit color of strawberry is an outward appearance, and is an important consideration factor for consumers who easily conflate fruit taste, aroma and nutrition with the color of fruit-skin. The major types and quantities of flavonoids, which contain flavonols, anthocyanins, and proanthocyanidins, control the color of strawberry fruit (He and Giusti 2010). Anthocyanins which were main pigments, were synthesized by flavonoid pathway, and determined the red, purple and blue colors of many fruits, leaves and flowers (Lin-Wang et al. 2014). Flavonoid compounds have multiple functions. They are potent antioxidants in vitro, and they effectively quench free radicals and terminate the chain reaction that is responsible for the oxidative damage (He and Giusti 2010). In grape, it influences the taste and quality of wine as well as preservation time. Additionally, flavonols also protect plants from ultraviolet (UV) and free-radical damage (Zoratti et al. 2014). Light is an important environment factor to not only regulate growth, development and metabolism of flowering plants like Arabidopsis, but also fruit trees (Li et al. 2012; Maier et al. 2013).
Plants have evolved extremely delicate light sensing and light signal transduction mechanism. It has been identified that plants can receive light by different photoreceptors, including blue/ultraviolet (UV)-A light receptors cryptochromes and phototropins, the red and far-red light perceiving phytochromes and the UV light-sensing photoreceptor UVR8 (Chaves et al. 2011; Demarsy and Fankhauser 2009; Heijde and Ulm 2012; Nagatani 2010). Light plays an important role in each stage of plants, such as seeds germination, hypocotyl elongation, cotyledon expansion, phototropism, chloroplast movement, stomatal opening and development, shade-avoidance response, flowering time, circadian rhythms, plant defense against UV-B and flavonoid and anthocyanin biosynthesis (Banaś et al. 2012; Briggs and Christie 2002; Chen and Chory 2011; Christie et al. 2012; Kang et al. 2009). In the light signal transduction, CONSTITUTIVE PHOTOMORPHOGENIC1 (COP1), an E3 ligase, is a switch of light-regulated plant growth and development. In the darkness, COP1 mainly localizes to the nucleus and interacts with many transcription factors, such as ELONGATED HYPOCOTYL5 (HY5), and its homolog HYH, plus LONG AFTER FAR-RED1 (LAF1), LONG HYPOCOTYL IN FAR- RED1 (HFR1), BLUE INSENSITIVE TRAIT1 (BIT1), and CONSTANS (CO), and subsequently mediates their ubiquitination and degradation by the 26S proteasome pathway (Hong et al. 2008; Liu et al. 2008; Osterlund et al. 2000; Seo et al. 2003; Yang et al. 2005). In the light, active photoreceptors transform from cytoplasm to nucleus, and interact with COP1, which promotes COP1 localization to cytoplasm, inhibits COP1 activity and allows the protein of transcription factors to reaccumulate (Lian et al. 2011; Liu et al. 2011; Lu et al. 2015; Zheng et al. 2013).
It has shown that anthocyanin biosynthesis is controlled by a series of key structural genes that encode for important enzymes in biosynthesis pathway and are regulated by the transcription factors containing MYB, bHLH, and WD-repeat proteins which consist of “MBW” complex (Allan et al. 2008; Baudry et al. 2006; Butelli et al. 2008; Jaakola 2013). In Arabidopsis, COP1 interacts with the PRODUCTION OF ANTHOCYANIN PIGMENT1 (PAP) protein family of R2R3 MYB transcription factors (PAP1, PAP2, MYB113 and MYB114) which regulate anthocyanin biosynthesis pathway by “MBW” complex including TTG1 and TTG8, and mediates its ubiquitination and degradation in darkness (Gonzalez et al. 2008; Zhang et al. 2003; Zimmermann et al. 2004). Conversely, PAP protein is accumulated under light and the expression of PAP1, PAP2 and TT8 is induced by light, while the expression of TTG1 is light-independent (Cominelli et al. 2008). Moreover, the over-expressing AtPAP1 and AtPAP2 transgenic tobaccos show purple plants and flowers, respectively (Borevitz et al. 2000). Among fruit trees, the MYB TFs of apple and grape are the most-studied species regard to the regulation of anthocyanin biosynthesis. As are reported that MdMYB1, MdMYBA, MdMYB10, VvMYBA1 and VvMYBA2 are relative to anthocyanin biosynthesis in fruit skin (Azuma et al. 2012; Ban et al. 2007; Feng et al. 2013; Jeong et al. 2004; Takos et al. 2006), and VvMYBF1, VvMYB12, VvMYB5a and VvMYB5b are reference to flavonol biosynthesis in fruit skin (Azuma et al. 2012; Matus et al. 2009, 2010). MdMYB9, MdMYB10 and VvMYBPA1, VvMYBPA2 regulate proanthocyanidin biosynthesis in leaves and fruit skin, respectively (Azuma et al. 2012; Gesell et al. 2014; Koyama et al. 2012). Additionally, overexpression of MdMYB10 in a white-fleshed, green-leaved cultivated species of RoyalGala, results in a red flesh and leave phenotype (Espley et al. 2007). Meanwhile, it is reported that the stability of MdMYB1 is regulated by light in accompany with the interaction between MdCOP1/2 and MdMYB1 (Li et al. 2012). Similarly, in Rosaceae, many MYB TFs have also been found to regulate anthocyanin accumulation and fruit coloration, such as pear, peach, blueberry and strawberry (Aharoni et al. 2001; Butelli et al. 2012; Lin-Wang et al. 2010; Zifkin et al. 2012). In the Fragaria genus, the accumulation of anthocyanin, which is the richest flavonoid-derived pigment in Fragaria fruits (Hannum 2004), controls the fruit color. As previous studies show that over-expression of MYB transcription factors FaMYB10 and FvMYB10 in F. ananassa and F. vesca, respectively, result in accumulation of fruit anthocyanin in plants (Lin-Wang et al. 2010). Furthermore, FvMYB10 can activate the expression of anthocyanin biosynthesis genes, such as Arabidopsis DFR, F. vesca DFR and UFGT, but only in the presence of FvbHLH33, which demonstrates FvMYB10 acts together with bHLH transcription factor FvbHLH33 (Lin-Wang et al. 2014). Besides the activator of anthocyanin biosynthesis, the repressors are also identified within the MYB TFs. For example, Arabidopsis MYB TFs AtMYB3, AtMYB4, and AtMYBL2(Dubos et al. 2008; Jin et al. 2000), MdMYB16, MdMYB17, and MdMYB111 from apple (Lin-Wang et al. 2011), and FaMYB1 and FcMYB1 in strawberry (Aharoni et al. 2001; Salvatierra et al. 2013) has been verified for negative regulation of anthocyanin biosynthesis. A recent study is published by Hawkins et al. and they sequence and compare the genomes of three F. vesca accessions: ‘Hawaii 4’, ‘Rügen’, and ‘Yellow Wonder’. Through systematic analysis of SNP, they find a SNP in FvMYB10 is responsible for the yellow color fruits, which shows that FvMYB10 is a key transcription factor of regulating anthocyanin biosynthesis (Hawkins et al. 2016). There are some evidences that light conditions not merely have effect on the accumulation of anthocyanin but also the sugar accumulation of fruits. The apple fruit bagged reduces the soluble solids in the fruit (Kikuchi et al. 1997). The sugar content in fruits of outward canopy is higher than fruits of inward canopy (Chen et al. 2001). Similarly, contrast to shading treatment, sugar quantity is more than 30% under normal light illumination (Giliberto et al. 2005).
In this study, the diploid strawberry F. Vesca is used as a model for exploring the effect of light on anthocyanin accumulation of strawberry fruit. We sequence and compare the transcriptome difference of fruits under light and darkness in different development phase. Meanwhile, we also analyze the co-regulated genes by light and FvMYB10 and find the major co-regulated genes are regulated in a similar manner, especially the key anthocyanin biosynthesis genes. Furthermore, the expression of FvMYB10 is not only up-regulated, but also the protein of FvMYB10 is stabilized by light. Moreover, we also find light promotes the accumulation of sugar in ripen fruit in association with change of sugar metabolism genes.
Materials and methods
Plant materials and growth conditions
Fragaria vesca seeds of RG (Ruegen, the first modern cultivar, i.e., runnerless, everbearing and red fruited, originated from Castle Putbus in Germany) were kindly provided by Dr Chunyin Kang from Huazhong Agricultural University. RG were grown in flowerpots and cultivated under continuous light. The fruits of growth for ten days after flowering were wrapped in three layers of foil for dark treatment. The fruits of light- and dark-grown were collected until the fruits under light grew to mature stage with a completely red coloration, and immediately frozen in liquid nitrogen, then stored at − 80 °C until use.
RNA-Seq and transcriptome analysis
Total RNA was extracted using a method modified from Chang et al. The DNase I was used for treating total RNA samples at 37 °C for 30 min. And then the mRNA was purified by Dynabeads® Oligo (dT)25 (Life Technologies, USA). The quality of RNA was detected by an Agilent 2100 Bioanalyzer (Agilent Technologies, Palo Alto, USA). The cDNA libraries were prepared by using NEBNext® Ultra™ RNA Library Prep kit and sequenced for paired-end reads on Illumina HiSeq™ 2500. Raw reads from each library were filtered separately. The adaptor sequences and reads with unknown sequences “N” larger than 5% were removed. Besides that, low quality reads that contained more than 20% (Q < 20 bases) were discarded. After filtering, all clean reads were quality assessed using software FastQC (Andrews 2014). The reads were mapped to the strawberry reference genome (Fragaria vesca v1.1) and genes expression level were estimated with RPKM (reads per kilo base per million mapped reads). Differential expressed genes (DEGs) between dark and light were analyzed through DEGseq (Wang et al. 2010) and the statistical tests were revised for multiple testing with the Benjamina–Hochberg false discovery rate (FDR < 0.05). Significantly differentially expressed genes were selected by fold change ≥ 2 and adjusted p value ≤ 0.05. GOseq (Young et al. 2010) was used to perform GO functional classification of DEGs to predict possible functions and the KEGG database was used to annotate the pathways involving DEGs with an E-value threshold of 10−5. The Illumina raw data sets have been deposited in the NCBI sequence read archive (SRA) under accession number SRP132167.
Total anthocyanin and soluble sugar contents analysis
Total anthocyanin content was measured following a previous method from Chory (Chory 1992) with some modifications as described by Li (Li et al. 2014). For the soluble sugar content analysis, the dark and light samples were ground to powder with liquid nitrogen. The 0.5 g powder were mixed with 20 ml sterile distilled water, incubated in a boiling-water bath for 30 min, and then filtered by three layers of filter paper; the residual was washed twice with 5 ml of sterile distilled water; the volume was increased to 50 ml which was as soluble sugar extraction. 1 ml extraction was diluted to 50, 1 ml diluent and 10 ml anthrone reagent (100 mg anthrone was dissolved in 100 ml concentrated sulfuric acid) were mixed and heated in boiling-water bath for 10 min, and then immediately cooled in ice bath. The absorbance of reaction mixture was detected at 620 nm and determined the soluble sugar content.
Quantitative RT-PCR (qRT-PCR) analysis
Total RNA from the dark and light samples were extracted following the manufacturer’s instructions of RNA plant Plus Reagent (Tiangen, Beijing, China, Cat.DP437). DNase I was used to remove the genomic DNA contamination and first-strand cDNA was synthesized using iScript™ Reverse Transcription Supermix for RT-qPCR (Bio-Rad, USA). Real-time qPCR was carried out using the CFX96 Touch™ Real-Time PCR System (Bio-Rad, USA) with SYBR® Premix Ex Taq™ II (Takara, China). The qPCR reaction was incubated 1 min at 95 °C, followed by 45 cycles of 5 s at 95 °C, 15 s at 60 °C, and 15 s at 72 °C, followed by 65–95 °C melting curve detection. Transcript abundance of target genes were calculated relative to the internal control gene of Fv26S. All primers sequences were listed in Supplementary Table S1.
Acquisition of transgenic lines and MYB10 protein accumulation
The full length CDS of FvMYB10 was obtained by PCR with primers MYB10-F (5′-ATGGAGGGTTATTTCGGTGTGAGAA-3′) and MYB10-R (5′-TACGTAGGAGATGTTGACTAGA-3′), and then inserted into the plant expression vector of PHB-YFP, which expressed MYB10-YFP fusion protein under 35S promotor. The reconstruct expression vector of 35S:strMYB10-YFP was transferred into Agrobacterium tumefaciens strain GV3101, which was used to infect Arabidopsis wild type (WT) by dipping flowers. For the FvMYB10 protein accumulation analysis, the seeds of T2 transgenic lines were sterilized with 20% bleaching water for 15 min and washed by sterile water with five times. Then the seeds were plated on 1/2 Murashige and Skoog basal medium (MS, Sigma-Aldrich) with 1% sucrose. After 4 days’ stratification at 4 °C, the seeds were exposed to 4 h white light followed by 5 days’ growth in darkness before moved into light for different time light treatment. The seedlings were collected in nitrogen for protein extraction. Western blotting was described previously (Li et al. 2014). GFP antibody was used to specifically detect MYB10-YFP protein and ACTIN protein was as internal control.
Results
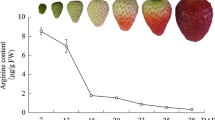
Determination the anthocyanins in dark- and light-grown fruits
Ten days’ fruits after flowering were treated with dark and light until ripening, and under continuous light, the fruits had red skin, compared with white skin in fruits under dark (Fig. 1a). By the colorimetric method, we detected the anthocyanin content, and significant variation in the amounts of anthocyanin were observed between dark- and light-grown fruits (Fig. 1b), which was consistent with the significant difference of skin color. These results indicated that light promoted anthocyanin accumulation in strawberry fruits.
Differently expressed genes analysis and go functional classification
To further explore the transcriptional regulatory mechanism of light on strawberry fruits ripening, strawberry fruits treated by dark and light were obtained for transcriptome sequencing. Over 94.37 million raw reads were generated and approximate 75% reads were mapped to F. vesca reference genome (Supplementary Table S2). By the DESeq software, we totally identified 4177 DEGs (differently expressed genes) between dark and light samples. Among the DEGs, 1756 genes in light samples were greater than two-fold differences than those in the dark samples, while 2421 genes were down-regulated by more than two-fold (Supplementary Table S3). GOseq was used to analysis distribution of DEGs in Gene Ontology, which illuminated gene functions of DEGs. In the 4177 DEGs, 2807 genes were successfully assigned to three categories including biological process, cellular component and molecular function with 47 groups (Fig. 2 and supplementary Table S4). In the biological process, the major DEGs were assigned into “metabolic process”, “cellular process” and “single-organism process”. Within the cellular component category, many DEGs were classified in “cell”, “cell part”, “membrane” and “membrane part”. For the molecular function category, the subcategories of “catalytic activity” and “binding” were mostly related. In the DEGs, we found the key structural enzyme genes controlling anthocyanin biosynthesis, such as CHS (gene26825, gene26826), CHI (gene23367), F3H (gene14611), DFR (gene15174, gene15176) and LDOX (gene32347), were up-regulated in light-treatment fruits more than two-fold change, which was consistent with anthocyanins accumulation promoted by light. The detailed information about gene expression ratios was presented in Supplementary Table S3.
Go analysis of all genes and DEGs in strawberry. The results are summarized in three main GO categories: cellular component, molecular function, and biological process. All Gene means the gene number base on all strawberry genes. DEG gene means the gene number base on the DEG of light versus dark. (Color figure online)
KEGG pathway analysis of differently expressed genes
In order to analyze the biochemical pathways which were affected by light signal, we mapped DEGs to KEGG database for further analysis. A total of 492 DEGs were assigned into 98 KEGG pathways, the major of which were related with metabolism pathways, and 16 KEGG pathways including 316 DEGs were enriched with corrected-p value < 0.05 (Fig. 3 and Supplementary Table S5). The KEGG pathways analysis revealed that the major genes were involved in “carbon metabolism (50 genes)”, “plant hormone signal transduction (42 genes)”, “phenylalanine metabolism (37 genes)”, and “starch and sucrose metabolism (35 genes)”. There were 14 genes were significantly enriched in flavonoid biosynthesis pathway, but this pathway had the highest enrichment factor of 3.98, and these genes were related to anthocyanin biosynthesis and mainly included structural flavonoid genes.
Validation DEGs involved in anthocyanin biosynthesis
To verify RNA-Seq results, we selected some flavonoid pathway genes related to anthocyanin biosynthesis and detected their expression level in dark and light strawberry fruits by qRT-PCR assay. As shown in Fig. 4, the transcript level of the five flavonoid pathway genes were significantly up-regulated in light fruits, which was identical with RNA-Seq results.
Soluble sugar analysis in fruits
The KEGG pathways enrichment analysis showed that there were 50 and 35 genes related with carbon metabolism and starch and sucrose metabolism, respectively. Given sugars had traditionally been regarded as the metabolic resources required for carbon skeleton construction and energy supply in plants, we analyzed total soluble sugar content in dark and light fruits by colorimetric method. The result showed that the ripe fruits under light contained 85.7 mg per g fresh weigh soluble sugar, which was higher than 17.3 mg per g fresh weigh of white fruits under dark (Fig. 5). The result indicated that light promoted accumulation of soluble sugar in mature fruits.
Classify transcription factor families of differently expressed genes
Transcription factors (TFs) played an important role in transcriptional regulation. In F. vesca, a total of 1485 TFs were identified and had been submitted to the Plant TFDB (plant transcription factors database). 251 differently expressed transcription factor genes which were regulated by light, were identified, and they were classified into 42 transcription factor families (Fig. 6 and Supplementary Table S6). In the 42 TF families, MYB and bHLH families respectively contained 29 and 27 TFs which were the most two families. Following five families including ERF, NAC, C2H2, WRKY and bZIP contained 10 TFs at least. The detailed quantity information was showed in the supplementary Table S6. The result of transcription factor classification suggested that light mainly regulated the expression level of MYB and bHLH TFs in mature fruits which was coincide with MBW (MYB-bHLH-WD40) complexs regulation of anthocyanin biosynthesis genes (Li 2014; Schaart et al. 2013).
Light and FvMYB10 synergistically regulate the expression of anthocyanin biosynthesis genes
Among the above mentioned 29 MYB TFs, the gene of FvMYB10, whose over-expression lines up-regulated anthocyanin biosynthesis genes including CHS, CHI, F3H, DFR and LDOX and promoted anthocyanin accumulation in strawberry fruits (Lin-Wang et al. 2014), was up-regulated with more than 16 folds in our RNA-Seq data, and we also confirm that light promotes the transcription level of FvMYB10 by qRT-PCR assay (Supplementary Fig. S1). Given light also enhanced anthocyanin content in fruits, light maybe regulated anthocyanin biosynthesis genes in synergistic action with FvMYB10. We acquired the FvMYB10-regulated genes from previous study (Lin-Wang et al. 2014), comparing the genes regulated by light and FvMYB10, and found 892 genes were co-regulated (Fig. 7a). Additionally, the heatmap showed that the major genes were regulated in the same way (Fig. 7b), which suggested that light and FvMYB10 participated same transcriptional process. Furthermore, the related to anthocyanin biosynthesis genes were co-regulated by light and FvMYB10 and the coregulated genes information was shown in Supplementary Table S7. These results, in addition to the up-regulation of FvMYB10 by light, suggested that light may regulate anthocyanin accumulation through FvMYB10.
Both light and FvMYB10 co-regulated genes and light promotes FvMYB10 protein accumulation. a The venn diagram shows the overlaps between light- and FvMYB10-regulated genes. b The heatmap analysis of co-regulated genes by light and FvMYB10. Red and green colors represent induced and repressed genes, respectively. Scale bar denotes the log2 value of fold change. c Light promotes FvMYB10 protein accumulation. Five-day-old dark-grown FvMYB10-OX transgenic Arabidopsis seedlings were exposed to white light (100 µmol/m2/s) for 6 h or 12 h, then MYB10 protein was analyzed by western blotting with GFP antibody. Actin serves as loading control. (Color figure online)
Light promotes the protein accumulation of FvMYB10
By blasting MYB10 protein sequence homology with Arabidopsis, we found that AtPAP1 and AtPAP2 was most similar with the FvMYB10. It had been reported that light promoted AtPAPs protein accumulation (Maier et al. 2013), and we also explored whether light promotes accumulation of FvMYB10. We over-expressed FvMYB10 tagged with YFP in Arabidopsis wild type (35S::MYB10-YFP/WT) and analyzed FvMYB10 protein content in transgenic lines under dark and light. As shown in Fig. 7c, 6 and 12 h white light illumination obviously promoted FvMYB10 accumulation in over-expression lines, which indicated that light had the ability to stabilize FvMYB10 protein. These results implied that light regulated FvMYB10 both at transcription level and post-translation level.
The influence of light in the different development period of strawberry
As we all known, the whole process of plant growth and development, from seeds germination to fruits ripening, had always been accompanied by light. In order to explore the effect of light for the growth of strawberry fruits, we analyzed transcriptome changes by comparing different development stage fruits including 15 days’ green fruits (RG15d) after pollination, turning-color stage (RGTurning) and red ripeness fruits with dark fruits. The RNA-Seq datas of RG15d and RGTurning comed from previous study (Hawkins et al. 2017). The DEGs analysis of RG15d and RGTurning compared with dark was same as light compared with dark. The results showed light regulated a large number of genes in different periods of development of fruit (Fig. 8a). In the green fruits, 6444 genes were regulated, and 3172 genes and 4177 genes were influenced in turning-color fruits and mature fruits by light, respectively. Among three stages of fruits, 627 genes were co-regulated by light with a strikingly similar manner (Fig. 8b). The detail genes expression level was presented in Supplementary Table 8. Gene Ontology (GO) analyses demonstrated that the co-controlled 627 genes were mainly relative to oxidation–reduction process, plant-type secondary cell wall biogenesis and anthocyanin-containing compound biosynthetic process (supplementary Table S9). Surprisingly, the anthocyanin biosynthesis relative genes which included CHS, CHI, F3H, DFR and LODX were up-regulated, which suggested that light involved in anthocyanin synthesis in different development stages.
DEGs regulated by light in different stages. a The venn diagram shows the overlaps light-regulated genes among different stages. b The heatmap analysis of co-regulated genes by light in different stages. Red and green colors represent induced and repressed genes, respectively. Scale bar denotes the log2 value of fold change. (Color figure online)
In the green stage, the fruits swelled quickly in association with cell expansin. Totally, 11 expansin genes were influenced at this period where 8 genes were only changed. Most of expansin genes were down-regulated by light which inhibited the transcription level of expansin genes in seedlings of Arabidopsis (He et al. 2015), and the detail information was showed in Supplementary Table S10. The results indicated light inhibited the expression of expansin genes at early-stage of fruit development. Usually, the fruits ripened in accompany with fruit softening and accumulation of sugar which were relative to cell wall degradation and sugar metabolism, respectively. And we analyzed expression change of genes including pectate lyase (PL) (Castillejo et al. 2004), polygalacturonase (PG) (Figueroa et al. 2008), pectin methylesterase (PME) (Brummell and Harpster 2001), acid invertase (AI), sucrose synthase (SS) and sucrose phosphate synthase (SPS) (Bhatia and Singh 2002). As shown in supplementary Table S10, the major cell wall metabolism genes were up-regulated at mature fruit. For the sugar metabolism, AI and SPS were also up-regulated at red ripen or had no obvious change except that gene32247 was down-regulated at green fruit rather than up-regulated at turning-color stage. In contrast to AI and SPS, SS were almost down-regulated at three development stages. These results indicated that light also regulated the expression level of genes involved in fruit softening and sugar accumulation. In addition to fruit ripening, tens if not hundreds of flavor and aroma compounds were generated during ripening phase. By the RNA-Seq data, we found alcohol acyltransferase (AAT), quinone oxidoreductase (QR) and sesquiterpene synthase (PINS), which were relative to flavor and aroma compounds of fruit (Aharoni et al. 2004; Cumplido-Laso et al. 2012; Raab et al. 2006), were up-regulated in ripen fruit and down-regulated or no change in green fruit (Supplementary Table S10).
Discussion
Effect of light on flavonoid biosynthesis in fruits
Phenolic compounds are a large group in plants secondary metabolites, including flavonoids and hydroxycinnamic acids. Anthocyanins, belonging to flavonoids subfamily, are main pigment in many flowers, fruits and vegetables (He and Giusti 2010). These pigments have multiple functions, such as providing protection from UV-damage, attracting animals and insets to plants which is important for pollination and seeds dispersal, and benefiting for human health (Zoratti et al. 2014). In our study, through the light and dark treatment for strawberry fruits, we find that the fruit under light has a red-skin rather than a white-skin under dark (Fig. 1a). Subsequently, the detection of anthocyanin content between light and dark treatment fruits also shows light promotes anthocyanin accumulation in fruits (Fig. 1b). By the RNA-Seq analysis, we analyze the regulation of light for fruit at transcriptome level. And we find light promotes many anthocyanin biosynthesis genes expression, such as CHS, F3H, CHI, DFR and LODX (Fig. 4 and supplementary Table S3). Given solar light has a widespread spectral range perceived by different photoreceptors, light quality and quantity have different effects on anthocyanin biosynthesis (Zoratti et al. 2014). Kadomura-Ishikawa et al. treat white-stage strawberry fruits by using different wavelength light including blue light, green light, red light, white light, and plus dark. They find light treatment, especial blue light, can increase anthocyanin content compared to darkness, and the expression of FaCHS is increased significantly after 4 days of treatment (Kadomura-Ishikawa et al. 2013), which is consistent with our RNA-Seq and qRT-PCR results. Interestingly, Kondo et al. also treat the plants with a red or blue light-emitting diode (LED) for 3 h before sunrise and 3 h after sunset. And in contrast, anthocyanin concentrations are highest in blue LED-treated skin, followed by red LED treatment. Meanwhile, the content of different pigments is detailedly quantified and especially the amount of malvidin-based anthocyanin increased toward harvest in blue and red-LED treated skin, unlike in untreated controls (Kondo et al. 2014). Anthocyanin content shows significant differences in strawberry fruits grown by using different colored light-quality selective plastic films. In the same study, significantly higher anthocyanin content in fruits treated with red and yellow films are observed than that of the control (white film), and significantly lower anthocyanin content after treatment with the green and blue films (Miao et al. 2016). Covering fruits with colored films, for example, red-colored film, improves the shortwave radiation and makes much more shortwave light penetrate and absorbed by fruits. These results imply that shorter wavelength shows the most prominent effect in the accumulation of flavonoids in fruits.
The mechanism of light-controlled flavonoid biosynthesis has some progress at molecular level in fruits. The plants of Tomato CRY2 overexpression show a high-pigment phenotype, resulting in overproduction of anthocyanins and chlorophyII in leaves and of flavonoids in fruits (Giliberto et al. 2005). In strawberry, FaPHOT2 knockdown results in decreased anthocyanin content; however, overexpression of FaPHOT2 increases the anthocyanin content (Kadomura-Ishikawa et al. 2013). These findings suggest blue light induced anthocyanin accumulation, and FaPHOT2 may play a role in perceiving blue light, and mediating anthocyanin biosynthesis in fruits. Interestingly, the regulation of light for flavonoid compounds has been shown to continue in post-harvest fruits. Xu et al. find that blue light treatment improved total anthocyanin content in strawberry fruit during storage at 40 µmol m−2 s−1 for 12 days at 5 °C. Meanwhile, the treatment increases the enzyme activities of glucose-6-phosphatase, phenylalanine ammonialyase, cinnamate-4-hydroxylase, chalcone synthase, flavanone-3-β-hydroxylase, anthocyanin synthase, and UDP-glycose flavonoid-3-O-glycosyltranferase (Xu et al. 2014). Our results and previous findings provide a method that increasing light illumination, especially blue light illumination, during fruit grown and storage may improve fruit quality in terms of nutritional value and content of bioactive compounds.
Regulation of fruit flavonoid biosynthesis by light
A multitude of well-characterized enzymes, whose expression are specifically induced by a ternary protein complex composed of R2R3-MYB, bHLH, and WD40 (MBW complexes), which are well-conserved in higher plants (Schaart et al. 2013). In Arabidopsis, three R2R3-MYB proteins (MYB11, MYB12, and MYB111) control flavonol biosynthesis via activating the early biosynthetic steps, whereas the production of anthocyanins and PAs requires the MYB-bHLH-WD40 (MBW, including AtTT2 (AtMYB123), AtTT8 (AtbHLH042), and AtTTG1) complex to activate the late biosynthetic genes (Li 2014). Our RNA-Seq data shows that light promotes the expression of many MYB and bHLH transcription factors (Fig. 6 and Supplementary Table S6), which are exactly members of MBW complex. The strawberry genes of FaMYB9/FaMYB11, FabHLH3 and FaTTG1 are the respective functional homologues of AtTT2, AtTT8 and AtTTG1, which specifically induce the expression of the proanthocyanidins (PAs) biosynthesis genes in Arabidopsis. In unripe strawberries, the flavonoids are mainly represented by PAs, while in ripe fruits the red-coloured anthocyanins also accumulate (Schaart et al. 2013). Transgenic strawberry ripe fruits of overexpression of the Arabidopsis PA biosynthesis regulators AtTT2, AtTT8 and AtTTG1 show a loss of red coloration in the flesh associated with an increased PA content and a decreased anthocyanin accumulation contrast to control fruits (Schaart et al. 2013). However, in these transgenic plants, the expression level of FaF3H is slightly increased, FaANS, FaANR and FaLAR, which are the key enzymes of PA biosynthesis, transcript accumulation was strongly increased (Schaart et al. 2013). The effect on the transcript abundance was consistent with those observed at the metabolic level. It has been reported that during ripening of Fragaria × ananassa fruits, FaMYB10 is a main anthocyanin accumulation regulation protein. And it specifically expresses in fruits and is not or lowly expressed in other strawberry vegetative tissues (Lin-Wang et al. 2014). Lin-Wang et al. find that the expression of FaMYB10 is very low in the early developmental stages of fruit under low light intensity, but increases substantially during the ripening stages, coinciding with the reddening of the fruit (Lin-Wang et al. 2010). Our transcriptome data also shows FvMYB10 transcript level is increased in mature fruits by light treatment rather than decreased in RG15d fruits and no change in RGTurning fruits (Supplementary Table S10). The AtPAP1 and AtPAP2, two members of a small protein family that is required for anthocyanin accumulation and for the expression of structural genes in the anthocyanin biosynthesis pathway and FaMYB10 and FvMYB10 are the homologs in strawberry (Lin-Wang et al. 2014), interact with COP1 and SPA proteins, a E3 ligase protein complex, which mediate the ubiqutilation and degradation of AtPAP1 and AtPAP2 in dark (Maier et al. 2013). Additionally, SPA genes are also required for reducing PAP1 and PAP2 transcript levels in dark-grown seedlings (Maier et al. 2013). These findings indicate that the COP1/SPA complex affects PAP1 and PAP2 both transcriptionally and post-translationally. For the fruit trees, whether there is same regulation mechanism as well as Arabidopsis? In apple, MdMYB10, MdMYB1 and MdMYBA whose transcription level is induced by light and which are allelic to each other, have been isolated and characterized as key regulatory genes for anthocyanin accumulation and fruit coloration (Ban et al. 2007; Espley et al. 2007; Takos et al. 2006). MdCOP1s also interact with MdMYB1 and function in the ubiquitination and degradation of MdMYB1 via the proteasome pathway and regulate fruit coloration in apple (Li et al. 2012). We analyze the regulated genes by light and FvMYB10 at transcriptome level, and find there are a large number of coregulated genes in strawberry with a same regulation direction (Fig. 7b), which suggests MYB10 in strawberry plays an important role in light signal pathway, and light may indirectly regulate multiple genes through MYB10. Following, we detect the abundance change of FvMYB10 in dark and light by overexpression transgenic plants of 35S::FvMYB10-YFP/WT. And surprisingly, we could observe light significantly promote FvMYB10 protein accumulation under light compared with dark (Fig. 7c), which indicates light may regulate FvMYB10 at transcriptionally level and post-translationally level. The exploration of concrete mechanism of anthocyanin biosynthesis provides the potential utilization value in the genetic improvement of fruit quality.
Effect of light on sugar and flavor in fruits
The fruits ripen not only accompanys with red-coloration and anthocyanin accumulation, but also sugar accumulation and flavor and aroma materials formation. At the earliest, the researchers find that the apple fruit bagged reduce the soluble sugar in the fruit (Kikuchi et al. 1997). And in citrus culture, Chen et al. find the sugar content in fruits of outward canopy is higher than fruits of inward canopy and shading treatment promotes sugar transformation from juice vesicle to citrus pericarp (Chen et al. 2001). In tomato, although no differences in sucrose concentration between light- and dark-grown fruit, light-grown fruit accumulates almost twice as much starch and hexose as do dark-grown fruit (Guan and Janes 1991). These findings are consistent with our result that the soluble sugar content is significantly higher in light-grown strawberry fruit than dark-grown fruit (Fig. 4). Furthermore, the expression level of genes, for example, AI and SPS are upregulated by light in ripening stage, which is relative to sugar accumulation, is coincided with sugar content. Interestingly, the RNA-Seq data also shows light up-regulated some genes relative to aroma compounds which indicates light may regulate the formation of flavor at fruit ripening. But the types of aroma compounds, included the types of sugar, regulated by light and the regulation mechanism are still needed further research. Furthermore, the KEGG pathway analysis reveals that there are 42 genes involved in plant hormone signal transduction (Fig. 3). And emerging evidence has suggested that relative functions of plant hormones are not restricted to a particular stage (Kumar et al. 2014), and a complex network of more than one plant hormone is involved in controlling various aspects of fruit development. Surprisingly, RNA-Seq data has shown that genes in the anthocyanin biosynthesis pathway were actively regulated by ABA (Li et al. 2015) and ABA also promoted the transcription level of FaMYB10 (Kadomura-Ishikawa et al. 2015). But more evidences indicted that light and abscisic acid independently regulated FaMYB10 expression level. Recently, some studies in Arabidopsis have demonstrated the crosstalk between light and phytohormones in regulating the development of seedlings (Xu et al. 2017; Yang et al. 2018). So, it is possible that light affect ABA signaling pathway in strawberry fruits, and exploring the crosstalk of light and phytohormones in regulating the development of strawberry fruits will give us a deeper insight of light signal transduction.
References
Aharoni A et al (2001) The strawberry FaMYB1 transcription factor suppresses anthocyanin and flavonol accumulation in transgenic tobacco. Plant J 28:319–332
Aharoni A et al (2004) Gain and loss of fruit flavor compounds produced by wild and cultivated strawberry species. Plant Cell 16:3110–3131
Allan AC, Hellens RP, Laing WA (2008) MYB transcription factors that colour our fruit. Trends Plant Sci 13:99–102
Andrews S (2014) FastQC: a quality control tool for high throughput sequence data. http://www.bioinformatics.babraham.ac.uk/projects/fastqc/. Accessed 15 June 2018
Azuma A, Yakushiji H, Koshita Y, Kobayashi S (2012) Flavonoid biosynthesis-related genes in grape skin are differentially regulated by temperature and light conditions. Planta 236:1067–1080
Ban Y, Honda C, Hatsuyama Y, Igarashi M, Bessho H, Moriguchi T (2007) Isolation and functional analysis of a MYB transcription factor gene that is a key regulator for the development of red coloration in apple skin. Plant Cell Physiol 48:958–970
Banaś AK, Aggarwal C, Łabuz J, Sztatelman O, Gabryś H (2012) Blue light signalling in chloroplast movements. J Exp Bot 63:1559–1574
Baudry A, Caboche M, Lepiniec L (2006) TT8 controls its own expression in a feedback regulation involving TTG1 and homologous MYB and bHLH factors, allowing a strong and cell-specific accumulation of flavonoids in Arabidopsis thaliana. Plant J 46:768–779
Bhatia S, Singh R (2002) Phytohormone-mediated transformation of sugars to starch in relation to the activities of amylases, sucrose-metabolising enzymes in sorghum grain. Plant Growth Regul 36:97–104
Borevitz JO, Xia YJ, Blount J, Dixon RA, Lamb C (2000) Activation tagging identifies a conserved MYB regulator of phenylpropanoid biosynthesis. Plant Cell 12:2383–2393
Briggs WR, Christie JM (2002) Phototropins 1 and 2: versatile plant blue-light receptors. Trends Plant Sci 7:204–210
Brummell DA, Harpster MH (2001) Cell wall metabolism in fruit softening and quality and its manipulation in transgenic plants. Plant Mol Biol 47:311–339
Butelli E et al (2008) Enrichment of tomato fruit with health-promoting anthocyanins by expression of select transcription factors. Nat Biotechnol 26:1301–1308
Butelli E et al (2012) Retrotransposons control fruit-specific, cold-dependent accumulation of anthocyanins in blood oranges. Plant Cell 24:1242–1255
Castillejo C, de la Fuente JI, Iannetta P, Botella M, Valpuesta V (2004) Pectin esterase gene family in strawberry fruit: study of FaPE1, a ripening-specific isoform. J Exp Bot 55:909–918
Chaves I et al (2011) The cryptochromes: blue light photoreceptors in plants and animals. Annu Rev Plant Biol 62:335–364
Chen M, Chory J (2011) Phytochrome signaling mechanisms and the control of plant development. Trends Cell Biol 21:664–671
Chen JW, Zhang SL, Zhang LC, Xu CJ, Chen KS (2001) Effects of shading on the distribution of photosynthate, metabolism and accumulation of sugar in developing citrus fruits. Acta Phytophysiol Sin 27:499–504
Chory J (1992) A genetic model for light-regulated seedling development in Arabidopsis. Development 115:337–354
Christie JM et al (2012) Plant UVR8 photoreceptor senses UV-B by tryptophan-mediated disruption of cross-dimer salt bridges. Science 335:1492–1496
Cominelli E, Gusmaroli G, Allegra D, Galbiati M, Wade HK, Jenkins GI, Tonelli C (2008) Expression analysis of anthocyanin regulatory genes in response to different light qualities in Arabidopsis thaliana. J Plant Physiol 165:886–894
Cumplido-Laso G et al (2012) The fruit ripening-related gene FaAAT2 encodes an acyl transferase involved in strawberry aroma biogenesis. J Exp Bot 63:4275–4290
Demarsy E, Fankhauser C (2009) Higher plants use LOV to perceive blue light. Curr Opin Plant Biol 12:69–74
Dubos C et al (2008) MYBL2 is a new regulator of flavonoid biosynthesis in Arabidopsis thaliana. Plant J 55:940–953
Espley RV, Hellens RP, Putterill J, Stevenson DE, Kutty-Amma S, Allan AC (2007) Red colouration in apple fruit is due to the activity of the MYB transcription factor, MdMYB10. Plant J 49:414–427
Feng F, Li M, Ma F, Cheng L (2013) Phenylpropanoid metabolites and expression of key genes involved in anthocyanin biosynthesis in the shaded peel of apple fruit in response to sun exposure. Plant Physiol Biochem 69:54–61
Figueroa CR, Pimentel P, Gaete-Eastman C, Moya M, Herrera R, Caligari PDS, Moya-León MA (2008) Softening rate of the Chilean strawberry (Fragaria chiloensis) fruit reflects the expression of polygalacturonase and pectate lyase genes. Postharvest Biol Technol 49:210–220
Gesell A, Yoshida K, Tran LT, Constabel CP (2014) Characterization of an apple TT2-type R2R3 MYB transcription factor functionally similar to the poplar proanthocyanidin regulator PtMYB134. Planta 240:497–511
Giliberto L et al (2005) Manipulation of the blue light photoreceptor cryptochrome 2 in tomato affects vegetative development, flowering time, and fruit antioxidant content. Plant Physiol 137:199–208
Gonzalez A, Zhao M, Leavitt JM, Lloyd AM (2008) Regulation of the anthocyanin biosynthetic pathway by the TTG1/bHLH/Myb transcriptional complex in Arabidopsis seedlings. Plant J 53:814–827
Guan HP, Janes HW (1991) Light regulation of sink metabolism in tomato fruit: I. Growth and sugar accumulation. Plant Physiol 96:916–921
Hannum SM (2004) Potential impact of strawberries on human health: a review of the science. Crit Rev Food Sci Nutr 44:1–17
Hawkins C, Caruana J, Schiksnis E, Liu Z (2016) Genome-scale DNA variant analysis and functional validation of a SNP underlying yellow fruit color in wild strawberry. Sci Rep 6:29017
Hawkins C, Caruana J, Li J, Zawora C, Darwish O, Wu J, Alkharouf N, Liu Z (2017) An eFP browser for visualizing strawberry fruit and flower transcriptomes. Hortic Res 4:17029. https://doi.org/10.1038/hortres.2017.29
He JA, Giusti MM (2010) Anthocyanins: natural colorants with health-promoting properties. Annu Rev Food Sci T 1:163–187
He S-B, Wang W-X, Zhang J-Y, Xu F, Lian H-L, Li L, Yang H-Q (2015) The CNT1 domain of Arabidopsis CRY1 alone is sufficient to mediate blue light inhibition of hypocotyl elongation. Mol Plant 8:822–825
Heijde M, Ulm R (2012) UV-B photoreceptor-mediated signalling in plants. Trends Plant Sci 17:230–237
Hong SH et al (2008) CRY1 inhibits COP1-mediated degradation of BIT1, a MYB transcription factor, to activate blue light-dependent gene expression in Arabidopsis. Plant J 55:361–371
Jaakola L (2013) New insights into the regulation of anthocyanin biosynthesis in fruits. Trends Plant Sci 18:477–483
Jeong ST, Goto-Yamamoto N, Kobayashi S, Esaka M (2004) Effects of plant hormones and shading on the accumulation of anthocyanins and the expression of anthocyanin biosynthetic genes in grape berry skins. Plant sci 167:247–252
Jin H et al (2000) Transcriptional repression by AtMYB4 controls production of UV-protecting sunscreens in Arabidopsis. EMBO J 19:6150–6161
Kadomura-Ishikawa Y, Miyawaki K, Noji S, Takahashi A (2013) Phototropin 2 is involved in blue light-induced anthocyanin accumulation in Fragaria x ananassa fruits. J Plant Res 126:847–857
Kadomura-Ishikawa Y, Miyawaki K, Takahashi A, Masuda T, Noji S (2015) Light and abscisic acid independently regulated FaMYB10 in Fragaria x ananassa fruit. Planta 241:953–965
Kang CY, Lian HL, Wang FF, Huang JR, Yang HQ (2009) Cryptochromes, phytochromes, and COP1 regulate light-controlled stomatal development in Arabidopsis. Plant Cell 21:2624–2641
Kikuchi T, Arakawa O, Norton RN (1997) Improving skin color of ‘Fuji’ apple in Japan. Fruit Var J 51:71–75
Kondo S et al (2014) Abscisic acid metabolism and anthocyanin synthesis in grape skin are affected by light emitting diode (LED) irradiation at night. J Plant Physiol 171:823–829
Koyama K, Ikeda H, Poudel PR, Goto-Yamamoto N (2012) Light quality affects flavonoid biosynthesis in young berries of Cabernet Sauvignon grape. Phytochemistry 78:54–64
Kumar R, Khurana A, Sharma AK (2014) Role of plant hormones and their interplay in development and ripening of fleshy fruits. J Exp Bot 65:4561–4575
Li S (2014) Transcriptional control of flavonoid biosynthesis. Plant Signal Behav 9:e27522
Li Y-Y, Mao K, Zhao C, Zhao X-Y, Zhang H-L, Shu H-R, Hao Y-J (2012) MdCOP1 ubiquitin E3 ligases interact with MdMYB1 to regulate light-Induced anthocyanin biosynthesis and red fruit coloration in apple. Plant Physiol 160:1011–1022
Li T, Jia K-P, Lian H-L, Yang X, Li L, Yang H-Q (2014) Jasmonic acid enhancement of anthocyanin accumulation is dependent on phytochrome A signaling pathway under far-red light in Arabidopsis. Biochem Biophys Res Commun 454:78–83
Li D, Li L, Luo Z, Mou W, Mao L, Ying T (2015) Comparative transcriptome analysis reveals the influence of abscisic acid on the metabolism of pigments, ascorbic acid and folic acid during strawberry fruit ripening. PLoS ONE 10:e0130037
Lian H-L et al (2011) Blue-light-dependent interaction of cryptochrome 1 with SPA1 defines a dynamic signaling mechanism. Gene Dev 25:1023–1028
Lin-Wang K et al (2010) An R2R3 MYB transcription factor associated with regulation of the anthocyanin biosynthetic pathway in rosaceae. BMC Plant Biol 10:50
Lin-Wang KUI et al (2011) High temperature reduces apple fruit colour via modulation of the anthocyanin regulatory complex. Plant Cell Environ 34:1176–1190
Lin-Wang K et al. (2014) Engineering the anthocyanin regulatory complex of strawberry (Fragaria vesca). Front Plant Sci. https://doi.org/10.3389/fpls.2014.00651
Liu L-J et al (2008) COP1-mediated ubiquitination of CONSTANS is implicated in cryptochrome regulation of flowering in Arabidopsis. Plant Cell 20:292–306
Liu B, Zuo Z, Liu H, Liu X, Lin C (2011) Arabidopsis cryptochrome 1 interacts with SPA1 to suppress COP1 activity in response to blue light. Gene Dev 25:1029–1034
Lu X-D, Zhou C-M, Xu P-B, Luo Q, Lian H-L, Yang H-Q (2015) Red-light-dependent interaction of phyB with SPA1 promotes COP1–SPA1 dissociation and photomorphogenic development in Arabidopsis. Mol Plant 8:467–478
Maier A et al (2013) Light and the E3 ubiquitin ligase COP1/SPA control the protein stability of the MYB transcription factors PAP1 and PAP2 involved in anthocyanin accumulation in Arabidopsis. Plant J 74:638–651
Matus JT, Loyola R, Vega A, Peña-Neira A, Bordeu E, Arce-Johnson P, Alcalde JA (2009) Post-veraison sunlight exposure induces MYB-mediated transcriptional regulation of anthocyanin and flavonol synthesis in berry skins of Vitis vinifera. J Exp Bot 60:853–867
Matus JT, Poupin MJ, Cañón P, Bordeu E, Alcalde JA, Arce-Johnson P (2010) Isolation of WDR and bHLH genes related to flavonoid synthesis in grapevine (Vitis vinifera L.). Plant Mol Biol 72:607–620
Miao L et al (2016) Colored light-quality selective plastic films affect anthocyanin content, enzyme activities, and the expression of flavonoid genes in strawberry (Fragaria × ananassa) fruit. Food Chem 207:93–100
Nagatani A (2010) Phytochrome: structural basis for its functions. Curr Opin Plant Biol 13:565–570
Osterlund MT, Hardtke CS, Wei N, Deng XW (2000) Targeted destabilization of HY5 during light-regulated development of Arabidopsis. Nature 405:462–466
Raab T, López-Ráez JA, Klein D, Caballero JL, Moyano E, Schwab W, Muñoz-Blanco J (2006) FaQR, required for the biosynthesis of the strawberry flavor compound 4-hydroxy-2,5-dimethyl-3(2H)-furanone, encodes an enone oxidoreductase. Plant Cell 18:1023–1037
Salvatierra A, Pimentel P, Moya-León MA, Herrera R (2013) Increased accumulation of anthocyanins in Fragaria chiloensis fruits by transient suppression of FcMYB1 gene. Phytochemistry 90:25–36
Schaart JG et al (2013) Identification and characterization of MYB-bHLH-WD40 regulatory complexes controlling proanthocyanidin biosynthesis in strawberry (Fragaria × ananassa) fruits. New Phytol 197:454–467
Seo HS, Yang JY, Ishikawa M, Bolle C, Ballesteros ML, Chua NH (2003) LAF1 ubiquitination by COP1 controls photomorphogenesis and is stimulated by SPA1. Nature 423:995–999
Takos AM, Jaffé FW, Jacob SR, Bogs J, Robinson SP, Walker AR (2006) Light-induced expression of a MYB gene regulates anthocyanin biosynthesis in red apples. Plant Physiol 142:1216–1232
Wang L, Feng Z, Wang X, Wang X, Zhang X (2010) DEGseq: an R package for identifying differentially expressed genes from RNA-seq data. Bioinformatics 26:136–138
Xu F, Cao S, Shi L, Chen W, Su X, Yang Z (2014) Blue light irradiation affects anthocyanin content and enzyme activities involved in postharvest strawberry fruit. J Agric Food Chem 62:4778–4783
Xu F et al (2017) Photoactivated CRY1 and phyB interact directly with AUX/IAA proteins to inhibit auxin signaling in Arabidopsis. Mol Plant. https://doi.org/10.1016/j.molp.2017.12.003
Yang J et al (2005) Light regulates COP1-mediated degradation of HFR1, a transcription factor essential for light signaling in Arabidopsis. Plant Cell 17:804–821
Yang C, Xie F, Jiang Y, Li Z, Huang X, Li L (2018) Phytochrome A negatively regulates the shade avoidance response by increasing auxin/indole acidic acid protein stability. Dev Cell 1:29–41
Young MD, Wakefield MJ, Smyth GK, Oshlack A (2010) Gene ontology analysis for RNA-seq: accounting for selection bias. Genome Biol. https://doi.org/10.1186/gb-2010-11-2-r14
Zhang F, Gonzalez A, Zhao MZ, Payne CT, Lloyd A (2003) A network of redundant bHLH proteins functions in all TTG1-dependent pathways of Arabidopsis. Development 130:4859–4869
Zheng X et al (2013) Arabidopsis Phytochrome B promotes SPA1 nuclear accumulation to repress photomorphogenesis under far-red light. Plant Cell 25:115–133
Zifkin M et al (2012) Gene expression and metabolite profiling of developing highbush blueberry fruit indicates transcriptional regulation of flavonoid metabolism and activation of abscisic acid metabolism. Plant Physiol 158:200–224
Zimmermann IM, Heim MA, Weisshaar B, Uhrig JF (2004) Comprehensive identification of Arabidopsis thaliana MYB transcription factors interacting with R/B-like BHLH proteins. Plant J 40:22–34
Zoratti L, Karppinen K, Luengo Escobar A, Häggman H, Jaakola L (2014) Light-controlled flavonoid biosynthesis in fruits. Front Plant Sci. https://doi.org/10.3389/fpls.2014.00534
Acknowledgements
This work was supported by the National Natural Science Foundation of China grants to H.-L.L (31570282 and 31170266).
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Xu, P., Zawora, C., Li, Y. et al. Transcriptome sequencing reveals role of light in promoting anthocyanin accumulation of strawberry fruit. Plant Growth Regul 86, 121–132 (2018). https://doi.org/10.1007/s10725-018-0415-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10725-018-0415-3