Abstract
Key message
Pp4ERF24 and Pp12ERF96 fine tune blue light-induced anthocyanin biosynthesis via interacting with PpMYB114 and promoting the interaction between PpMYB114 and PpbHLH3, which enhances the expression of PpMYB114-induced PpUFGT.
Abstract
The red coloration of pear fruit is attributed to anthocyanin accumulation, which is transcriptionally regulated by the MYB-bHLH-WD40 complex. A number of ethylene response factors (ERF) have been identified to regulate anthocyanin biosynthesis in different plants. In pear, several ERF transcription factor genes were identified to be potentially involved in the light-induced anthocyanin biosynthesis according to transcriptome data. But the molecular mechanism of these ERFs underlying the regulation of anthocyanin accumulation is unknown. In this study, exposure of ‘Red Zaosu’ pear, a mutant of ‘Zaosu’ pear, to blue light significantly induced the anthocyanin accumulation by increasing the expression levels of anthocyanin biosynthetic genes. Gene expression analysis confirmed that the expression of Pp4ERF24 and Pp12ERF96 genes were up-regulated in the process of blue light-induced anthocyanin biosynthesis. Yeast two-hybrid and bimolecular fluorescence complementation assay revealed that Pp4ERF24 and Pp12ERF96 interacted with PpMYB114, but not with PpMYB10. Bimolecular fluorescence complementation assay demonstrated that the interaction between these two ERFs and PpMYB114 enhanced the interaction between PpMYB114 and PpbHLH3. Further analysis by dual luciferase assay verified that these two ERFs increased the up-regulation of PpMYB114-mediated PpUFGT expression. Furthermore, co-transformation of Pp12ERF96 with PpMYB114 and PpbHLH3 in tobacco leaves led to enhanced anthocyanin accumulation. Transient overexpression of Pp4ERF24 or Pp12ERF96 alone in ‘Red Zaosu’ pear fruit also induced anthocyanin biosynthesis in pear peel. Our findings provide insights into a mechanism involving the synergistic interaction of ERFs with PpMYB114 to regulate light-dependent coloration and anthocyanin biosynthesis in pear fruits.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Pear (Pyrus spp.) is an economically important fruit in China. The pear fruit peel appears red because of the accumulation of anthocyanins (Huang et al. 2009; Steyn et al. 2005), which are important bioactive compounds responsible for red, blue, and purple color. Thus, anthocyanins affect fruit quality and commercial value. Anthocyanins are biosynthesized via the phenylpropanoid and flavonoid pathway, which involves several enzyme-encoding structural genes. The early biosynthetic genes [i.e., PAL (phenylalanine ammonia lyase), CHS (chalcone synthase), CHI (chalcone isomerase), and F3′H (flavanone 3′-hydroxylase)] are involved in the production of common precursors of the flavonoid pathway. In contrast, the late biosynthetic genes [i.e., DFR (dihydroflavonol 4-reductase), ANS (anthocyanin synthase), and UFGT (UDP-glucose: flavonoid 3-glucosyltransferase)] are specific for the anthocyanin biosynthesis pathway (Winkel-Shirley 2001).
The R2R3-MYB proteins interact with a basic helix-loop-helix (bHLH) and a WD-repeat protein to form a MYB-bHLH-WD40 (MBW) complex to transcriptionally regulate anthocyanin biosynthesis (Allan et al. 2008; Ballester et al. 2010; Xie et al. 2012; Xu et al. 2015). Of the MBW components, the MYB transcription factors are the most important regulators of anthocyanin biosynthesis by directly binding to the promoters of anthocyanin structural genes (Allan et al. 2008). In apple, MdMYB1 cosegregates with the loci for skin color and is the genetic basis for apple skin color, while MdMYB10-R6 and MdMYB110 are responsible for the development of red fruit flesh (Takos et al. 2006). In grapevine, VvMYB1 and VvMYB2 regulate anthocyanin biosynthesis via the expression of VvUFGT (Kobayashi et al. 2002, 2004; Walker et al. 2007). In pear, PpMYB10 and PpMYB114 enhance anthocyanin biosynthesis by regulating the expression of structural genes (Feng et al. 2010; Yao et al. 2017).
The ethylene response factor (ERF) family is a large gene family and ERFs are plant-specific transcription factors which play critical roles in various biological processes and responses to environmental stresses (Nakano et al. 2006). For example, MdERF2 negatively regulates ethylene biosynthesis during apple fruit ripening by suppressing MdACS1 transcription (Li et al. 2016b). In loquat fruit, EjAP2-1, an AP2/ERF factor, directly represses lignin biosynthesis via interaction with EjMYB1 (Zeng et al. 2015). Several ERFs are also identified to regulate anthocyanin biosynthesis in different plants. In Arabidopsis, aterf4aterf8 double mutant reduces light-responsive anthocyanin accumulation, which indicates that these two ERFs positively regulate light-induced anthocyanin biosynthesis (Koyama and Sato 2018). In apple, MdERF3 is transcriptionally regulated by MdMYB1 and involved in anthocyanin biosynthesis (An et al. 2018). In pear, PpERF3 interacts with PpMYB114 and PpbHLH3 to form an ERF3-MYB114-bHLH3 complex, which co-regulate the coloration of red pear fruits (Yao et al. 2017). In our previously studies, several ERF transcription factors were found to respond to light which might be potentially involved in anthocyanin biosynthesis (Bai et al. 2017). However, it is currently unknown how these ERF genes regulate light-induced anthocyanin biosynthesis in pear.
In this study, four PpERFs were selected as candidate genes which might regulate anthocyanin biosynthesis according to transcriptome analysis in pear (Bai et al. 2017). Then we investigated the regulation mechanism of these PpERFs on fruit coloration using ‘Red Zaosu’ pear, a red sport of the green-skinned ‘Zaosu’ pear which is a hybrid of the Asian pear and Pyrus communis. Subsequently, we found that Pp4ERF24 and Pp12ERF96 interacted with PpMYB114 to induce anthocyanin biosynthesis in ‘Red Zaosu’ pear by enhancing the interaction between PpMYB114 and PpbHLH3. The results presented herein showed a previously uncharacterized anthocyanin biosynthesis regulatory module which provides new insights into the molecular mechanism underlying the red coloration of pear fruits.
Materials and methods
Plant materials and treatment
‘Red Zaosu’ pear fruits were obtained from the orchard at the Research Institute of Horticulture, Henan Academy of Agricultural Sciences in 2016. A total of 150 fruitlets were covered with paper bags (consisting of yellow and black outer and inner layers, respectively) 22 days after full bloom (DAFB). Bagged and uncolored fruits with a similar size and quality were harvested at maturity (155 DAFB) and immediately transported to the laboratory.
The blue light irradiation experiments were performed as described previously (Tao et al. 2018), but different samples were used. Briefly, fruits were removed from bags in a darkroom. Fruits lacking mechanical injuries were randomly divided into three groups. The fruits were then transferred to shelves in a phytotron and continuously irradiated with a blue LED (453.2 nm, 21 µmol m− 2 s− 1) at 17 °C and 80% relative humidity. Fruit peels collected prior to irradiation (i.e., 0 h) and after 12, 24, 72, 144, and 240 h were immediately frozen in liquid nitrogen and stored at − 80 °C until use.
RNA extraction and cDNA synthesis
Total RNA was extracted using a modified CTAB method (Zhang et al. 2012), and the concentration was measured with a BioDrop spectrophotometer (Biochrom, Cambridge, UK). First-strand cDNA was synthesized from 2 µg RNA using the PrimeScript™ RT reagent Kit with gDNA Eraser (Takara, Dalian, China). The cDNA was diluted twofold, after which a 1-µl aliquot was used as the template for gene cloning and real-time quantitative PCR (qPCR) analysis.
Real-time quantitative PCR
Real-time quantitative PCR primers were designed using an online tool (https://www.ncbi.nlm.nih.gov/tools/primer-blast/) (Tables S1, S2). Primer efficiency and specificity were verified using melting curves and by sequencing PCR products. The 15-µl qPCR reaction mixture consisted of 7.5 µl SYBR Premix Ex Taq™ (Takara, Ohtsu, Japan), 0.5 µl each primer (10 µM), 1 µl cDNA, and 5.5 µl RNase-free water. The qPCR was completed using a CFX Connect™ real-time PCR system (Bio-Rad, Hercules, CA, USA) with the following program: 95 °C for 3 min; 40 cycles of 95 °C for 10 s and 60 °C for 30 s. A template-free control was included for each run. Relative transcript levels were determined according to the 2− ΔΔCt algorithm (Livak and Schmittgen 2001), with the pear Actin gene (PpActin, JN684184) used for normalizing data.
Yeast two-hybrid assay
A yeast two-hybrid (Y2H) assay was conducted using the Matchmaker™ Gold yeast two-hybrid system (Clontech). The full-length coding sequences (CDSs) encoding the prey and bait proteins were cloned into the AD (pGADT7) and BD (pGBKT7) vectors (Clontech) using the ClonExpress™ II One Step Cloning Kit (Vazyme). The primers for constructing vectors are listed in Table S3. The gene-BD plasmid was inserted into Y2HGold cells alone for an auto-activation analysis. The Y2HGold cells were then co-transformed with the AD and BD vectors and added to synthetically defined (SD) medium lacking leucine (Leu) and tryptophan (Trp) (DDO, SD/−Leu/−Trp). The cells were screened for possible physical interactions using SD medium lacking Leu, Trp, adenine (Ade), and histidine (His), but supplemented with 5-bromo-4-chloro-3-indolyl α-D-galactopyranoside (X-α-gal) and aureobasidin A (AbA) (QDO, SD/−Leu/−Trp/−Ade/−His/X-α-gal/AbA).
Bimolecular fluorescence complementation assay
The full-length PpMYB114 and ERF gene CDSs were inserted into the p2YN and p2YC vectors using the ClonExpress™ II One Step Cloning Kit. Agrobacterium tumefaciens GV3101 cells were transformed with the resulting plasmids according to a freeze–thaw method. The cells were incubated overnight and then re-suspended with infiltration buffer (0.2 mM acetosyringone, 10 mM MES, and 10 mM MgCl2) for an optical density at 600 nm of 1.0. Equal volumes of the different combinations were mixed and used to infiltrate tobacco leaves with a needle-free syringe. The YFP fluorescence was detected using a confocal laser scanning microscope 60 h later.
Dual luciferase assay
The full-length trans gene CDSs were inserted into the pGreenII 0029 62-SK vector, while the promoter sequence of the cis gene was cloned into the pGreenII 0800-LUC (firefly luciferase) vector. The resulting plasmids were separately inserted into A. tumefaciens GV3101 cells according to a freeze-thaw method. A solution comprising a 10 (SK):1(LUC) ratio was used to infiltrate tobacco leaves with a needle-free syringe. The LUC and Renilla luciferase enzyme activities were analyzed 60 h later using the Dual-Luciferase Reporter Assay System (Promega) with Modulus Luminometers (Promega). Three independent experiments with six biological replicates were conducted for each assay.
Firefly luciferase complementation imaging assay
A firefly luciferase complementation imaging assay was conducted as previously described (Li et al. 2017). The PpMYB114 and PpbHLH3 CDSs were cloned into separate pCAMBIA1300-nLuc and pCAMBIA1300-cLuc vectors. Additionally, Pp4ERF24 and Pp12ERF96 were inserted into the pGreenII 0029 62-SK vector. The prepared plasmids were inserted into A. tumefaciens GV3101 cells, after which different combinations comprising equal volumes of each vector were used to infiltrate tobacco leaves. After 3 days, luciferase activities were detected using the NightOWL II LB983 low-light cooled CCD imaging apparatus by spraying 0.5 mM luciferin (Promega) on the infiltrated position followed by a 5-min incubation in darkness.
Transient transformation assay of gene function in tobacco leaves and pear fruits
To verify the gene function of Pp4ERF24 and Pp12ERF96 in inducing anthocyanin biosynthesis, the full-length CDSs of PpMYB114, PpbHLH3, Pp4ERF24 and Pp12ERF96 were inserted into the pGreenII 0029 62-SK vector with the control of the 35S promoter. The prepared plasmids were inserted into A. tumefaciens GV3101 cells. Then different combinations comprising equal volumes of each vector were used to infiltrate tobacco leaves as for the dual luciferase assay or fruits of ‘Red Zaosu’ pear. After infiltration, the pear fruits were put in darkness overnight and moved onto shelves under continuous light (60 µmol m− 2 s− 1). The phenotypes of tobacco leaves were observed 6 days after infiltration, while the phenotypes of pear fruits were observed 3 days after infiltration.
Statistical analysis
Student’s t-test and one way ANOVA (Tukey) were completed using the Statistical Product and Service Solutions program (version 19) (SPSS Inc., Chicago, IL, USA).
Results
Identification of candidate ERF genes according to transcriptome data
Using the previously generated RNA-seq data (Bai et al. 2017), two PpERF candidate genes whose expression were correlated with that of PpMYB10 were identified. These two genes were named as Pp4ERF24 and Pp12ERF100 following the nomenclature of an AP2/ERF gene family analysis by Li et al. (2018). Further BLAST analysis suggested that another two ERF genes, Pp12ERF96 and Pp1ERF1, were paralogs of Pp4ERF24 and Pp12ERF100 (Fig. S1A), respectively. The transcriptome data suggested that Pp4ERF24, Pp12ERF100, and Pp1ERF1 expression levels increased at 24 h after the removal of bags, while Pp12ERF96 expression remained low throughout the experiment (Fig. S1B).
Expression analysis of genes related to anthocyanin biosynthesis in ‘Red Zaosu’ pear during red coloration under blue light treatment
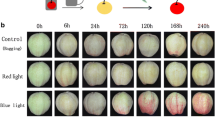
The ‘Red Zaosu’ pear fruits started coloration obviously at 72 h after initiating the blue light treatment, and then continuously turned red until 240 h (Fig. 1a). The transcript abundances of PpUFGT exhibited a tendency that initially increased and peaked at 72 h, and then subsequently decreased. Additionally, the expression levels of the anthocyanin biosynthetic regulatory genes, PpMYB10 and PpMYB114, exhibited the similar expression trends, and peaked at 24 h and then gradually decreased. The expression levels of PpbHLH3, PpbHLH33 and PpWD40 slightly increased firstly and then decreased (Fig. 1b).
Blue light induces anthocyanin biosynthesis in ‘Red Zaosu’ pear fruits. a ‘Red Zaosu’ fruit peel color at different time points during a blue light treatment. b Anthocyanin biosynthetic structural and regulatory genes expression levels at different time points during pear coloration. Data are presented as the mean ± standard error of three biological replicates
Expression analysis of candidate PpERF genes in ‘Red Zaosu’ pear during red coloration
Furthermore, we analyzed the transcript levels of four ERF genes in ‘Red Zaosu’ pear samples treated with blue light. The expression levels of Pp4ERF24 and its paralog, Pp12ERF96, initially increased at 12 h after starting the blue light treatment and then decreased, but increased again at 144 h. In contrast, Pp12ERF100 and Pp1ERF1 expression levels exhibited the opposite pattern, with a decrease at 12 h followed by an increase, and then decreased again at 240 h (Fig. 2).
Interactions between MYB and identified ERF transcription factors
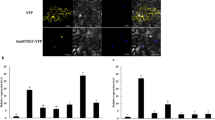
According to the expression pattern, we analyzed the interactions between MYB transcription factors and these four ERF transcription factors. An Y2H assay indicated that the encoded ERF proteins did not interact with PpMYB10 (Fig. S2A), but the physical interaction between Pp4ERF24 and PpMYB114 were detected (Fig. 3a). Unfortunately, because of the auto-activation of PpMYB114, Pp12ERF96, Pp12ERF100, and Pp1ERF1, we were unable to analyze any potential physical interactions among these proteins in an Y2H assay (Fig. S2B). We subsequently investigated the in vivo interactions between the ERF proteins and PpMYB114 using bimolecular fluorescence complementation (BiFC) assay. As presented in Fig. 3b, green fluorescence was detected in the nuclei when tobacco leaves were co-infiltrated with PpMYB114-YFPN and Pp4ERF24-YFPC or PpMYB114-YFPN and Pp12ERF96-YFPC. In contrast, fluorescence was not observed for co-infiltrations with PpMYB114-YFPN and Pp12ERF100-YFPC or PpMYB114-YFPN and Pp1ERF1-YFPC. There was also a lack of fluorescence with the control combinations. Therefore, PpMYB114 may interact with Pp4ERF24 and Pp12ERF96.
Pp4ERF24 and Pp12ERF96 interact with PpMYB114. a A yeast two-hybrid (Y2H) assay revealed that Pp4ERF24 interacted with PpMYB114. Pp4ERF24 gene fragments were inserted into the pGBKT7 vector (BD), while PpMYB114 was incorporated into the pGADT7 vector (AD). QDO, SD medium lacking Trp, Leu, His, and Ade; QDO/X-α-gal/AbA, QDO medium containing X-α-Gal and AbA. The pGADT7-T and pGBKT7-53 vectors were used as the positive control, while the combination of AD and Pp4ERF24-BD served as the negative control. Blue plaques indicated an interaction between two proteins. The basal AbA concentration was 300 ng ml− 1. b A bimolecular fluorescence complementation (BiFC) assay in N. benthamiana leaves revealed that PpMYB114 interacted with Pp4ERF24 and Pp12ERF96, but not with Pp12ERF100 and Pp1ERF1. 2YN and 2YC, PpMYB114-2YN and 2YC (negative control)
Effects of Pp4ERF24 and Pp12ERF96 on the interaction between PpMYB114 and PpbHLH3
To examine how the interactions between PpMYB114 and Pp4ERF24 or between PpMYB114 and Pp12ERF96 influence the interaction between PpMYB114 and PpbHLH3, we conducted a firefly luciferase complementation imaging assay. Plasmids were prepared for the production of PpMYB114 and PpbHLH3 fused to the N and C termini of Luciferase, respectively (i.e., PpMYB114-nLuc and PpbHLH3-cLuc). Additionally, Pp4ERF24 and Pp12ERF96 were inserted into the pGreenII 0029 62-SK vector for a subsequent overexpression. Tobacco leaves were co-infiltrated with different combinations of the prepared plasmids for the production of fusion proteins. A slight luminescence signal was detected in regions containing PpMYB114-nLuc and PpbHLH3-cLuc, while strong luminescence signals were observed when Pp4ERF24-SK or Pp12ERF96-SK were also present (Fig. 4a). These results suggested that Pp4ERF24 and Pp12ERF96 enhanced the interaction between PpMYB114 and PpbHLH3.
Pp4ERF24 and Pp12ERF96 enhance the interaction between PpMYB114 and PpbHLH3. a A luciferase complementation imaging assay revealed that Pp4ERF24 and Pp12ERF96 enhanced the interaction between PpMYB114 and PpbHLH3 in tobacco leaves. Agrobacterium tumefaciens GV3101 cells harboring different combinations were used to infiltrate different wild tobacco leaf regions. Luciferase activities were recorded in these regions 60 h after infiltration. b A dual luciferase assay verified that the co-transformation with different combinations of PpMYB114, PpbHLH3, Pp4ERF24, and Pp12ERF96 affected PpUFGT promoter activity. The PpUFGT promoter activity is expressed as a ratio of LUC and REN activities. Asterisks indicate significant difference at P < 0.01 by t tests. Data are presented as the mean ± standard error of three biological replicates
Effects of Pp4ERF24 and Pp12ERF96 on the transcriptional activation of PpMYB114-mediated PpUFGT
Our dual luciferase assay results indicated that PpMYB114 activated the expression of PpUFGT (Fig. 4b). Furthermore, the combined presence of PpMYB114 and bHLH3 increased the activation effect (Fig. 4b). Therefore, the PpMYB114–Pp4ERF24 and PpMYB114–Pp12ERF96 interactions might affect the binding of PpMYB114 to the PpUFGT promoter. To verify this possibility, we conducted a dual luciferase assay in which PpMYB114, PpbHLH3, Pp4ERF24, and Pp12ERF96 were incorporated into the pGreenII 0029 62-SK vector for a subsequent overexpression, while the PpUFGT promoter was inserted into the pGreenII 0800-LUC vector. Different plasmid combinations were used to co-transform tobacco leaves (Fig. 4b). Both Pp4ERF24 and Pp12ERF96 were observed to up-regulate PpUFGT expression in the presence of PpMYB114 and PpbHLH3. However, PpUFGT expression levels in tobacco leaves with Pp4ERF24 and PpMYB114 or Pp12ERF96 and PpMYB114 were similar to those in leaves with PpMYB114 alone (Fig. 4b). These results implied that Pp4ERF24 and Pp12ERF96 may indirectly enhance PpUFGT expression by enhancing the interaction between PpMYB114 and PpbHLH3.
Heterologous overexpression of Pp4ERF24 and Pp12ERF96 with PpMYB114 and PpbHLH3 in tobacco leaves
Transient transformation assay in tobacco leaves was performed to verify the function of the PpERF transcription factors. No red pigmentation was observed when PpMYB114 was overexpressed alone, while the tobacco leaves turned red when PpMYB114 and PpbHLH3 were co-overexpressed (Fig. 5a). However, when PpMYB114 and Pp12ERF96 were co-overexpressed, the tobacco leaves also remained green (Fig. 5a). When Pp12ERF96 was co-transformed with PpMYB114 and PpbHLH3, the tobacco leaves turned red (Fig. 5a). Pigment accumulation was quantitatively analyzed, the hue angle decreased (more redness) while total anthocyanin content increased when three TFs were co-expressed compared with both PpMYB114 and PpbHLH3 were over-expressed (Fig. 5b, c). These results indicated that Pp12ERF96 promoted anthocyanin accumulation through PpMYB114/PpbHLH3, probably due to enhancing the interaction between PpMYB114 and PpbHLH3. Unfortunately, no more pigment accumulation was observed when Pp4ERF24 was co-transformed with PpMYB114 and PpbHLH3, probably because of the instability of heterogeneous overexpression (data not shown).
Transient expression assays confirm the function of Pp12ERF96 in regulating anthocyanin in tobacco leaves. a The phenotypes of tobacco leaves 6 days after infiltration. a, PpMYB114; b, PpMYB114 + PpbHLH3; c, PpMYB114 + Pp12ERF96; d, PpMYB114 + PpbHLH3 + Pp12ERF96. b The hue angle of tobacco leaves 6 days after infiltration. c Measurement of total anthocyanin contents. Lowercase letters indicate significant difference at P < 0.05 by Tukey test. Data are presented as the mean ± standard error of three biological replicates
Homologous overexpression of Pp4ERF24 and Pp12ERF96 in ‘Red Zaosu’ pear fruits
To further confirm that Pp4ERF24 and Pp12ERF96 could promote anthocyanin accumulation in pear, the transient transformation of Pp4ERF24 and Pp12ERF96 was performed in ‘Red Zaosu’ pear upon maturity. The vectors of Pp4ERF24-SK, Pp12ERF96-SK were obtained, and the empty vector of pGreenII 62-SK was used as control. As shown in Fig. 6a and b, overexpression of Pp4ERF24 and Pp12ERF96 promoted anthocyanin accumulation in pear skin around the injection sites compared with control. RT-qPCR analysis showed that the expression levels of PpPAL, PpCHS, PpCHI, PpDFR, PpANS, PpUFGT, PpMYB10, PpbHLH33 and PpWD40 were up-regulated in the Pp4ERF24 or Pp12ERF96 injection areas compared with control (Fig. 6c). Furthermore, overexpression of Pp12ERF96 also promoted the expression levels of PpF3H, PpMYB114 and PpbHLH3 (Fig. 6c). Therefore, these results illustrated that Pp4ERF24 and Pp12ERF96 played positive roles in regulating anthocyanin biosynthesis in ‘Red Zaosu’ pear fruit.
Transient expression assays confirm the function of Pp4ERF24 and Pp12ERF96 in regulating anthocyanin in ‘Red Zaosu’ pear skins. a The phenotypes of ‘Red Zaosu’ pear 3 days after infiltration. a, pGreenII 62-SK; b, Pp4ERF24; c, Pp12ERF96. b Measurement of total anthocyanin contents. Two asterisks indicated significant difference at P < 0.01 by t tests. Data are presented as the mean ± standard error of three biological replicates. c Analysis of expression levels of anthocyanin biosynthesis genes in ‘Red Zaosu’ pear skins using RT-qPCR. Asterisk indicates significant difference at P < 0.05 by t tests. Data are presented as the mean ± standard error of three biological replicates
Discussion
The anthocyanin biosynthesis pathway is controlled by various regulatory factors in addition to the MBW complex. For example, several transcriptional regulators, such as MdJAZ (An et al. 2015), MdHXK1 (Hu et al. 2016), MdCOL11 (Bai et al. 2014), MdEIL1 (An et al. 2017a), MdHY5 (An et al. 2017b), and MdSIZ1 (Zhou et al. 2017), function as either activators or repressors in fine tuning anthocyanin biosynthesis in apple, indicating the essential roles of transcription factors in the coloration of fruits. Several ERF transcription factors have also been shown to regulate anthocyanin biosynthesis in different plants. In apple, MdERF3 is involved in ethylene-induced anthocyanin accumulation via the following pathway: MdEIL1 directly binds to the promoter of MdMYB1 and transcriptionally activated its expression, while MdMYB1 interacts with the promoter of MdERF3, thereby promoting ethylene biosynthesis, which finally forming a positive feedback (An et al. 2018). In Arabidopsis, the class II repressors, AtERF4 and AtERF8, positively regulate anthocyanin biosynthesis in light responses, probably via repressing the expression of a negative regulator of the anthocyanin biosynthesis (Koyama and Sato 2018). In pear, Yao et al. (2017) found that PpMYB114 interacts with PpERF3 to form a new complex (ERF3-MYB114-bHLH3) and promotes anthocyanin biosynthesis by binding to the promoters of the anthocyanin biosynthetic structural genes (Yao et al. 2017). In our study, Pp4ERF24 and Pp12ERF96 also participated in fine tuning anthocyanin biosynthesis (Figs. 3, 4, 5, 6). Based on the phylogenetic analysis, we confirmed that the sequences of these two ERF proteins differed from PpERF3 (Fig. S1A). Furthermore, in the pear genome database, Pp4ERF24 and Pp12ERF96 locate on chromosomes 4 and 12, separately, while PpERF3 locates on chromosome 5. Pp4ERF24 and Pp12ERF96 were not indispensable for pear red coloration, but they acted as accessory factors by interacting with PpMYB114 to enhance the interaction between PpMYB114 and PpbHLH3 (Figs. 3, 4a), which led to the enhanced expression of MYB114-induced anthocyanin biosynthetic structural genes (Fig. 4b), and finally promoted anthocyanin accumulation. Heterologous transient assay in tobacco leaves also confirmed that Pp12ERF96 significantly induced anthocyanin accumulation when co-expressed with PpMYB114 and PpbHLH3 (Fig. 5). However, overexpression of Pp4ERF24 with PpMYB114 and PpbHLH3 failed to induce redder coloration in tobacco leaves, which might be attribute to the specificity of this ERF in pear, and the instability of heterogeneous overexpression. Furthermore, when Pp4ERF24 or Pp12ERF96 was overexpressed in ‘Red Zaosu’ pear fruits, anthocyanin accumulation was promoted, respectively (Fig. 6). These results implied that Pp4ERF24 and Pp12ERF96 are positive regulators of anthocyanin biosynthesis and confirmed the existence of a previously uncharacterized regulatory module.
It has been reported that several factors posttranscriptionally regulate MBW complex by interacting with MYB protein and then indirectly regulate anthocyanin biosynthesis. In A. thaliana, light-induced mitogen-activated protein kinase 4 positively regulates anthocyanin biosynthesis by interacting with AtMYB75 and increasing its stability (Li et al. 2016a). In Arabidopsis, the bHLH transcription factor TCP3 interacts with several flavonoid-regulating R2R3-MYBs (AtMYB75/AtMYB90/AtMYB113/AtMYB114/AtMYB12/AtMYB111) to promote flavonoid biosynthesis (Li and Zachgo 2013). In this study, two ERF genes (Pp4ERF24 and Pp12ERF100) that were highly co-expressed with PpMYB10 (correlation coefficiency of 0.95) were identified according to the light-induced anthocyanin biosynthesis transcriptome data (Bai et al. 2017). Additionally, we identified another two ERF genes, Pp12ERF96 and Pp1ERF1, which are paralogs of Pp4ERF24 and Pp12ERF100 (Fig. S1A), respectively. Due to the high correlation coefficiency between these ERFs and PpMYB10, we hypothesized that these ERF proteins might interact with PpMYB10 and co-regulate anthocyanin biosynthesis. However, an Y2H assay revealed that none of these proteins interacted with PpMYB10 (Fig. S2A). Additionally, PpMYB114, which was co-expressed with PpMYB10, was not identified as a differentially expressed gene because of inaccuracies in its predicted transcript sequence (Pbr042132.1), which lacked an important 222-bp R3 domain, resulting in neglect of this gene. Thus, we analyzed the interaction between PpMYB114 and these four ERF proteins, and found that Pp4ERF24 and Pp12ERF96 interacted with PpMYB114 (Fig. 3a, b). In pear, PpMYB10 is an ortholog of MdMYB1 gene (Feng et al. 2010), while PpMYB114 is probably unique, which may lead to the different interaction between these two MYBs and ERFs (Figs. S2, 3).
The anthocyanin biosynthesis pathway is controlled by various factors including environmental factors and developmental factors (Jaakola 2013). Light is one of the most important environmental factors which regulates anthocyanin biosynthesis. The light-responsive factors, which contain the photoreceptors and their downstream regulators [e.g., CONSTITUTIVELY PHOTOMORPHOGENIC 1 (COP1) and ELONGATED HYPOCOTYL 5 (HY5)], negatively or positively influence light-induced anthocyanin biosynthesis (Li et al. 2012; Maier et al. 2013; Shin et al. 2013; Zhang et al. 2011). In apple, MdCOP1 inhibits anthocyanin biosynthesis by interacting with MdMYB1, resulting in ubiquitination and degradation (Li et al. 2012). MdHY5 induces anthocyanin biosynthesis by binding to the promoter of MdMYB1 (An et al. 2017b). According to these findings, light regulates anthocyanin biosynthesis via transcriptionally or post-transcriptionally regulating MdMYB1. In this study, the expression patterns of four ERF genes in transcriptome data of pear fruit and ‘Red Zaosu’ pear suggested their possible involvement in the light-response (Figs. S1B, 2). We analyzed the promoters of Pp4ERF24 and Pp12ERF96, which consisted of many light-responsive elements, including G-box, Sp1, and L-box (Fig. S3). The G-box is a well-known light-responsive element that is regulated by HY5, which positively mediates photomorphogenesis (Chattopadhyay et al. 1998). Thus, we speculated that Pp4ERF24 and Pp12ERF96 expression levels might be regulated by blue light via HY5 or other transcription factors. Our findings may be associated with light-induced anthocyanin biosynthesis, which remains to be explored.
Previous studies confirmed that PpMYB10 and PpMYB114 induce the expression of PpUFGT by binding to its promoter, with PpUFGT expression levels correlated with those of PpMYB10 and PpMYB114 (Qian et al. 2014; Yao et al. 2017). In grape, VvUFGT expression is regulated by VvMYBA1, and its expression level is correlated with that of VvMYBA1 (Zheng et al. 2013). In apple, MdUFGT and MdMYB1 exhibit similar expression patterns (Takos et al. 2006). However, in our study, the PpUFGT expression level rapidly increased at 12 h after starting the blue light treatment, while PpMYB10 and PpMYB114 expression levels exhibited a slow increase over the first 24 h. Obviously, PpUFGT expression was not closely correlated with PpMYB10 or PpMYB114 expression (Fig. 1b), implying other factors may directly or indirectly regulate PpUFGT transcription. We also observed that the Pp4ERF24 and Pp12ERF96 expression levels quickly increased at 12 h after initiating the light treatment (Fig. 2). In addition, luciferase assay confirmed the indirectly positive regulation of PpUFGT by Pp4ERF24 and Pp12ERF96 (Fig. 4b). Therefore, we speculated that the synergistic regulation by these two ERF transcription factors and two MYB proteins led to the inconsistent expression patterns of PpUFGT and PpMYB genes.
In addition, the Pp4ERF24 and Pp12ERF96 expression patterns in ‘Red Zaosu’ pear fruits were different from those in ‘Meirensu’ fruits according to an analysis of transcriptome data (Fig. 2, S1B). These differences were probably due to the genetic background of red coloration in different pear cultivars. ‘Meirensu’ pear is a hybrid of ‘Kosui’ pear and ‘Huoba’ pear, of which the red color is inherited from ‘Huoba’ pear. While ‘Red Zaosu’ pear is a sport from ‘Zaosu’ pear, of which the bud mutation mechanism remains unknown. As to the coloration profiles, ‘Meirensu’ pear only accumulates anthocyanin in mature fruits, but ‘Red Zaosu’ pear accumulates anthocyanin in the young leaves, flowers, fruitlets and mature fruits (Qian et al. 2014). Therefore, the regulatory patterns of red coloration in ‘Meirensu’ and ‘Red Zaosu’ pears differ from each other. Furthermore, it has been well characterized that the rearrangement of multiple repeats in the promoter of MdMYB10 produces a red foliage and fruit core phenotypes in apple (Espley et al. 2009). However, no difference of the promoter of PpMYB10 was found between ‘Red Zaosu’ pear and ‘Zaosu’ pear (Qian et al. 2014) and there was no coloration in the fruit core of ‘Red Zaosu’, indicating that the red coloration of ‘Red Zaosu’ was also different from MYB10-R6 apple and may specifically be regulated by some other patterns. On the other hand, the different expression patterns of Pp4ERF24 and Pp12ERF96 in ‘Red Zaosu’ and ‘Meirensu’ were potentially due to the different treatments in these two experiments, as we exposed ‘Red Zaosu’ pear fruits to continuous blue light, while the ‘Meirensu’ pear fruits examined in the previous study were treated with natural sunlight (i.e., circadian rhythm) (Bai et al. 2017).
Overall, the data presented herein indicate that Pp4ERF24 and Pp12ERF96 fine tune blue light-induced anthocyanin biosynthesis partly through the following pathway: Pp4ERF24 and Pp12ERF96 interact with PpMYB114 and promote the interaction between PpMYB114 and PpbHLH3, which enhances the transcriptional activation of PpUFGT expression by PpMYB114, ultimately inducing anthocyanin biosynthesis (Fig. 7). These findings suggest that there is a previously uncharacterized regulatory module of light-induced anthocyanin biosynthesis which is mediated by ERF transcription factors, and provide new evidences that the red coloration of pear fruits is regulated by a complex network.
A model showing Ethylene Response Factors Pp4ERF24 and Pp12ERF96 regulate blue light-induced anthocyanin biosynthesis in ‘Red Zaosu’ pear fruits by interacting with PpMYB114. Pp4ERF24 and Pp12ERF96 interact with PpMYB114 and promote the interaction between PpMYB114 and PpbHLH3, thus, enhancing the transactivation of PpMYB114 to PpUFGT and inducing more anthocyanin biosynthesis. Overlap of PpMYB114 and PpbHLH3 means enhanced interacting effect
Abbreviations
- ERF:
-
Ethylene response factor
- UFGT:
-
UDP-glucose: flavonoid 3-glucosyltransferase
- EBG:
-
Early biosynthetic gene
- LBG:
-
Late biosynthetic gene
- PAL:
-
Phenylalanine ammonia lyase
- CHS:
-
Chalcone synthase
- CHI:
-
Chalcone isomerase
- F3′H:
-
Flavanone 3′-hydroxylase
- DFR:
-
Dihydroflavonol 4-reductase
- ANS:
-
Anthocyanin synthase
- bHLH:
-
Basic helix-loop-helix
- DAFB:
-
Days after full bloom
References
Allan AC, Hellens RP, Laing WA (2008) MYB transcription factors that colour our fruit. Trends Plant Sci 13:99–102
An X, Tian Y, Chen K, Liu XJ, Liu DD, Xie X, Cheng CG, Cong P, Hao Y (2015) MdMYB9 and MdMYB11 are involved in the regulation of the JA-induced biosynthesis of anthocyanin and proanthocyanidin in apples. Plant Cell Physiol 56:650–662
An J, Liu X, Li H, You C, Wang X, Hao Y (2017a) Apple RING E3 ligase MdMIEL1 inhibits anthocyanin accumulation by ubiquitinating and degrading MdMYB1 protein. Plant Cell Physiol 58:1953–1962
An J, Qu F, Yao J, Wang X, You C, Wang X, Hao Y (2017b) The bZIP transcription factor MdHY5 regulates anthocyanin accumulation and nitrate assimilation in apple. Hortic Res 4:17023
An J, Wang X, Li Y, Song L, Zhao L, You C, Hao Y (2018) EIN3-LIKE1, MYB1, and ETHYLENE RESPONSE FACTOR3 act in a regulatory loop that synergistically modulates ethylene biosynthesis and anthocyanin accumulation. Plant Physiol 178:808
Bai S, Saito T, Honda C, Hatsuyama Y, Ito A, Moriguchi T (2014) An apple B-box protein, MdCOL11, is involved in UV-B- and temperature-induced anthocyanin biosynthesis. Planta 240:1051–1062
Bai S, Sun Y, Qian M, Yang F, Ni J, Tao R, Li L, Shu Q, Zhang D, Teng Y (2017) Transcriptome analysis of bagging-treated red Chinese sand pear peels reveals light-responsive pathway functions in anthocyanin accumulation. Sci Rep 7:63. https://doi.org/10.1038/s41598-017-00069-z
Ballester A, Molthoff J, de Vos R, Hekkert B, Orzaez D, Fernandez-Moreno J, Tripodi P, Grandillo S, Martin C, Heldens J, Ykema M, Granell A, Bovy A (2010) Biochemical and molecular analysis of pink tomatoes: deregulated expression of the gene encoding transcription factor SlMYB12 leads to pink tomato fruit color. Plant Physiol 152:71–84
Chattopadhyay S, Ang L, Puente P, Deng X, Wei N (1998) Arabidopsis bZIP protein HY5 directly interacts with light-responsive promoters in mediating light control of gene expression. Plant Cell 10:673
Espley RV, Brendolise C, Chagne D, Kutty-Amma S, Green S, Volz R, Putterill J, Schouten HJ, Gardiner SE, Hellens RP, Allan AC (2009) Multiple repeats of a promoter segment causes transcription factor autoregulation in red apples. Plant Cell 21:168–183
Feng S, Wang Y, Yang S, Xu Y, Chen X (2010) Anthocyanin biosynthesis in pears is regulated by a R2R3-MYB transcription factor PyMYB10. Planta 232:245–255
Hu D, Sun C, Zhang Q, An J, You C, Hao Y (2016) Glucose sensor MdHXK1 phosphorylates and stabilizes MdbHLH3 to promote anthocyanin biosynthesis in apple. PLoS Genet 12:e1006273. https://doi.org/10.1371/journal.pgen.1006273
Huang C, Yu B, Teng Y, Su J, Shu Q, Cheng Z, Zeng L (2009) Effects of fruit bagging on coloring and related physiology, and qualities of red Chinese sand pears during fruit maturation. Sci Hortic 121:149–158. https://doi.org/10.1016/j.scienta.2009.01.031
Jaakola L (2013) New insights into the regulation of anthocyanin biosynthesis in fruits. Trends Plant Sci 18:477–483
Kobayashi S, Ishimaru M, Hiraoka K, Honda C (2002) Myb-related genes of the Kyoho grape (Vitis labruscana) regulate anthocyanin biosynthesis. Planta 215:924–933. https://doi.org/10.1007/s00425-002-0830-5
Kobayashi S, Goto-Yamamoto N, Hirochika H (2004) Retrotransposon-induced mutations in grape skin color. Science 304:982
Koyama T, Sato F (2018) The function of ETHYLENE RESPONSE FACTOR genes in the light-induced anthocyanin production of Arabidopsis thaliana leaves. Plant Biotechnol 35:87–91. https://doi.org/10.5511/plantbiotechnology.18.0122b
Li S, Zachgo S (2013) TCP3 interacts with R2R3-MYB proteins, promotes flavonoid biosynthesis and negatively regulates the auxin response in Arabidopsis thaliana. Plant J 76:901–913. https://doi.org/10.1111/tpj.12348
Li Y, Mao K, Zhao C, Zhao X, Zhang H, Shu H, Hao Y (2012) MdCOP1 ubiquitin E3 ligases interact with MdMYB1 to regulate light-induced anthocyanin biosynthesis and red fruit coloration in apple. Plant Physiol 160:1011–1022
Li S, Wang W, Gao J, Yin K, Wang R, Wang C, Petersen M, Mundy J, Qiu J (2016a) MYB75 phosphorylation by MPK4 is required for light-induced anthocyanin accumulation in Arabidopsis. Plant Cell 28:2866–2883
Li T, Jiang Z, Zhang L, Tan D, Wei Y, Yuan H, Li T, Wang A (2016b) Apple (Malus domestica) MdERF2 negatively affects ethylene biosynthesis during fruit ripening by suppressing MdACS1 transcription. Plant J 88:735–748
Li T, Xu Y, Zhang L, Ji Y, Tan D, Yuan H, Wang A (2017) The jasmonate-activated transcription factor MdMYC2 regulates ETHYLENE RESPONSE FACTOR and ethylene biosynthetic genes to promote ethylene biosynthesis during apple fruit ripening. Plant Cell 29:1316–1334. https://doi.org/10.1105/tpc.17.00349
Li X, Tao S, Wei S, Ming M, Huang X, Zhang S, Wu J (2018) The mining and evolutionary investigation of AP2/ERF genes in pear (Pyrus). BMC Plant Biol 18:46. https://doi.org/10.1186/s12870-018-1265-x
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2−∆∆CT method. Methods 25:402–408. https://doi.org/10.1006/meth.2001.1262
Maier A, Schrader A, Kokkelink L, Falke C, Welter B, Iniesto E, Rubio V, Uhrig JF, Hülskamp M, Hoecker U (2013) Light and the E3 ubiquitin ligase COP1/SPA control the protein stability of the MYB transcription factors PAP1 and PAP2 involved in anthocyanin accumulation in Arabidopsis. Plant J 74:638–651
Nakano T, Suzuki K, Fujimura T, Shinshi H (2006) Genome-wide analysis of the ERF gene family in Arabidopsis and rice. Plant Physiol 140:411–432
Qian M, Sun Y, Allan AC, Teng Y, Zhang D (2014) The red sport of ‘Zaosu’ pear and its red-striped pigmentation pattern are associated with demethylation of the PyMYB10 promoter. Phytochemistry 107:16–23
Shin DH, Choi M, Kim K, Bang G, Cho M, Choi S, Choi G, Park YI (2013) HY5 regulates anthocyanin biosynthesis by inducing the transcriptional activation of the MYB75/PAP1 transcription factor in Arabidopsis. FEBS Lett 587:1543–1547
Steyn W, Wand S, Holcroft D, Jacobs G (2005) Red colour development and loss in pears. Acta Hortic 671:79–85. https://doi.org/10.17660/ActaHortic.2005.671.9
Takos AM, Jaffé FW, Jacob SR, Bogs J, Robinson SP, Walker AR (2006) Light-induced expression of a MYB gene regulates anthocyanin biosynthesis in red apples. Plant Physiol 142:1216
Tao R, Bai S, Ni J, Yang Q, Zhao Y, Teng Y (2018) The blue light signal transduction pathway is involved in anthocyanin accumulation in ‘Red Zaosu’ pear. Planta 248:37–48. https://doi.org/10.1007/s00425-018-2877-y
Walker AR, Lee E, Bogs J, McDavid DAJ, Thomas MR, Robinson SP (2007) White grapes arose through the mutation of two similar and adjacent regulatory genes. Plant J 49:772–785. https://doi.org/10.1111/j.1365-313x.2006.02997.x
Winkel-Shirley B (2001) Flavonoid biosynthesis. A colorful model for genetics, biochemistry, cell biology, and biotechnology. Plant Physiol 126:485–493
Xie X, Li S, Zhang R, Zhao J, Chen Y, Zhao Q, Yao Y, You C, Zhang X, Hao Y (2012) The bHLH transcription factor MdbHLH3 promotes anthocyanin accumulation and fruit colouration in response to low temperature in apples. Plant Cell Environ 35:1884–1897
Xu W, Dubos C, Lepiniec L (2015) Transcriptional control of flavonoid biosynthesis by MYB-bHLH-WDR complexes. Trends Plant Sci 20:176–185
Yao G, Ming M, Allan AC, Gu C, Li L, Wu X, Wang R, Chang Y, Qi K, Zhang S, Wu J (2017) Map-based cloning of the pear gene MYB114 identifies an interaction with other transcription factors to coordinately regulate fruit anthocyanin biosynthesis. Plant J 92:437–451
Zeng J, Li X, Xu Q, Chen J, Yin X, Ferguson IB, Chen K (2015) /ERF gene, is a novel regulator of fruit lignification induced by chilling injury, via interaction with EjMYB transcription factors. Plant Biotechnol J 13:1325–1334.
Zhang Y, Zheng S, Liu Z, Wang L, Bi Y (2011) Both HY5 and HYH are necessary regulators for low temperature-induced anthocyanin accumulation in Arabidopsis seedlings. J Plant Physiol 168:367–374
Zhang D, Yu B, Bai JH, Qian MJ, Shu Q, Su J, Teng YW (2012) Effects of high temperatures on UV-B/visible irradiation induced postharvest anthocyanin accumulation in ‘Yunhongli No. 1’ (Pyrus pyrifolia Nakai) pears. Sci Hortic 134:53–59
Zheng Y, Li J, Xin H, Wang N, Guan L, Wu B, Li S (2013) Anthocyanin profile and gene expression in berry skin of two red Vitis vinifera grape cultivars that are sunlight dependent versus sunlight independent. Aust J Grape Wine Res 19:238–248
Zhou L, Li Y, Zhang R, Zhang C, Xie X, Zhao C, Hao Y (2017) The small ubiquitin-like modifier E3 ligase MdSIZ1 promotes anthocyanin accumulation by sumoylating MdMYB1 under low-temperature conditions in apple. Plant Cell Environ 40:2068–2080
Acknowledgements
This study was supported by the National Natural Science Foundation of China (Grant Nos. 31471852 to YT and 31772272 to SB) and the Earmarked Fund for China Agriculture Research System (CARS-28).
Author information
Authors and Affiliations
Contributions
JN, SB, and YT conceived and planned the study. RT and LY helped collect samples and extracted the total RNA. JN and YZ completed the qPCR, vector construction, and bioinformatics analysis. JN and LG conducted the Y2H, BiFC, dual luciferase, firefly luciferase complementation imaging assays and the transient transformation assay of gene function in tobacco leaves and ‘Red Zaosu’ fruits. JN, SB, MQ and YT wrote the manuscript. All of the authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no competing interest.
Electronic supplementary material
Below is the link to the electronic supplementary material.
11103_2018_802_MOESM4_ESM.pdf
(A) Phylogenetic analysis of ERF proteins from ‘Red Zaosu’ pear (PpERF) and Prunus persica (PpeERF). (B) Pp4ERF24, Pp12ERF96, Pp12ERF100, and Pp1ERF1 expression patterns based on ‘Meirensu’ transcriptome data. Supplementary material 4 (PDF 721 KB)
11103_2018_802_MOESM5_ESM.pdf
(A) A yeast two-hybrid (Y2H) assay revealed that Pp4ERF24, Pp12ERF96, Pp12ERF100, and Pp1ERF1 did not interact with PpMYB10. The pGADT7-T and pGBKT7-53 vectors were used as the positive control, while the AD and MYB10-BD combination served as the negative control. Blue plaques indicated an interaction between two proteins. The basal AbA concentration was 200 ng ml−1. (B) A Y2H assay confirmed that PpMYB114, Pp12ERF96, Pp12ERF100, and Pp1ERF1 underwent an auto-activation. The BD vector was used as the negative control. Supplementary material 5 (PDF 4485 KB)
11103_2018_802_MOESM6_ESM.pdf
Analysis of the Pp4ERF24 and Pp12ERF96 promoters revealed several light-responsive elements. Supplementary material 6 (PDF 713 KB)
Rights and permissions
About this article
Cite this article
Ni, J., Bai, S., Zhao, Y. et al. Ethylene response factors Pp4ERF24 and Pp12ERF96 regulate blue light-induced anthocyanin biosynthesis in ‘Red Zaosu’ pear fruits by interacting with MYB114. Plant Mol Biol 99, 67–78 (2019). https://doi.org/10.1007/s11103-018-0802-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11103-018-0802-1