Abstract
Anthocyanins, natural pigments with high antioxidant activities, are widely distributed in the plant kingdom and play important roles in various physiological processes. Much effort has been committed to enhancing the anthocyanin content of these health-promoting pigments in vegetables and grains, for their eye-catching colors and special nutrients. Previously, we reported that the SmMYB1 gene encoding a R2R3 MYB transcription factor participated in the regulation of anthocyanin biosynthesis in the peel of eggplant. In this work, we introduced SmMYB1 into a non-anthocyanin-accumulating eggplant cultivar (Solanum aethiopicum group Gilo) via Agrobacterium-mediated transformation. Genetically engineered plants exhibited high concentrations of anthocyanin in leaves, petals, stamens, and fruit peels under normal growth conditions, especially in fruit flesh. Furthermore, highly methylated anthocyanins, malvidin 3-(p-coumaroyl)rhamnoside(glucoside)-5-glucoside and malvidin 3-(feruloyl)rhamnoside(glucoside)-5-glucoside, were separated from the purple fruit flesh and identified by HPLC–ESI–MS/MS. qRT-PCR analysis revealed that most anthocyanin structural genes were dramatically up-regulated in the tissues of transgenic lines compared with non-transformed plants. In addition, the transgenic seedlings had greater tolerance to freezing stress and better recovery under rewarming conditions. These results provide a good foundation for the breeding of new eggplant cultivars with more healthy agronomic traits in future studies.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Anthocyanins, an important subgroup of flavonoids, are the main natural water-soluble pigments widely distributed in the plant kingdom. They are responsible for the red, blue, and purple colors found in plant tissues in a wide range of flowers, fruits, foliage, seeds, and roots (Koes et al. 2005; Tanaka et al. 2008). Large-scale studies indicate that the colorful pigments furnish flowers and fruits with eye-catching colors to attract insects, birds, and animals for pollination and seed dispersal (Bradshaw and Schemske 2003). Anthocyanins also play important roles in protecting plants against damage from UV radiation, freezing stress, drought stress, and microbial agents (Grotewold 2006; Harborne and Williams 2000; Mendez et al. 1999). Earlier studies proved that regular intake of foods rich in anthocyanins could reduce the risks of suffering from chronic illnesses such as cancer, atherosclerosis, and degenerative disease (de Pascual-Teresa and Sanchez-Ballesta 2007; Espley et al. 2013). Vegetables, grains, and fruits rich in anthocyanins are therefore receiving more and more attention from consumers.
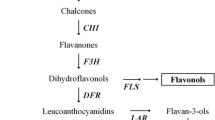
To date, anthocyanin biosynthesis has been studied extensively at the metabolic and molecular levels (Harborne and Williams 2000). Most of the structural genes encoding the enzymes responsible for each step have been isolated from various plants, for example Arabidopsis (Arabidopsis thaliana), apple (Malus ×domestica), petunia (Petunia hybrida), grape (Vitis vinifera), and other species (Honda et al. 2002; Shi and Xie 2014; Tsuda et al. 2000; Van der Krol et al. 1990). As Fig. 1 shows, in the multienzymatic pathway, the initial precursor phenylalanine is catalyzed stepwise by phenylalanine ammonia lyase (PAL), cinnamate 4-hydroxylase (C4H), 4-coumarate-CoA ligase (4CL), chalcone synthase (CHS), chalcone isomerase (CHI), flavanone 3-hydroxylase (F3H), flavonoid 3-hydroxylase (F3′H), flavonoid 3′,5′-hydroxylase (F3′5′H), dihydroflavonol 4-reductase (DFR), anthocyanidin synthase (ANS), and O-methyltransferase (OMT) (Li et al. 2006). The colorful anthocyanindins are then catalyzed by flavonoid-3-O-glucosyltransferase (UFGT) for glycosylation and form more stable molecules, anthocyanins (Jaakola 2013; Winkel-Shirley 2001). In addition, dihydrokaempferol can be hydroxylated by F3′H or F3′5′H to produce dihydroquercetin and dihydromyricetin, respectively. DFR and the subsequent enzymes of the anthocyanin pathway will then catalyze dihydrokaempferol, dihydroquercetin, and dihydromyricetin to produce of the brick–red pelargonidin, red cyanidin, and violet delphinidin compounds, respectively. However, cyanidin and delphinidin can be further methylated at positions 3′ and/or 5′ on the B ring, eventually leading to the production of peonidin, petunidin, or malvidin.
Schematic representation of the biosynthetic pathway of the anthocyanins. The names of the compounds are indicated in boxes. The enzyme names are PAL phenylalanine ammonia lyase, C4H cinnamate 4-hydroxylase, 4CL 4-coumarate-CoA ligase, CHS chalcone synthase, CHI chalcone isomerase, F3H flavanone 3-hydroxylase, F3′H flavanone 3′-hydroxylase, F3′5′H flavanone 3′,5′-hydroxylase, DFR dihydroflavonol 4-reductase, ANS anthocyanidin synthase, and OMT O-methyltransferase
Recently, a large number of studies have shown that the anthocyanin biosynthesis genes are regulated mainly at the mRNA level by transcription factors (TF) of several families. The TFs are within the MADS box, MYB, basic helix–loop–helix (bHLH), and WD40 repeat protein families (Goff et al. 1992; Jaakola 2013; Koes et al. 2005; Lalusin et al. 2006). A number of anthocyanin biosynthesis regulatory genes have been cloned from various plants, such as Arabidopsis, petunia, maize, apple, tomato, and other species (Ban et al. 2007; Bernhardt et al. 2005; Borevitz et al. 2000; Chiu et al. 2010; Jaakola et al. 2010; Lalusin et al. 2006; Mathews 2003; Morita et al. 2006; Park et al. 2007; Schwinn et al. 2006; Spelt et al. 2000; Takos et al. 2006). By analyzing mutants with abnormal levels of anthocyanins in model plants with genetic and molecular technology, it has been proved that the transcription of anthocyanin biosynthesis genes is directly regulated by a transcriptional activation of the MYB–bHLH–WD40 complex (MBW) that consists of R2R3 MYB, Bhlh, and WD40 proteins (Albert et al. 2011; Baudry et al. 2004; Broun 2005; Gonzalez et al. 2008; Lepiniec et al. 2006). However, MADS box genes probably function on the upstream pathway of MYB proteins in bilberry (Jaakola et al. 2010). Previous studies showed that purple leaves or fruits rich in anthocyanin arise from the transcriptional activation of MYB transcription factors (Ban et al. 2007; Chiu et al. 2010; Mano et al. 2007; Mathews 2003; Takos et al. 2006). These studies centered on the molecular analysis of anthocyanin biosynthesis provide essential knowledge for technologists to genetically enhance the anthocyanin contents of crops and vegetables (Butelli et al. 2008; Gonzali et al. 2009).
In previous study, we reported that the SmMYB1 gene encoding a R2R3 MYB transcription factor participated in the process of anthocyanin biosynthesis regulation in the peel of purple eggplant cultivar (Solanum melongena L) (Zhang et al. 2014c). A binary vector containing the full-length coding sequence of SmMYB1 driven by the 35S promoter was then created. In this article, a non-anthocyanin-accumulating eggplant cultivar (Solanum aethiopicum group Gilo) with medicinal and ornamental value was used for transformation via Agrobacterium-mediated T-DNA transfer (Toppino et al. 2008). The transgenic eggplants showed intense purple pigments in most tissues, especially in the flesh of tender fruits. Anthocyanins in the sarcocarp of transgenic eggplant were separated and identified by high-performance liquid chromatography–electrospray ionization tandem mass spectrometry (HPLC–ESI–MS/MS). Furthermore, the expression of anthocyanin biosynthesis and regulatory genes was analyzed in the tissues which displayed the accumulation of purple pigments. In addition, the freezing stress tolerance of transgenic and wild-type seedlings was analyzed.
Materials and methods
Plant materials and growth conditions
The seeds of eggplant (Solanum aethiopicum group Gilo) were obtained from Chongqing Academy of Agriculture Sciences. Transgenic and wild-type seedlings were grown in the greenhouse with 16-h/8-h light/dark photoperiod (28 °C) and watered daily. The leaves, petals, stamens, peels, and flesh were collected, immediately frozen with liquid nitrogen, and stored at −80 °C for RNA extraction and other analyses. In addition, the peels and flesh of tender fruits (immature fruits) were collected at 15 days after anthesis.
Vector construction and plant transformation
The full-length open reading frame of SmMYB1 (GenBank: KF727476) was amplified using primers SmMYB1-con-F, 5′-GCGATCTAGAAATAAAATGAATAATCCTCC-3′, and SmMYB1-con-R, 5′-GACTGAGCTCCGAAAACAGACAATATTACTA-3′. The PCR products were linked into plasmid (pBI121) to yield an overexpression plasmid pBI121-SmMYB1, containing the SmMYB1 cDNA driven by the 35S promoter. The construct was then transferred into Agrobacterium tumefaciens strain LBA4404 and used to infect eggplant cotyledon explants (Filippone and Lurquin 1989; Zhang et al. 2014c). Transformed lines were selected for kanamycin (80 mg L−1 final concentration) resistance and then confirmed by PCR using the primers NPTII-F/R (NPTII-F GACAATCGGCTGCTCTGA and NPTII-R AACTCCAGCATGAGATCC).
RNA extraction and quantitative real-time PCR analysis
Total RNA of all samples was isolated using RNAiso Plus (TaKaRa, Dalian, PRC) according to the manufacturer’s instructions. Then, 1 μg total RNA was used as a template for reverse transcription PCR with an oligo(dT)20 primer and M-MLV reverse transcriptase (Promega, Madison, WI, USA) following the manufacturer’s protocol. The synthesized cDNAs were diluted twice in RNase-/DNase-free water for qRT-PCR analysis.
qRT-PCR was performed using the CFX96™ Real-Time System (C1000™ Thermal Cycler, Bio-Rad). The qRT-PCR mixture (10 μL of total volume) consisted of 5 μL of GoTaq® qRT-PCR Master Mix (Promega), 0.5 μL of mixed primers, 1 μL of diluted cDNA, and 3.5 μL of RNase-/DNase-free water. PCR amplification was carried out using two-step cycling conditions of 98 °C for 3 min, followed by 40 cycles of 98 °C for 2 s and 60 °C for 10 s. NRT (no reverse transcription control) and NTC (no template control) were performed as negative controls. Amplification was followed by a melting curve analysis with continual fluorescence data acquisition during the 60–95 °C melts. Melting curve analysis of qRT-PCR samples revealed that there was only one product for each gene primer reaction. Three replicates for each sample were used, and SmGAPDH was used as internal standard. The nucleotide sequences of primer pairs used for qRT-PCR analysis are listed in Supplementary Table 1 (Zhang et al. 2014c).
Chemicals
The external standards, anthocyanin (delphinidin 3-rutinoside chloride and malvidin 3-glucoside chloride), were purchased from Phytolab (Germany). The HPLC-grade formic acid and methanol (MeOH) were bought from Sigma. All of the other solvents were provided by Aldrich (St. Louis, MO, USA).
Anthocyanin extraction and HPLC–ESI–MS/MS analysis
Anthocyanins were extracted and separated by HPLC according to the protocols described by Park and others with minor modifications (Park et al. 2011; Zhang et al. 2014c). Each sample (100 mg) was extracted with 1 mL of methanol/H2O/acetic acid (85:15:0.5) at room temperature. The solution was vigorously vortexed for 5 min, sonicated for 5 min, and then centrifuged at 12,000g for 8 min. The extraction for each sample was performed twice, and the supernatants were mixed for the following HPLC analysis. The extract was filtered through a 0.2-μm PTFE syringe filter to remove cell debris. The filtrate was analyzed by an Agilent Technologies 1200 Series HPLC (Agilent Technologies, Palo Alto, CA, USA), equipped with an Agilent 1200 HPLC variable wavelength detector. The data were analyzed by Agilent 1200 HPLC ChemStation software. The chromatographic separation was carry out on a Zorbax Stablebond Analytical SB-C18 column (4.6 mm × 250 mm, 5 μm, Agilent Technologies, Rising Sun, MD, USA). The injection sling was 10 μL. Elution was performed using mobile phase A (aqueous 5 % formic acid solution) and mobile phase B (methanol). The detection was set at a wavelength of 520 nm, and the column oven temperature was set at 40 °C. The flow rate was 1 mL/min. The gradient program was as follows: 0–5 min, 10–20 % B; 5–10 min, 20–30 % B; 10–15 min, 30–40 % B; 15–25 min, 40 % B; 25–30 min, 40–60 % B; 31–32 min, 90 % B; and 32–35 min, 10 % B. Quantification of the different anthocyanins was based on peak areas and calculated as equivalents of the external standards. All contents were expressed as milligrams per gram of dry weight. Low-resolution electrospray mass spectrometry was performed with a solariX ion trap mass spectrometer (Bruker Daltoniks, Billerica, MA, USA). The experimental conditions were as follows: ESI interface, nebulizer, 50 psi; dry gas, 15.0 psi; dry temperature, 320 °C; MS/MS, scan from m/z 200 to 2000; ion trap, scan from m/z 200 to 2000; source accumulation, 50 ms; ion accumulation time, 300 ms; flight time to acq. cell, 1 ms; smart parameter setting (SPS), compound stability, 50 %; and trap drive level, 60 %.
Total anthocyanin content analysis
The total anthocyanin contents were measured by a pH-differential spectrum method (Park et al. 2011). Frozen samples (100 mg) were crushed into powder in liquid nitrogen and then treated separately with 2 mL of pH 1.0 buffer (50 mM KCl and 150 mM HCl) and 2 mL of pH 4.5 buffer (400 mM sodium acetate and 240 mM HCl). The solution was vigorously vortexed for 10 min, sonicated for 10 min, and placed at 4 °C for 12 h. The mixtures were centrifuged at 12,000g for 20 min at 4 °C. Absorbance of the supernatants was then measured at 510 nm. The total anthocyanin content was calculated according to the equation:
where A1 represents the absorbance of supernatants gathered from pH 1.0 buffer solution at 510 nm, and A2 represents the absorbance of supernatants gathered from pH 4.5 buffer solution at 510 nm. The value 484.8 represents the molecular mass of cyanidin-3-glucoside chloride, while 24,825 reflects its molar absorptivity at 510 nm. Each sample was analyzed in triplicate, and the results were presented as the average of the three measurements.
Dehydration assay of leaves of transgenic and control plants
For the dehydration assay of leaves, 35-day-old seedlings of wild-type and transgenic plants were harvested for analysis. Leaves of a similar size, age, and position were detached and placed on dry filter papers for the measurement of the water loss rate. The leaves were weighed at 15 min, 30 min, 1 h, 2 h, 4 h, 8 h, and 16 h and then placed in the oven until constant weight. Water loss of leaf was presented as a percentage of initial fresh weight.
Freezing tolerance assay of transgenic and control plants
For the freezing treatment, 35-day-old seedlings of wild-type and transgenic plants were incubated at −5 ± 1 °C for 0 h, 2 h, 4 h, and 8 h. The leaves were then harvested for analysis. Twenty leaf disks (each 1 cm2) were placed in a transparent container containing 15 ml deionized water and shaked for 2 h before measurement of the initial electric conductance (S1). The transparent container was then heated in boiling water for 20 min and cooled to room temperature to measure the ultimate electric conductance (S2). The electric conductance of deionized water was used as the blank (S0) (Rapisarda et al. 2000). The relative electrolyte leakage (REL) was calculated from the following equation:
Statistical analyses
SPSS version 17.0 (SPSS Inc., Chicago, IL, USA) was used for the data analysis. One-way analysis of variance (ANOVA) was performed with post hoc Tukey’s honestly significant difference (HSD) test, with significance set at p < 0.05 and p < 0.01.
Results
SmMYB1 ubiquitously promotes anthocyanin biosynthesis in eggplant
To obtain transgenic eggplant with high levels of anthocyanin accumulation, a construction containing the full-length coding sequence of SmMYB1 driven by the 35S promoter was created and transformed into wild-type eggplant (Solanum aethiopicum group Gilo) via Agrobacterium tumefaciens-mediated T-DNA transfer (Supplementary Figure 1A). Eight independent kanamycin-resistant seedlings were generated and were confirmed as transgenic eggplant by PCR. Three independent transgenic lines (lines N, P, and R) exhibiting extensive production of purple pigments in the whole plants were selected for further characterization. The transgenic plants showed pigmented leaves, stems, flowers, fruit peels, and flesh with prominent purple or violet colors (Fig. 2; Supplementary Figure 1). In addition, the pigments in the dark purple fruit of transgenic lines gradually faded as the process of ripening starts (data not shown). The phenotypes referred to above were also confirmed in T1 and T2 generations of transgenic eggplant plants.
Overexpression of SmMYB1 promotes ubiquitous anthocyanin accumulation in transgenic eggplants. N, P, and R indicate T1 progeny of independent transgenic lines transformed with binary vector pBI121-SmMYB1, while WT indicates non-transformed plants. a The 35-day-old eggplant seedlings of non-transformed plants. b The 35-day-old eggplant seedling of transgenic lines. c Anthocyanin accumulates ubiquitously in leaves, petals, stamen, fruit peels, and fruit flesh of transgenic eggplants with WT as control
Identification and quantitative analyses of the purple pigments in the fruit flesh of transgenic eggplant
A total of seven different anthocyanins were separated and characterized from the extracts of the purple fruit flesh of transgenic eggplant using the HPLC–ESI–MS/MS method (Fig. 3). For the purpose of verifying the identity of anthocyanins in transgenic eggplant, we analyzed the fragmentation patterns of MS/MS (m/z) of the compounds displayed in HPLC profiles with reference to the information of radical groups reported previously (Supplementary Figure 2A; Table 1). Consequently, seven different anthocyanins were identified as the major ingredients of the purple pigments in the fruit flesh of transgenic eggplant (Table 1). It is worthy of note that most of the anthocyanins identified were delphinidin and delphinidin derivative-based pigments. Highly methylated delphinidin-based anthocyanins, malvidin-based pigments, malvidin 3-(feruloyl)rhamnoside(glucoside)-5-glucoside, and malvidin 3-(p-coumaroyl)rhamnoside(glucoside)-5-glucoside occupied 13 and 68 % of the total amount of anthocyanins, respectively. However, it is strange that only trace amount of the cyanidin-based anthocyanin, cyanidin 3-(p-coumaroyl)rhamnoside(glucoside)-5-glucoside (0.012 mg/g dry weight), in reference to peak 3 were detected in the purple flesh (Fig. 3; Table 1). Moreover, pelargonidin-based anthocyanins were not detected in the corresponding tissue in transgenic eggplant.
Identification of anthocyanins in the purple fruit flesh of transgenic eggplants. a HPLC profiles of anthocyanins of the purple fruit flesh. The purple fruit flesh framed by the red circle in the upper part indicates the tissues collected for HPLC–ESI–MS/MS analysis. Peak numbers refer to the anthocyanins listed in Table 1. b Structure and major cleavage of malvidin 3-(feruloyl)rhamnoside(glucoside)-5-glucoside in reference to peak 5 of Fig. 3a. c Structure and major cleavage of malvidin 3-(p-coumaroyl)rhamnoside(glucoside)-5-glucoside in reference to peak 6 of Fig. 3a. (Color figure online)
Total content of anthocyanins in fruit flesh was found to be 2.026 mg/g dry weight using HPLC (Table 1), while anthocyanins were absent in the corresponding tissue of wild-type plants. However, the total content of anthocyanins in the purple flesh of transgenic eggplant was not as high as that found in fruits of transgenic tomato with fruit-specific expression of two anthocyanin biosynthesis regulators (ROSEA and DEL) (Butelli et al. 2008). The methylated anthocyanin, malvidin 3-(p-coumaroyl)rhamnoside(glucoside)-5-glucoside in reference to peak 6 (Fig. 3a, c), showed the highest level (1.373 mg/g dry weight) in the flesh of transgenic eggplant (Table 1), while the content of malvidin 3-(feruloyl)rhamnoside(glucoside)-5-glucoside in reference to peak 5 (Fig. 3a, b) ranked second.
Total anthocyanin contents of various tissues in wild-type and transgenic eggplants
The anthocyanin contents of leaves, petals, stamens, fruit peels, and flesh in wild-type and transgenic lines were measured with a pH-differential spectrum method. Almost no anthocyanins were detected in various tissues of wild-type plants, while large amount of anthocyanins were extracted from leaves, petals, stamens, fruit peels, and flesh of transgenic lines. As shown in Supplementary Figure 3A, the highest level of anthocyanin accumulation was 2.26 mg/g fresh weight in the stamens of transgenic line P, while the total anthocyanin contents of petals and leaves ranked second and third, respectively. It is worth noting that considerable amounts of anthocyanin accumulation were also detected in fruit peels and flesh of transgenic eggplants (Supplementary Figure 3A).
Transcriptional analysis of SmMYB1 gene in wild-type and transgenic eggplants
To investigate the intrinsic molecular mechanisms underlying the specific pattern of anthocyanin accumulation, the expression profile of SmMYB1 and total contents of anthocyanins were analyzed in leaves, petals, stamens, fruit peels, and flesh of wild-type and transgenic eggplants. Supplementary Figure 3A shows that anthocyanins accumulate ubiquitously at high levels in transgenic eggplants compared with the background value in non-transformed eggplants. Supplementary Figure 3B shows the dramatically high expression of SmMYB1 in various plant tissues of transgenic lines compared with wild-type eggplant. However, the times of increase vary in different tissues of transgenic lines. In leaves, stamens, and flesh, SmMYB1 was up-regulated more than 1000-fold, while SmMYB1 was only up-regulated 50-fold in peels. In addition, the times of increase of SmMYB1 gene were found to be merely 4–5-fold in transgenic petals. It is worth mentioning that the times of increase depend on the background levels of SmMYB1 in different tissues of wild-type eggplant. However, the total contents of anthocyanins were closely in accordance with the expression levels of the SmMYB1 gene in various tissues of transgenic eggplant. These results demonstrate that overexpression of SmMYB1 promotes abundant anthocyanin biosynthesis ubiquitously in eggplant (Solanum aethiopicum group Gilo).
Expression profiles of anthocyanin biosynthesis genes in various tissues of wild-type and transgenic eggplant
In the earlier study, we showed that SmMYB1 lies in the same cluster as the other anthocyanin synthesis activators from various species and SmMYB1 could stimulate anthocyanin accumulation in kanamycin-resistant callus. To further study the molecular mechanisms underlying anthocyanin accumulation in transgenic eggplants, we investigated the expression profiles of anthocyanin structural genes SmPAL, SmCHS, SmCHI, SmF3H, SmF3′5′H, SmDFR, SmANS, and SmUFGT in leaves, petals, stamens, fruit peels, and flesh between wild-type and transgenic eggplant (Supplementary Figure 4A–H). Overall, these results showed that the expression levels of all structural genes, except SmPAL and SmCHI, were significantly up-regulated in different transgenic eggplant tissues compared with wild-type eggplant.
In detail, the expression levels of late structural genes SmF3′H, SmF3′5′H, SmDFR, SmANS, and SmUFGT were all dramatically up-regulated in transgenic tissues, especially in fruit flesh (Supplementary Figure 4D, E, F, G), whereas of the early structural genes, only SmCHS was significantly up-regulated in various transgenic tissues, with the exception of stamens. In the fruit flesh of transgenic lines, SmCHI was also up-regulated to a level which is comparable to the expression in other tissues. It is noteworthy that most structural genes showed the highest expression levels in petal and stamen tissues of wild-type plants. Therefore, the times of increase of these genes in transgenic petal and stamen tissues were limited. Even so, most late structural genes showed the highest levels of transcripts in petal and stamen tissues of transgenic lines. Thus, it can be seen that the expression profiles of most structural genes coincide well with the expression pattern of SmMYB1. Therefore, all the results mentioned above prove that the overexpression of transcription factor SmMYB1 in transgenic eggplants promotes anthocyanin accumulation by transcriptional activation of most anthocyanin biosynthesis genes.
Expression profiles of anthocyanin biosynthesis regulatory genes in various tissues of wild-type and transgenic eggplants
To investigate the mechanisms underlying the transcriptional regulation of structural genes, the expression levels of transcriptional factors SmbHLH1 and SmAN11 were analyzed in various tissues of wild-type and transgenic lines by qRT-PCR. Compared with the wild type, the expression of SmbHLH1 and SmAN11 both remained stable in various tissues of the transgenic lines, except in stamens (Supplementary Figure 5A and B). In the highly complicated regulatory network of anthocyanin biosynthesis, R2R3 MYB proteins participate in the transcriptional activation of the AtTT8 homologs and promote pigment production coordinately with bHLH proteins in many species (Butelli et al. 2008; Chiu et al. 2010; Yuan et al. 2009; Zhang et al. 2015). However, it is evident that the suppressed expression of SmbHLH1 did not arise directly from the overexpression of SmMYB1 in transgenic stamens. Similarity and phylogenetic analysis of SmbHLH1 with other bHLH proteins from various species suggests that SmbHLH1 should be a functional anthocyanin biosynthesis activator in eggplant. Here, we inferred that the decreased expression of SmbHLH1 for unknown reasons might be responsible for the down-regulation of SmPAL and SmCHS in transgenic stamens compared with non-transformed plants, whereas the slightly increase in SmAN11 indicates that the WD40 gene is induced by SmMYB1 in a non-stringent manner.
Overexpression of SmMYB1 gene enhances tolerance to freezing stress
The production of anthocyanins in vegetative tissues often increases the tolerance of plants to various environmental stressors (Holton and Cornish 1995; Klaper et al. 1996; Yakimchuk and Hoddinott 1994). To investigate whether high amounts of anthocyanins confer defense against freezing stress, the effects of freezing stress on the transgenic and non-transformed eggplants were tested in 35-day-old seedlings. No significant differences in growth rates were observed between the transgenic plants and wild-type plants cultured at 25 °C up to 35 days (Fig. 4a). By examining the dry weight of leaves at the same developmental stage, the results showed that there were no obvious differences between transgenic and wild-type plants (data not shown). However, we checked the detached-leaf water loss rate, and the results showed that the leaves from the transgenic plants had lower water loss rates than wild-type plants at each dehydration time point (Fig. 4d).
The appearance of wild-type (WT) and transgenic seedlings of eggplants under freezing stress. a The 35-day-old eggplant seedlings of WT and transgenic lines grown at 25 °C. b The 35-day-old eggplant seedlings of WT and transgenic lines after 8-h freezing treatment (−5 ± 1 °C). c The 35-day-old eggplant seedlings of WT and transgenic lines under rewarming conditions. d The water loss rates of WT and transgenic plants. e The electric conductance of leaves at each freezing treatment time point. The individual plants shown are representative of the three plants grown per treatment
For the freezing treatment, wild-type and transgenic plants of 35 days were incubated at −5 ± 1 °C for 0 h, 2 h, 4 h, and 8 h. After incubation under freezing stress for 8 h, all the leaves of the plants were injured, but the leaves of wild-type plants were injured much more seriously than those of the transgenic lines (Fig. 4b). In addition, the transgenic plants showed a good recovery upon rewarming conditions, while the wild-type plants eventually died because of the serious damage in leaves and buds (Fig. 4c). To further study the mechanisms underlying the increased freezing tolerance, the electric conductance of leaves was measured at each time point in the freezing treatment experiment. Figure 4e shows that the relative electrolyte leakage (REL) increased markedly slower in transgenic plants than in wild-type plants. These results indicate that the cell membrane of transgenic leaves was less damaged by the freezing treatment.
Discussion
Eggplant (Solanum melongena L.), an important agronomical solanaceous crop, is cultivated worldwide for its tender fruits (immature fruits). It was ranked as one of the top ten vegetables for its capacity of oxygen radical scavenging due to phenolic constituents in the fruit (Cao et al. 1996). Eggplant cultivars differ in fruit size, color, and shape, but the cultivars with dark purple pigments in fruit skin are apparently more popular for their high nutritional value. However, it is a pity that to date no anthocyanins have been detected in the flesh of the cultivars grown all over the world. Therefore, we attempted to cultivate new purple-fleshed eggplant cultivars by genetic engineering.
In a previous article, a functional MYB transcription factor, SmMYB1, was isolated from the edible cultivar (Zi Chang) of eggplant. In this study, a wild eggplant cultivar (Solanum aethiopicum group Gilo) with no visible anthocyanins was used for transformation. Fortunately, transgenic plants with high expression levels of SmMYB1 display abundant anthocyanin accumulation ubiquitously in various tissues including the flesh of fruits. Even stigmas also show light purple pigments at the top. These results suggest that anthocyanin production can in fact be induced in any tissues in wild eggplant.
By analyzing the purple pigments in flesh of transgenic eggplant with HPLC–ESI–MS/MS, malvidin-based pigments were identified as the chief components of the separated anthocyanins. In the biosynthesis pathway of anthocyanin, flavonoid 3′,5′-hydroxylase catalyzed DHK to produce DHM at positions 3′ and 5′ in the B ring with extremely high efficiency. Then, in subsequent reactions, O-methyltransferase (OMT) catalyzes methylation at the 3′ and/or 5′ hydroxyl groups of most delphinidins to produce petunidin or malvidin-based anthocyanins, respectively. Combined with the expression analysis of anthocyanin structural genes in various tissues between wild-type and transgenic lines, it can be seen that the dramatic up-regulation of SmF3′5′H was responsible for the abundant production of delphinidin. Meanwhile, it is not strange that pelargonidin- and cyanidin-based anthocyanins were seldom detected in the corresponding tissue in transgenic eggplant. Compared with the results previously reported, the anthocyanins identified in transgenic eggplant are evidently different from the pigments characterized in the non-transformed cultivars (Wu and Prior 2005). The anthocyanins were methylated in a great extent at the position of the B ring in fruit flesh of transgenic eggplants, whereas that is very rare in edible eggplant. Perhaps the high expression level of SmMYB1 activated the transcription of some uncharacterized OMTs in transgenic eggplant. It can be seen that all the anthocyanins in transgenic eggplant were glycosylated pigments which arise from glycosylation at the C5 and C3 positions of anthocyanins, while the acylated form of modification at the C3 position of the C ring in anthocyanin precursors seems quite common in this study. These results indicate that glycosylation, acylation, methylation, and hydroxylation of anthocyanin precursors in flavonoid metabolism were the main patterns of modification in fruit flesh of transgenic eggplant.
Although no visible anthocyanin pigmentation was apparently observed in the wild-type tissues studied above, a certain amount of transcripts of all the anthocyanin structural genes were detected in the corresponding tissues, especially in petals and stamens. These results indicate that adequate transcripts of most structural genes should be a prerequisite for large amounts of anthocyanin accumulation in various tissues of transgenic eggplant. This phenomenon seems quite common in many species which showed visible anthocyanin accumulation (Borevitz et al. 2000; Chiu et al. 2010; Mathews 2003; Yuan et al. 2009; Zhang et al. 2014b, 2015). Obviously, the threshold of structural genes for activating abundant anthocyanin production should be much higher than the expression levels in petals and stamens of non-transformed eggplant.
In the phenylpropanoid pathway, the transcripts of SmPAL were almost unaffected in transgenic tissues except for a visible decrease in stamens (Supplementary Figure 4A). In addition, the transcript levels of SmCHS were down-regulated by nearly 80 % in stamens, compared with those significantly up-regulated in leaves, petals, fruit peels, and flesh of transgenic lines (Supplementary Figure 4B). As we have mentioned above, the total anthocyanin contents in stamens were apparently higher than all the other tissues of the transgenic lines, so it seems that large amounts of pigment production might impact negatively on the normal metabolism and development of transgenic stamens. Therefore, the transcription of SmPAL and SmCHS might be adjusted to a lower level to accommodate the global metabolism in purple stamens, the so-called feedback effect (Baudry et al. 2006).
Expression analysis of anthocyanin structural and regulatory genes in various tissues between wild-type and transgenic lines indicates that most anthocyanin biosynthetic genes were up-regulated accordingly by the transcriptional activation of the MBW complex consisting of SmMYB1 and other interaction partners. R2R3 MYB TFs have been implicated in regulation of anthocyanin biosynthesis by activating the transcription of anthocyanin structural genes in many plants (Albert et al. 2011; Borevitz et al. 2000; Bovy 2002; Jung et al. 2009; Mathews 2003; Schwinn et al. 2006; Shin et al. 2006; Takos et al. 2006). In addition, overexpression of bHLH transcription factor solely can result in efficient anthocyanin accumulation in transgenic tomato (Albert et al. 2011; Bovy 2002; Mooney et al. 1995; Xie et al. 2012; Zhang et al. 2014b). In addition, the expression levels of most structural genes are dramatically up-regulated in transgenic eggplant tissues, while the expression of SmCHS and SmPAL was evidently suppressed in transgenic stamens. This abnormal phenomenon suggests that excessive production of anthocyanins may be toxic for the normal development and growth of stamens. The plants probably adjusted the anthocyanin production by inhibiting the transcription of several structural genes in the stamens themselves. Moreover, the suppression of SmCHS and SmPAL might arise from the slight down-regulation of SmbHLH1 for unknown reasons in transgenic stamens.
Previous studies indicate a close link between freezing stress and anthocyanin accumulation in plants (Ahmed et al. 2014; Bovy 2002; Hannah et al. 2006; Xie et al. 2012). The high content of anthocyanin confers better tolerance to chilling stress in seedlings. Heterologous expression of LeAN2 induces anthocyanin accumulation and also enhances resistance to chilling stress in Arabidopsis (Meng et al. 2014). In this study, we investigated the tolerance of transgenic and wild-type seedlings of eggplant under freezing stress. The results showed that the transgenic seedlings were less injured than wild-type under freezing stress for 8 h. The results of the relative electrolyte leakage (REL) assay demonstrated that membrane damage was less serious in transgenic seedlings. The lower water loss rates also indicated that transgenic seedlings may possess better tolerance to stressors (Harborne and Williams 2000). In addition, transgenic seedlings have exerted better recovery under rewarming conditions. These results suggest that a high concentration of anthocyanins provides transgenic eggplant seedlings with more tolerance to freezing stress.
Colorful vegetables and fruits rich in anthocyanins are attracting increasing attention from consumers for their eye-catching colors and health-promoting effects. Many technologists are focusing on breeding new cultivars of vegetables with high contents of anthocyanins with plant biotechnology (Butelli et al. 2008; Zhang et al. 2014a). In this study, transgenic eggplants with purple-fleshed fruit were generated. However, the distribution of anthocyanins was not uniform in the flesh, and the total content was not comparable to that of transgenic tomato engineered by the combined expression of an R2R3MYB protein and a bHLH protein from snap-dragon driven by the fruit-specific promoter E8, since the expression levels of most anthocyanin biosynthesis genes in fruit flesh are much lower than other tissues including fruit peels, stamens, and leaves. These results in this article show that overexpression of SmMYB1 only was insufficient to drive the abundant biosynthesis of anthocyanin in fruit flesh. More efforts should be given to the creation of transgenic eggplant with more anthocyanin biosynthesis regulators driven by fruit-specific promoters. Nonetheless, we provide a good foundation for the generation of genome-modified eggplant with higher content of anthocyanins. Moreover, the transgenic eggplant seedlings displayed a greater tolerance to freezing stress than wild-type. In conclusion, this work provides important information for breeding vegetables with health-promoting ingredients and better resistance to abiotic stress.
Abbreviations
- PAL:
-
Phenylalanine ammonia lyase
- C4H:
-
Cinnamate 4-hydroxylase
- 4CL:
-
4-Coumarate-CoA ligase
- CHS:
-
Chalcone synthase
- CHI:
-
Chalcone isomerase
- F3H:
-
Flavone 3-hydroxylase
- F3′H:
-
Flavonoid 3′-hydroxylase
- DFR:
-
Dihydroflavonol reductase
- ANS:
-
Anthocyanidin synthase
- UFGT:
-
Flavonoid-3-O-glucosyltransferase
- GST:
-
Glutathione S-transferase
- OMT:
-
O-Methyltransferase
- DHK:
-
Dihydrokaempferol
- DHQ:
-
Dihydroquercetin
- DHM:
-
Dihydromyricetin
- HPLC:
-
High-performance liquid chromatography
- ESI–MS/MS:
-
Electrospray ionization tandem mass spectrometry
- qRT-PCR:
-
Quantitative real-time PCR
References
Ahmed NU, Park J-I, Jung H-J, Hur Y, Nou I-S (2014) Anthocyanin biosynthesis for cold and freezing stress tolerance and desirable color in Brassica rapa. Funct Integr Genomic 15:383–394
Albert NW, Lewis DH, Zhang H, Schwinn KE, Jameson PE, Davies KM (2011) Members of an R2R3-MYB transcription factor family in Petunia are developmentally and environmentally regulated to control complex floral and vegetative pigmentation patterning. Plant J Cell Mol Biol 65:771–784
Ban Y, Honda C, Hatsuyama Y, Igarashi M, Bessho H, Moriguchi T (2007) Isolation and functional analysis of a MYB transcription factor gene that is a key regulator for the development of red coloration in apple skin. Plant Cell Physiol 48:958–970
Baudry A, Heim MA, Dubreucq B, Caboche M, Weisshaar B, Lepiniec L (2004) TT2, TT8, and TTG1 synergistically specify the expression of BANYULS and proanthocyanidin biosynthesis in Arabidopsis thaliana. Plant J 39:366–380
Baudry A, Caboche M, Lepiniec L (2006) TT8 controls its own expression in a feedback regulation involving TTG1 and homologous MYB and bHLH factors, allowing a strong and cell-specific accumulation of flavonoids in Arabidopsis thaliana. Plant J Cell Mol Biol 46:768–779
Bernhardt C, Zhao M, Gonzalez A, Lloyd A, Schiefelbein J (2005) The bHLH genes GL3 and EGL3 participate in an intercellular regulatory circuit that controls cell patterning in the Arabidopsis root epidermis. Development 132:291–298
Borevitz JO, Xia Y, Blount J, Dixon RA, Lamb C (2000) Activation tagging identifies a conserved MYB regulator of phenylpropanoid biosynthesis. Plant Cell 12:2383–2393
Bovy A (2002) High-flavonol tomatoes resulting from the heterologous expression of the maize transcription factor genes LC and C1. Plant Cell Online 14:2509–2526
Bradshaw HD, Schemske DW (2003) Allele substitution at a flower colour locus produces a pollinator shift in monkeyflowers. Nature 426:176–178
Broun P (2005) Transcriptional control of flavonoid biosynthesis: a complex network of conserved regulators involved in multiple aspects of differentiation in Arabidopsis. Curr Opin Plant Biol 8:272–279
Butelli E, Titta L, Giorgio M, Mock HP, Matros A, Peterek S, Schijlen EG, Hall RD, Bovy AG, Luo J, Martin C (2008) Enrichment of tomato fruit with health-promoting anthocyanins by expression of select transcription factors. Nat Biotechnol 26:1301–1308
Cao G, Sofic E, Prior RL (1996) Antioxidant capacity of tea and common vegetables. J Agric Food Chem 44:3426–3431
Chiu LW, Zhou X, Burke S, Wu X, Prior RL, Li L (2010) The purple cauliflower arises from activation of a MYB transcription factor. Plant Physiol 154:1470–1480
de Pascual-Teresa S, Sanchez-Ballesta MT (2007) Anthocyanins: from plant to health. Phytochem Rev 7:281–299
Espley RV, Bovy A, Bava C, Jaeger SR, Tomes S, Norling C, Crawford J, Rowan D, McGhie TK, Brendolise C, Putterill J, Schouten HJ, Hellens RP, Allan AC (2013) Analysis of genetically modified red-fleshed apples reveals effects on growth and consumer attributes. Plant Biotechnol J 11:408–419
Filippone E, Lurquin P (1989) Stable transformation of eggplant (Solanum melongena L.) by cocultivation of tissues with Agrobacterium tumefaciens carrying a binary plasmid vector. Plant Cell Rep 8:370–373
Goff SA, Cone KC, Chandler VL (1992) Functional analysis of the transcriptional activator encoded by the maize B gene: evidence for a direct functional interaction between two classes of regulatory proteins. Gene Dev 6:864–875
Gonzalez A, Zhao M, Leavitt JM, Lloyd AM (2008) Regulation of the anthocyanin biosynthetic pathway by the TTG1/bHLH/Myb transcriptional complex in Arabidopsis seedlings. Plant J Cell Mol Biol 53:814–827
Gonzali S, Mazzucato A, Perata P (2009) Purple as a tomato: towards high anthocyanin tomatoes. Trends Plant Sci 14:237–241
Grotewold E (2006) The genetics and biochemistry of floral pigments. Annu Rev Plant Biol 57:761–780
Hannah MA, Wiese D, Freund S, Fiehn O, Heyer AG, Hincha DK (2006) Natural genetic variation of freezing tolerance in Arabidopsis. Plant Physiol 142:98–112
Harborne JB, Williams CA (2000) Advances in flavonoid research since 1992. Phytochemistry 55:481–504
Holton TA, Cornish EC (1995) Genetics and biochemistry of anthocyanin biosynthesis. Plant Cell 7:1071–1083
Honda C, Kotoda N, Wada M, Kondo S, Kobayashi S, Soejima J, Zhang Z, Tsuda T, Moriguchi T (2002) Anthocyanin biosynthetic genes are coordinately expressed during red coloration in apple skin. Plant Physiol Biochem 40:955–962
Jaakola L (2013) New insights into the regulation of anthocyanin biosynthesis in fruits. Trends Plant Sci 18:477–483
Jaakola L, Poole M, Jones MO, Kamarainen-Karppinen T, Koskimaki JJ, Hohtola A, Haggman H, Fraser PD, Manning K, King GJ, Thomson H, Seymour GB (2010) A SQUAMOSA MADS box gene involved in the regulation of anthocyanin accumulation in bilberry fruits. Plant Physiol 153:1619–1629
Jung CS, Griffiths HM, De Jong DM, Cheng S, Bodis M, Kim TS, De Jong WS (2009) The potato developer (D) locus encodes an R2R3 MYB transcription factor that regulates expression of multiple anthocyanin structural genes in tuber skin. Theor Appl Genet 120:45–57
Klaper R, Frankel S, Berenbaum MR (1996) Anthocyanin content and UVB sensitivity in Brassica rap. Photochem Photobiol 63:811–813
Koes R, Verweij W, Quattrocchio F (2005) Flavonoids: a colorful model for the regulation and evolution of biochemical pathways. Trends Plant Sci 10:236–242
Lalusin A, Nishita K, Kim S-H, Ohta M, Fujimura T (2006) A new MADS-box gene (IbMADS10) from sweet potato (Ipomoea batatas (L.) Lam) is involved in the accumulation of anthocyanin. Mol Genet Genomics 275:44–54
Lepiniec L, Debeaujon I, Routaboul JM, Baudry A, Pourcel L, Nesi N, Caboche M (2006) Genetics and biochemistry of seed flavonoids. Annu Rev Plant Biol 57:405–430
Li HM, Rotter D, Hartman TG, Pak FE, Havkin-Frenkel D, Belanger FC (2006) Evolution of novel O-methyltransferases from the Vanilla planifolia caffeic acid O-methyltransferase. Plant Mol Biol 61:537–552
Mano H, Ogasawara F, Sato K, Higo H, Minobe Y (2007) Isolation of a regulatory gene of anthocyanin biosynthesis in tuberous roots of purple-fleshed sweet potato. Plant Physiol 143:1252–1268
Mathews H (2003) Activation tagging in tomato identifies a transcriptional regulator of anthocyanin biosynthesis, modification, and transport. Plant Cell Online 15:1689–1703
Mendez M, Jones DG, Manetas Y (1999) Enhanced UV-B radiation under field conditions increases anthocyanin and reduces the risk of photoinhibition but does not affect growth in the carnivorous plant Pinguicula vulgaris. New Phytol 144:275–282
Meng X, Yin B, Feng H-L, Zhang S, Liang X-Q, Meng Q-W (2014) Overexpression of R2R3-MYB gene leads to accumulation of anthocyanin and enhanced resistance to chilling and oxidative stress. Biol Plantarum 58:121–130
Mooney M, Desnos T, Harrison K, Jones J, Carpenter R, Coen E (1995) Altered regulation of tomato and tobacco pigmentation genes caused by the delila gene of Antirrhinum. Plant J 7:333–339
Morita Y, Saitoh M, Hoshino A, Nitasaka E, Iida S (2006) Isolation of cDNAs for R2R3-MYB, bHLH and WDR transcriptional regulators and identification of c and ca mutations conferring white flowers in the Japanese morning glory. Plant Cell Physiol 47:457–470
Park KI, Ishikawa N, Morita Y, Choi JD, Hoshino A, Iida S (2007) A bHLH regulatory gene in the common morning glory, Ipomoea purpurea, controls anthocyanin biosynthesis in flowers, proanthocyanidin and phytomelanin pigmentation in seeds, and seed trichome formation. Plant J Cell Mol Biol 49:641–654
Park NI, Xu H, Li X, Jang IH, Park S, Ahn GH, Lim YP, Kim SJ, Park SU (2011) Anthocyanin accumulation and expression of anthocyanin biosynthetic genes in radish (Raphanus sativus). J Agric Food Chem 59:6034–6039
Rapisarda P, Fanella F, Maccarone E (2000) Reliability of analytical methods for determining anthocyanins in blood orange juices. J Agric Food Chem 48:2249–2252
Schwinn K, Venail J, Shang Y, Mackay S, Alm V, Butelli E, Oyama R, Bailey P, Davies K, Martin C (2006) A small family of MYB-regulatory genes controls floral pigmentation intensity and patterning in the genus Antirrhinum. Plant Cell 18:831–851
Shi MZ, Xie DY (2014) Biosynthesis and metabolic engineering of anthocyanins in Arabidopsis thaliana. Recent Pat Biotechnol 8:47–60
Shin Y-M, Park H-J, Yim S-D, Baek N-I, Lee C-H, An G, Woo Y-M (2006) Transgenic rice lines expressing maize C1 and R-S regulatory genes produce various flavonoids in the endosperm. Plant Biotechnol J 4:303–315
Spelt C, Quattrocchio F, Mol JN, Koes R (2000) Anthocyanin1 of petunia encodes a basic helix–loop–helix protein that directly activates transcription of structural anthocyanin genes. Plant Cell 12:1619–1632
Takos AM, Jaffe FW, Jacob SR, Bogs J, Robinson SP, Walker AR (2006) Light-induced expression of a MYB gene regulates anthocyanin biosynthesis in red apples. Plant Physiol 142:1216–1232
Tanaka Y, Sasaki N, Ohmiya A (2008) Biosynthesis of plant pigments: anthocyanins, betalains and carotenoids. Plant J 54:733–749
Toppino L, Valè G, Rotino GL (2008) Inheritance of Fusarium wilt resistance introgressed from Solanum aethiopicum Gilo and Aculeatum groups into cultivated eggplant (S. melongena) and development of associated PCR-based markers. Mol Breed 22:237–250
Tsuda T, Horio F, Osawa T (2000) The role of anthocyanins as an antioxidant under oxidative stress in rats. BioFactors 13:133–139
Van der Krol AR, Mur LA, Beld M, Mol J, Stuitje AR (1990) Flavonoid genes in petunia: addition of a limited number of gene copies may lead to a suppression of gene expression. Plant Cell Online 2:291–299
Winkel-Shirley B (2001) Flavonoid biosynthesis. A colorful model for genetics, biochemistry, cell biology, and biotechnology. Plant Physiol 126:485–493
Wu X, Prior RL (2005) Identification and characterization of anthocyanins by high-performance liquid chromatography–electrospray ionization-tandem mass spectrometry in common foods in the United States: vegetables, nuts, and grains. J Agric Food Chem 53:3101–3113
Xie XB, Li S, Zhang RF, Zhao J, Chen YC, Zhao Q, Yao YX, You CX, Zhang XS, Hao YJ (2012) The bHLH transcription factor MdbHLH3 promotes anthocyanin accumulation and fruit colouration in response to low temperature in apples. Plant Cell Environ 35:1884–1897
Yakimchuk R, Hoddinott J (1994) The influence of ultraviolet-B light and carbon dioxide enrichment on the growth and physiology of seedlings of three conifer species. Can J For Res 24:1–8
Yuan Y, Chiu LW, Li L (2009) Transcriptional regulation of anthocyanin biosynthesis in red cabbage. Planta 230:1141–1153
Zhang Y, Butelli E, Martin C (2014a) Engineering anthocyanin biosynthesis in plants. Curr Opin Plant Biol 19:81–90
Zhang Y, Chen G, Dong T, Pan Y, Zhao Z, Tian S, Hu Z (2014b) Anthocyanin accumulation and transcriptional regulation of anthocyanin biosynthesis in Purple Bok Choy (Brassica rapa var. Chinensis). J Agric Food Chem 62:12366–12376
Zhang Y, Hu Z, Chu G, Huang C, Tian S, Zhao Z, Chen G (2014c) Anthocyanin accumulation and molecular analysis of anthocyanin biosynthesis-associated genes in eggplant (Solanum melongena L.). J Agric Food Chem 62:2906–2912
Zhang Y, Hu Z, Zhu M, Zhu Z, Wang Z, Tian S, Chen G (2015) Anthocyanin accumulation and molecular analysis of correlated genes in Purple Kohlrabi (Brassica oleracea var. gongylodes L.). J Agric Food Chem 63:4160–4169
Acknowledgments
This work was supported by National Natural Science Foundation of China (No. 31572129), the Fundamental Research Funds for the Central Universities (No. 106112015CDJZR235504), and Technology System of National Bulk Vegetable Industry–Eggplant Breeding Position (CARS-25-A-06).
Author information
Authors and Affiliations
Corresponding author
Additional information
Yanjie Zhang and Guihua Chu have contributed equally to this work.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Zhang, Y., Chu, G., Hu, Z. et al. Genetically engineered anthocyanin pathway for high health-promoting pigment production in eggplant. Mol Breeding 36, 54 (2016). https://doi.org/10.1007/s11032-016-0454-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11032-016-0454-2