Abstract
Invertases (EC 3.2.1.26, INV) are indispensable for plant metabolism, development and stress response. Carbohydrate partitioning and the irreversible hydrolysis of sucrose into glucose and fructose in plants are driven by invertases. We performed a comprehensive analysis on the physico-chemical characteristics, chromosome localisation, exon–intron structures, motif distributions, evolutionary divergence and phylogenetic analysis, and tissue-specific spatio-temporal expression profiles of invertase genes under varied abiotic stressors and phytohormones. We identified twenty-four invertase genes (comprising nine cell wall (CWINV), two cell membrane (CMINV), eleven chloroplast (ChlINV), one cytosol (CyINV) and one vacuolar (VaINV)) in the tomato genome distributed on eight of the twelve chromosomes. The phylogenetic analysis clustered the invertase genes into two major clades. Segmental duplication contributed to the alkaline/neutral sub-domain expansion. The invertase genes were differentially expressed in fruits, roots, stems, leaves, flower buds and responded differentially to light, cold, drought, salinity stress and phytohormones (abscisic acid (ABA), gibberellic acid (GA), and indole-3 acetic acid (IAA)). Majority of the genes that were significantly up-regulated under the phytohormone treatments were consistently down-regulated in the stem, flowerbud and roots. The expression data reveals most of the cell wall and chloroplast-localised invertases were induced by phytohormones; while vacuolar and cytosol-localised invertases were markedly induced by abiotic stresses in the diverse tissues. The network interaction complex evidences that, most of the proteins were implicated in glycolysis, sucrose synthesis and/or pyrophosphate-fructose 6-phosphate 1-phosphotransferase activity. This study provides the groundwork for future functional analysis of invertase genes in tomato and other Solanaceae species.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Tomato (Solanum lycopersicum) is among the few healthful and commercially farmed vegetable crops, the third most widely produced and consumed vegetable globally after potato (Solanum tuberosum L.) and onion (Allium cepa L.) (Ludewig and Flügge 2013). It is also a major vegetable crop with over 153 million metric tonnes of global production (FAO 2017). It is well adapted to diverse agro-environments and serves as a source of useful genes for Solanaceae breeding programmes (Gur and Zamir 2014). It is a fairly short duration crop with high yield and economically attractive for both smallholder and commercial agriculture (Julius et al. 2017). Tomato is a major industrial crop reputed for its unsurpassable carotenoids especially beta-carotene and antioxidants (Zhang et al. 2013), vitamins A, B, C and E; nicotinic acid and lycopene (Li et al. 2019; Sahu and Chattopadhyay 2017) and other pharmaceutical compounds (Zanor et al. 2009). In molecular biology, tomato is a model plant for the study of fleshy fruit biology and physiology (Klee and Giovannoni 2011), phyto-compounds and metabolites, floral anatomy and plant architecture (Kimura and Sinha 2008), and plant response to stress (Chen et al. 2015a, b; Tomato Genome Consortium 2012; Wan et al. 2018).
Invertases are important enzymes in sucrose metabolism in higher plants. Crop growth and development is predominantly controlled by phyto-assimilation and translocation of photosynthates through assimilates partitioning into distinctive sink tissues (Bihmidine et al. 2015; Braun 2014; Yadav et al. 2015). Invertases hydrolyse sucrose, the major end-product of photosynthesis, into glucose and fructose for subsequent utilisation as nutrients, energy source and signalling molecules for crop growth, and stress responses (Wang et al. 2019a, b). Alkaline/neutral, cell wall and vacuolar invertases constitute the main types of invertase isoenzymes in higher plants. These useful enzymes are usually differentiated by their optimum pH, isoelectric points, subcellular localisation, and solubility (Yao et al. 2014). The enzymatic and biochemical properties of the cell wall and vacuolar invertases are in near similitude: thus, they have an acidic pH optimum of 4.5–5.5 and can cleave raffinose and stachyose. Glycoproteins are the prime constituents of acid invertases with three conserved sequence motifs: β-fructofuranosidase motif [NDPNG(A)], RDP and WECP(V)D motifs’ (Ji et al. 2005; Juárez-Colunga et al. 2018; Nonis et al. 2008; Yao et al. 2014). The cell wall invertases are usually formed as tightly bound-to-the-cell wall or as soluble molecules residing in the vacuole (Juárez-Colunga et al. 2018; Shen et al. 2019). It regulates sucrose partitioning (Tang et al. 1999), wounding and pathogen infection response (Schaarschmidt et al. 2006), seed formation and pollen growth (Jain et al. 2010). Conversely, the vacuolar invertase regulates fruits and sugar constituent of storage organs (Yu et al. 2008), cell expansion and osmotic stress response (Qi et al. 2007), and drought stress response (Roitsch and González 2004). Cell wall invertases limit transfer of carbon from source organs (leaves) and regulate sucrose to hexose in tomato apoplasts (Kocal et al. 2008). Cell wall invertases have also been implicated in apoplastic cleavage of sucrose in maize seed development (Chourey et al. 2006) and regulate sucrose metabolism in source and sink organs. However, a detailed characterisation of invertase gene family in tomato remains uncharted.
In tomato, the Glycoside hydrolase family 32 (GH32) comprises enzymes exhibiting invertase/fructofuranosidase (EC: 3.2.1.26); inulinase (EC: 3.2.1.7); levanase (EC: 3.2.1.65); exo-inulinase (EC: 3.2.1.80); sucrose: sucrose 1-fructosyltransferase (EC: 2.4.1.99); and fructan: fructan 1-fructosyltransferase (EC: 2.4.1.100) activities (Slugina et al. 2018). In tomato, a number of cell wall invertase genes and their functions have been reported (Fridman 2003; Fotopoulos 2005; Zhang et al. 2015). However, climate variability in recent years has exacerbated environmental stresses and thus, significantly impacting crop growth and development, resulting in their limited terrestrial distribution and production (Hirakawa et al. 2013). Soluble sugar accretion can stabilise cellular components and membranes under cold stress (Julius et al. 2017; Ruan et al. 2010). For instance, Le Hir et al. (2015) and Guo et al. (2018) reported that, varying temperatures significantly influence fructose homeostasis in the fructose-specific transporter, AtSWEET17 in Arabidopsis thaliana. The tomato genome is one of the smallest diploid genomes (950 Mb) within the Solanum genus; with a highly conserved gene order and thus, considered a reference genome (Tomato Genome Consortium 2012). Gene families originate from a common ancestry either through duplication or evolutionary divergence. Several genes reportedly emanate from gene families; about 41 and 77% of predicted proteins are members of distinct gene families in A. thaliana (Tomato Genome Consortium 2012) and Oryza sativa (Goff et al. 2002), respectively. Genome-wide profiling of genes in plant genomes is a valuable tool for identifying pathway components and for characterising gene-specific functions (Wan et al. 2018; Zhang et al. 2019a). Invertase gene family have been previously studied in the model species, Arabidopsis (Tymowska-Lalanne and Kreis 1998), rice (Ji et al. 2005), cassava (Yao et al. 2014), populus (Chen et al. 2015a), and recently in sugarcane (Wang et al. 2017), maize (Juárez-Colunga et al. 2018) and pepper (Shen et al. 2019). The study of tomato invertase genes is key to advancing new knowledge and deepening understanding of invertase gene-specific functions and their underlying molecular mechanisms. Genes involved in the same biological processes tend to regulate similar biological mechanisms. Thus, by analysing variations in gene expressions associated with biological processes, expression kinetics and spatio-temporal localisation under varying conditions may expedite identification of common gene set (Ogawa et al. 2018; Zhang et al. 2019a). Yet, invertase gene family and their roles in abiotic stress and phytohormone response in tomato remains unexplored. Thus, exploring the roles of invertases in abiotic stress and phytohormone response is essential for understanding stress response.
In this study, genome-wide identification, characterisation and spatio-temporal expression profile of invertase genes of the tomato genome were undertaken. Our results provide insights into the functions of invertase family genes from an evolutionary perspective and their potential roles in phytohormone and abiotic stress response.
Materials and Methods
Identification of Invertase Genes in S. lycopersicum and Other Species
Genomic sequences of invertase genes from A. thaliana (http://www.arabidopsis.org/), rice (O. sativa) (Wan et al. 2018), maize (Zea mays) (Juárez-Colunga et al. 2018), grape (Vitis vinifera) (Chen et al. 2015a, b), pawpaw (Carica papaya) (Ding et al. 2018), and Irish potato (S. tuberosum) (Liu et al. 2019) were used to search for complete invertase genes from phytozome (http://www.phytozome.net/) and PIECEII databases (http://www.bioinfogenome.net/piece/). Invertase gene information from Brachypodium distachyon, C. papaya, Glycine max, S. tuberosum, V. vinifera, and Z. mays were downloaded from the PLAZA (v2.5) database (http://bioinformatics.psb.ugent.be/plaza/news/index (Van Bel et al. 2012). BLAST results with similarity indices of > 50 % and < 10−5 probability scores were retrieved as candidate sequences and further validated using BLAST and BLASTX in the Solanaceae Genomic Network (SGN) database (https://solgenomics.net/organism/genome).
Additionally, candidate protein motifs were validated using the Hidden Markov Model (HMM) corresponding to specific binding domains from the protein families database (Pfam 31.0, http://pfam.xfam.org/) and putative invertase proteins with ‘reliable thresholds’ were identified (Jin et al. 2009; Finn et al. 2016) using the conserved domain database (CDD) (https://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi). The open reading frames (ORF) were retrieved using the NCBI ORFfinder (https://www.ncbi.nlm.nih.gov/orffinder/). The subcellular localisation of the invertase genes were predicted using the Plant-PLoc: Predicting plant protein subcellular location server (http://www.csbio.sjtu.edu.cn/bioinf/plant-multi/). The invertase genes were named according to previously reported nomenclature (Zhang et al. 2019a, b) and were clustered into six subcellular-specific clades (chloroplast (Chl-I & II), cell wall (CW-I, II & II), and vacuole (Va).
Multiple Sequence Alignment, Conserved Motifs and Invertase Gene Structures
The ProtParam tool (https://web.expasy.org/protparam/) was employed to determine the molecular weight (Mw), amino acids length (aa) and theoretical isoelectric points (pI). The intron–exon gene structure was visualised using the Gene Structure Display Server (GSDS, http://gsds.cbi.pku.edu.cn/index.php) based on the genomic sequences (GS) and the corresponding coding sequences (CDS) of each invertase gene (Hu et al. 2015). The conserved protein motifs were identified using the Multiple EM for Motif Elicitation (MEME) online tool (http://meme-suite.org/tools/meme) based on default settings (Bailey et al. 2009). The SignalP 4.1 server (http://www.cbs.dtu.dk/ser-vices/SignalP/) was used to predict the presence and location of signal peptide cleavage sites in the amino acid sequences. Multiple alignment of all the invertase proteins was performed using ClustalW (Larkin et al. 2007), and phylogenetic tree was constructed by MEGA X10 with a bootstrap of 1000 replicates using the neighbour-joining (NJ) method (Kumar et al. 2016).
Chromosomal Localisation and Gene Duplication in S. lycopersicum
The chromosomal localisation of the invertase genes were visualised using the MapChart software (https://www.wur.nl/en/show/Mapchart.html) based on positional information (Cho et al. 1999). The phytozome database (https://phytozome.jgi.doe.gov/pz/portal.html) was used to identify the orthologues and paralogues of the invertase proteins in S. lycopersicum and A. thaliana. The gene duplication events and divergence time were estimated as synonymous substitution (Ks) and non-synonymous substitution rates (Ka) using the KaKs_Calculator Toolbox (ver. 2.0) (Zhang et al. 2006). The divergence time was computed as, T = Ks/(2 × r × 10−8) × 10−6 million years ago (MYA) with the r = 15 × 10−8 synonymous substitutions per site per year for dicotyledonous plants (Rozas et al. 2017).
Homology Modelling, Protein–Protein Interaction and Gene Enrichment Analysis
We performed a three-dimensional homology modelling on all the 24 invertase proteins using the Protein Homology/analogY Recognition Engine (Phyre2 server, V 2.0; http://www.sbg.bio.ic.ac.uk/~phyre2/html/page.cgi?id=index) (Kelley et al. 2015) in the intensive mode. Fitted models were validated using Ramachandran plots. Models were superimposed to assess similarity or divergence of generated models and estimate fractions of the structure coverage. The STRING database (https://string-db.org/cgi/) was used to obtain putative protein–protein interaction among the invertase proteins and related proteins. Output from the STRING database was subsequently visualised in the standalone version of Cytoscape software (https://cytoscape.org/; Lopes et al. 2010). We ran a BLASTX search against the UNIPROT database using the protein sequences at a 1e−30 significance level. The matches were mined and compared to the GO annotation generated against UNIPROT hits. The GO annotation of the invertase genes were retrieved from the WEGO 2.0 database (http://wego.genomics.org.cn/).
In-Silico Expression Analyses and Prediction of Cis-Acting Regulatory Elements
Publicly available expression data for the invertase genes were downloaded from the tomato Expression Atlas database (TEA; http://tea.solgenomics.net/) for further analyses. The TEA database holds expression profiles of diverse organs/tissues including roots, leaf, flower buds and fruits at varied developmental stages from the tomato cultivar, Heinz 1706 (http://tea.solgenomics.net/). The expression data was log2 transformed and heatmapped using the Toolkit for Biologists integrating various biological data handling tools (TBtools) (Chen et al. 2018). Sequences from the promoter region (1.5 kb upstream of the start codon) of each gene was retrieved from the SGN database (https://solgenomics.net/organism/genome) in Generic File Format (GFF) to identify putative cis-acting regulatory elements (CAREs) using the PlantCare database (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/).
Expression Profiles of Invertase Genes Under Abiotic Stress and Phytohormonal Treatments
Response of the invertase genes to cold, drought, light, salinity, phytohormones including abscisic acid (ABA), gibberellic acid (GA), and indole-3 acetic acid (IAA (auxin)) were studied using the advanced inbred tomato line (Ailsa Craig). Seeds were grown in sterilised soil at 25 °C day/18 °C night, with a photoperiod of 12 h light/10 h darkness under greenhouse conditions. After 4 weeks, cold, drought, salinity and light stress were imposed on seedlings. Seedlings were placed under 5 °C for cold stress and samples were collected at 0, 3, 6, 12, 24 and 48 h after imposition of cold stress. In the salinity treatment, 200 mM NaCl was sprayed on seedlings to impose salt stress. The first fully expanded leaves from the apical buds were harvested at 0, 1, 3, 6, 12 and 24 h after treatments. For drought treatment, selected seedlings were cautiously pulled out from the soil and soil attached to the roots were gently washed off. Subsequently, the seedlings were placed on clean desks at room temperature. Samples were collected at 0, 1, 3, 6, 12, and 24 h. Plants without any treatment were considered as controls.
The 4 week old seedlings were grown in a greenhouse under a 16 h/8 h light/dark cycle at 25 °C. Plants were exposed to continuous light at 25 °C for 16 h followed by 8 h continuous dark under 25 °C. In a 48 h photoperiod, fruits were harvested to determine light stress on tomato fruits. In the phytohormonal treatments, seedlings were sprayed with 100 mM abscisic acid (ABA), 100 mM gibberellic acid (GA), 100 mM indole-3 acetic acid (IAA) and the first fully expanded leaves from the apical buds were harvested at 0, 1, 3, 6, 12 and 24 h for each treatment, respectively. Seedlings without any treatments were regarded as controls. Harvested leaves and fruits were subsequently collected and lyophilised in liquid nitrogen and stored in − 80 °C refrigerator for RNA extraction.
Total RNA was extracted using the Vazyme Plant RNA Extraction Kit according to the manufacturer’s instructions (Vazyme, China). The first-strand cDNA was synthesised with the HiScript™ II first-strand cDNA synthesis kit (Vazyme, China) and qRT-PCR reactions were performed using HiScript II QRT SuperMix (Vazyme, China) on a Roche LightCycler® 480II PCR System. Gene specific primers were designed using the Perl Primer standalone software (Premier Biosoft, CA, USA). Primer sequences of selected genes are presented in Table S1. β-actin was used as a reference gene (Li et al. 2014). All reactions were performed in triplicates. The 2−△△CT method (Livak and Schmittgen 2001) was employed to compute relative gene expressions. The expression data was log2 transformed and heatmapped using TBtools (Chen et al. 2018).
Results
Invertase Genes in S. lycopersicum and Their Physico-Chemical Characteristics
Overall, 24 invertase genes were detected in the genome of S. lycopersicum (ITAG release 4), comprising 9 cell wall (SlCWINV5-8, 11, 13, 16–18), 2 cell membrane (SlCMINV23-24), 11 chloroplast (SlChlINV1-4, 12, 14–15, 19–22), 1 cytosol (SlCINV10) and 1 vacuolar (SlVaINV9) genes with an unannotated member (SlCMINV24) as a potential pseudogene (Table 1). Additionally, 91.67 % of the invertase genes identified had a homologue in Arabidopsis. Further searches against the genomes of S. lycopersicum and Nicotiana benthamiana indicated that, four homologues of Arabidopsis were absent in the ancestral genome of tomato. Thus, we posit that, the homologues of these four Arabidopsis genes may have been lost through evolutionary divergence.
Amino acid lengths (aa) of the invertase genes spanned from 52 bp (SlCMINV24) to 655 bp (SlChlINV22; Table 1). The Mw and pI varied from 8.25 (SlChlINV15) to 74.46 kDa (SlChlINV2); and 5.14 (SlCWINV13) to 9.51 (SlChlINV4, Table 1), respectively. Sequence alignment of the amino acids indicated that, the SlCWINVs share 58.25 to 83.83 % identities among all the gene members (Figure S1). Except SlCMINV24, all identified invertase proteins had either alkaline and neutral invertase (Glyco_hydro_100) or glycosyl hydrolases family 32 N-terminal (Glyco_hydro_32N/C) conserved domain. SlCMINV24, perhaps lost its functional domain through evolutionary divergence. The number of ORF ranged from 6 (SlCMINV23) to 36 (SlChlINV19 and SlChlINV20) among the invertase genes identified (Table 1).
Chromosomal Location and Gene Duplication
Majority of the invertase genes have the Glyco_hydro_32 N & C termini domain (glycosyl hydrolases family 32 N&C-terminal), while the rest of the gene members possess alkaline and neutral invertase functional domain (Glyco_hydro_100 domain; Fig. 1). Ten distinct motifs were identified among the 24 invertase proteins via the MEME online database. Motif 2 was detected among all the genes, except SlCMINV24, SlChlINV4, SlCMINV23, SlChlINV15 and SlChlINV19 (Fig. 1a–c). Gene SlCMINV24, a potential pseudo-gene member that may have retained only motif 5 was consistently found in almost all the sub-domains. However, motifs 1, 3, 4 and 6 are unique to clade 1, while clade 2 is highly conserved with motifs 7, 8, 9 and 10 (Fig. 1a–c). The gene structure analysis shows high diversity and varied exon and introns. The highest number of exons (9) and introns (8) were recorded in SlChlINV19; the while lowest number of exons and introns were identified in SlChlINV12 and SlChlINV1 (Fig. 1d).
Phylogenetic relationships, gene structure and chromosome map of invertase genes. a Phylogenetic tree was constructed in MEGA 7 using the maximum likelihood method (1000 bootstrap). At the bottom of the figure, the relative position is proportionally displayed based on the kilobase scale. b Ten conserved motif distribution according to c motif consensus sequences. d Exon–intron structure of the invertase genes constructed using the gene structure display server (GSDS); exons are coloured in blue and introns are indicated by the grey colour (Color figure online)
The 24 invertase genes were identified on 8 out of the 12 chromosomes. The genes were unevenly distributed with the highest number of genes located on Chr 10 and the lowest number of genes on Chr 09 (Fig. 2). Seven (7) pairs of genes were identified to have undergone segmental duplication events with 3 pairs on chromosome Chr10 while the remaining 4 genes occurred on Chr01, 6, 9 and 11. However, 5 genes were tandemly duplicated (Fig. 2; Table 1). We juxtaposed the distributions of invertase genes with the genomes of A. thaliana, O. sativa, Z. mays, V. vinifera, C. papaya, and S. tuberosum, which revealed the conserved synteny of the invertase genes (Table 1). Six tandemly duplicated genes (SlChlINV3, SlChlINV4, SlCWINV17, SlCWINV18, SlChlINV19 and SlChlINV20) located on chromosomes, Chr 10 and Chr 01 were also identified (Table 1). Three of those tandem pairs (SlChlINV3 and SlChlINV4, SlCWINV17 and SlCWINV18, and SlChlINV19 and SlChlINV20) are copies corresponding to individual Arabidopsis homologues, indicating possible recent tandem gene duplication events (Tables S2–3). The homologues of SlChlINV3 and SlChlINV4 tandem pair was also found to be located contiguously to each other in Arabidopsis, suggesting that these clustering genes may have descended from a common progenitor. The remaining genes located in the non-syntenic regions are likely associated with whole-genome duplication (Tables 1, S3).
Chromosome map of invertase genes within the tomato genome. Twenty four (24) SlINV genes were mapped onto the 8 tomato chromosomes. Gene identities are designated based on their locations and their respective chromosome numbers indicated below each chromosome. Genome-wide or segmentally duplicated genes are indicated with red arrow. Tandemly duplicated genes are indicated with grey arrow (Color figure online)
Evolution of Invertase Genes Among Nine Plant Species
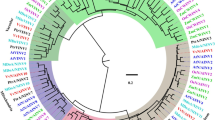
We identified all putative invertase genes in S. lycopersicum together with eight different plant species (Fig. 3). A phylogenetic tree of all the 137 invertase genes from nine species was constructed to study the evolutionary patterns of the invertase genes in the plant kingdom (Figs. 3 and S2). Clade I was formed first followed by clades II, III and Va as evidenced by the inter-cluster evolutionary distance (Fig. 3). The 137 genes were grouped into six clades from 5 dicotyledonous (A. thaliana, C. papaya, S. tuberosum, S. lycopersicum, and V. vinifera) and 4 monocotyledonous species (B. distachyon, G. max, O. sativa, and Z. mays) (Fig. 3). With 5 genes (SlCWINV21, SlCWINV22, SlChlINV2, SlChlINV3 and SlChlINV4) from tomato, clade I contains genes from four monocotyledonous and dicotyledonous species, while clades II, III, IV and V predominantly had monocotyledonous species only. Clade V and VI were mainly dicotyledonous species (Solanaceae), respectively. Genes in Clade VI were vacuolarly localised with potential pseudogenes from a monot (B. distachyon) and tomato (SlCMINV24).
Phylogenetic relationship of invertase genes in 9 plant species. The invertases were clustered into six different groups based on FastTree. The SGN database was used to extract the total invertases from the tomato genome and similarity among invertases of the tomato genome were clustered using MUSCLE alignment values. The diagonals with 100 MUSCLE values (the same proteins that are compared) were not considered for plotting the graph. Most of invertases had a similarity value less than 20 which show the diversity of invertases in tomato. Chl-I & II coloured in green refers to invertase proteins from 9 different species localised in the chloroplast. The light blue coloured cluster indicate invertase proteins from 9 different species localised in the cell wall (CWI-II); while the blue coloured cluster indicate only Solanaceae invertase proteins localised in the cell wall (CW-III). The purple cluster refers to invertase proteins from Solanaceae predominantly localised in the vacuole (Va). The 9 different plant species include Arabidopsis thaliana (AT), Solanum lycopersicum (Sl), Carica papaya (Cp), Oryza sativa (Os), Zea mays (Zm), Solanum tuberosum (AE), Vitis vinifera (GSV), Brachypodium distachyon (Bd), and Glycine max (XP) (Color figure online)
We constructed a phylogenetic tree using the FastTree MAFFT plugin in Geneious 11, and clustered the 24 invertase genes into six clades with a possible outgroup containing a potential pseudogene (Fig. 3). Thus, invertase genes from other dicots and monocots were distributed across all clades, whereas Solanaceae invertases (SL, AD & AE) were distributed across the six clades. However, all invertase genes in sub-clade CW-II (purple coloured, Fig. 3) were clade-specific, indicating that, this sub-clade may be Solanaceae-specific invertases. These unique invertase genes formed a distinct clade with other dicot invertases distinct from invertase genes in other plant species (Figure S2) and thus, affirming their Solanaceae specificity. Furthermore, 12 of the 24 invertase genes were detected to contain autophagy sequences (http://bioinformatics.psb.ugent.be/beg/sofware; not shown), and thus, further corroborating their Solanaceae specificity (Ullah et al. 2019).
Putative CAREs in the Promoter Regions of Invertase Genes
CAREs are generally essential in gene expression; and thus identifying them in invertases will be key to unearth their potential roles in gene expression. Sequences of 1.5 kb from the start codon (ATG) were used to identify putative CAREs from the promoter regions using the PlantCare database. Five thousand and fifty seven (5057) putative CAREs were identified on the promoters of the invertase genes, with a minimum and maximum length of 4 bp and 22 bp (s), respectively (Table 2). Our analysis revealed that, most of the CAREs contributed to phytohormonal response, biotic and abiotic stress regulation. Majority of the CAREs (67.80 %) were essential elements involved in stress response. About 9.83 % of the CAREs (3-AF1 binding site, ACE, AE-BOX, AT1-motif, ATCT-motif, BOX-4, TCCC-motif) were highly responsive to abiotic and biotic stress, followed by light responsive cis-elements (7.58 %) and phytohormones (6.06 %) (ABRE, AT-Rich elements, TGA-element, TGACG-motif, TCA-element, P-box, GARE-motif, CAT-BOX, A-Box, ERE). However, fewer CAREs (5.39, 3.20 and 0.14 %) were mainly enhancer elements, unknown elements with diverse functions and circadian control elements (CCAAT, CAAAT, CCCAATTT, CGTCA, TGA, TCA, AuxRe, GARE-motif), respectively (Figure S3).
More than 20 CAREs were duplicated in the promoter regions of all the genes. Twenty four (24) non-duplicated CAREs of the invertase genes were clustered into four functionally related groups (Table 2). The first unique group elements; AT ~ TATA-box (SlChlINV3), STRE (SlChlINV4), ATCT-motif (SlCyINV10), GATA-motif (SlChlINV15), CTAG-motif (SlCWINV8), and MBS (SlChlINV22) functionally respond to light and drought stresses (Priest et al. 2009). Box 5 (SlCWINV6), MYB (SlVaINV9), and CAT-Box (SlCMINV23) were the second unique CAREs functionally involved in palisade mesophyll cells differentiation, meristem expression, and circadian control (Hernandez-Garcia and Finer 2014). The third uniquely clustered CAREs functionally respond to ABA, IAA (auxin) and GA. These include ABRE (SlChlINV1, SlCWINV7), ERE (SlCINV11), P-box (SlCWINV16, SlCWINV17), AE-box (SlCWINV18), MYC (SlChlINV19), ABRE (SlCWINV20), chs-CMA1a (SlChlINV21), AAGAA-motif (SlCMINV24) which have been reported in Arabidopsis, barley and rice (Schmiesing et al. 2016). The fourth group of CAREs, LTR (SlChlINV2, SlChlINV12), MBSI (SlCWINV5), TC-rich repeats (SlChlINV13) are either involved in low-temperature responsiveness, flavonoid biosynthetic genes regulation or defense and stress responsiveness. Our analysis of the various CAREs indicates a regulatory complexity of invertase genes as most of them were intricately involved in environmental stress, tomato growth and development (Figure S3).
Invertase Gene Expression Profiles in Diverse Tissues
Publicly available RNA-Seq data were retrieved and analysed to assess organs/tissue-specific expression profiles of tomato fruit (at the varied developmental stages) and results were presented in heatmaps using the TBtools (Chen et al. 2018). 11 genes (SlChlINV2, SlCWINV5, SlCWINV8, SlVaINV9, SlCyINV10, SlChlINV12, SlCWINV13, SlChlINV14, SlCWINV16, SlCWINV17, SlCWINV18, and SlCMINV23) showed relatively significant expression levels in the various tissues of the cultivar, Heinz (Fig. 4). Conversely, SlChlINV1, SlCWhlINV3, SlCWINV5 and SlCWINV6 exhibited relatively moderate expression levels in the studied tissues (Fig. 4). Nevertheless, the SlCWINV6, SlCWINV8, SlVaINV9, SlCWINV16, SlCWINV17, SlChlINV19, SlChlINV20, SlChlINV21, SlChlINV22, and SlCMINV23 genes exhibited fairly lower expression profiles in most of the tissues of the tomato cultivar Heinz. The chloroplast (SlChlINV2, SlChlINV3, SlChlINV12, SlChlINV14, and SlChlINV15) and cell wall invertase genes (SlCyINV10 and SlCWINV13) recorded significantly higher expressions than the cytosol and vacuolar invertase genes. Thus, tissue-specific expression profiles of the invertase genes evidence divergence of gene functions in tomato growth and development. Majority of the invertases showed varied expressions in different tissues (Fig. 4).

accessed from the Tomato Expression Atlas database (http://tea.solgenomics.net/) (Color figure online)
Tissue-specific expression patterns of invertase genes. Hierarchical clustering of tissue-based expression profiles for invertase genes in organs of the cultivar tomato Heinz. The coloured bars represent the log2 expression values. The red colour indicates high expression levels and green colour shows low expression levels in the tissues and organs used for the expression profiling (as indicated at the bottom of each column). The cluster dendrogram is shown on the top and left side of the expression heatmap, and the gene names are indicated on the right side. Tissues used include young root (YR), mature root (RM), young leaf (YL), mature leaf (ML), flower bud (FB), stem (ST), immature green (IMG), mature green (MG), breaker (BR), red ripe fruits (RR) and seed (SD) as indicated in the heatmap. The data were
Expression Profiles of Invertase Genes Under Abiotic Stress and Phytohormone Response
Based on the prediction of CAREs, we validated the expression profiles of invertase genes under drought, cold, light, salinity stresses and phytohormones (ABA, GA, IAA). Results indicate that, most of the invertase genes were either induced or repressed by abiotic stress or phytohormones. Majority of the invertase genes were highly expressed, nonetheless SlCWINV6, SlCWINV7, SlCWINV8, SlVaINV9, SlChlINV11, SlChlINV16, SlCWINV17, SlCWINV18, SlChlINV20, SlChlINV23, SlCMINV24 were not expressed under the drought and salinity stress, respectively (Fig. 5). The invertase genes exhibited high transcript levels after exposure to the various treatments, but a few of them were induced by one or more treatments. The cold stress recorded the dominant portion of down-regulated genes (54.17 %). However, drought and light stress showed that, 91 and 83 % of genes were up-regulated, while 9 and 17 % decrease in transcript levels, respectively (Fig. 5). The GA phytohormonal treatment recorded 75 % up-regulated genes and 25 % down-regulated genes, while exposure to IAA recorded 66.67 % up-regulated genes and 33.33 % down-regulated genes, followed by ABA recording 62.5 % up-regulated genes and 37.5 % were down-regulated genes. Expression of invertase genes were significantly repressed in red ripe, breaker, matured green, immature green fruits and leaves under cold and salinity stress at the 1–6 h durations (Fig. 5b). However, moderate expression levels were recorded for most of the genes under both abiotic stress and phytohormonal treatments (Fig. 5). For instance, a number of light related cis-elements (4 G-boxes, 4 Box 4, 1 LAMP-element, 5 GATA-motifs, 1 GT1-motif and 1 AE-box), phytohormone related cis-elements (2 TCA-elements, 3 ABRE, 2 CGTCA-motifs, 2 TGACG-motifs, 2 P-boxes, and 1 TGA-box) and stress-related cis-elements (1 TC-rich repeats, 3 ARE and 1 GC-motif) were found on the promoter of SlChlINV15. These cis-elements may have accounted for the marked inducement of SlChlINV15 under the various abiotic stress and phytohormone treatments.
Expression profiles of selected invertase genes under four abiotic stresses (cold, drought, light, salinity) and three phytohormone (ABA, GA and IAA) treatments. The expression levels were normalised by log2 transformation. Values are means and SD obtained from 4 biological replicates. The error bars indicate standard error of mean at a statistical significance (*P < 0.05 and **P < 0.01) compared with the corresponding controls. a Refers to cold stress at 0, 1, 3, 6, 12 & 24 h in the leaf; b refers to drought stress at 0, 1, 3, 6, 12 & 24 h in the leaf; c refers to salinity stress at 0, 1, 3, 6, 12 & 24 h in the leaf; d refers to salinity stress in the flowerbud, stem and root; e refers to ABA response at 0, 1, 3, 6, 12 & 24 h in the leaf; f refers to GA response at 0, 1, 3, 6, 12 & 24 h in the leaf g; refers to IAA response at 0, 1, 3, 6, 12 & 24 h in the leaf; h refers to drought stress at 24 h in the flowerbud, root and stem; i refers to ABA response at 24 h in the flowerbud, root and stem; j refers to GA response at 24 h in the flowerbud, root and stem; k refers to IAA response at 24 h in the flowerbud, root and stem; (l) refers to light response at the immature green, mature green, breaker and red ripe stages of fruit (Color figure online)
Interestingly, SlCWINV4 and SlChlINV2 recorded the highest gene expression in the stem (under salinity stress) and breaker fruits under the light exposure (Fig. 5). Genes SlChlINV12 and SlChlINV14 were highly expressed in leaves 1 and 12 h under cold stress (Fig. 5a). Genes SlChlINV15 and SlChlINV20 were not expressed under the IAA phytohormonal treatment. Genes SlCWINV1, SlChlINV3, SlCWINV5, SlChlINV10, SlCWINV13, SlChlINV14, SlCWINV17, SlCWINV18, SlChlINV20, SlChlINV21, SlChlINV22, and SlChlINV23 were upregulated under the abiotic stresses (Fig. 5); while SlChlINV2, SlCWINV4, SlChlINV7, SlVaINV9, SlCWINV11 and SlChlINV16 were significantly induced and upregulated under the phytohormonal treatments (Fig. 5). Another classical example of how cis-elements modulate gene expression was evident in SICyINV10. The promoter of this gene habours the MYB binding site involved in drought-inducibility (MBS) and ABA responsiveness element (ABRE) and may have accounted for the high inducibility of SICyINV10 by drought and ABA treatments (Fig. 5e and h). All the 24 genes were induced by the different abiotic stresses and phytohormonal treatments. Thus, the diversity in their expression profiles indicate their response to abiotic stress and phytohormones.
Protein Modelling, Gene Ontology Enrichment and Network Analyses
We performed a three-dimensional homology modelling on all the 24 invertase proteins using the Phyre2 server, V 2.0; http://www.sbg.bio.ic.ac.uk/~phyre2/html/page.cgi?id=index) (Kelley et al. 2015) in the intensive mode. The Phyre2 server permitted selection of appropriate templates for each protein and heuristically maximised alignment coverage, percentage identity, and confidence score for each protein. The templates, c5gorE (SlCWINV1, SlChlINV2, SlChlINV3, SlChlINV4, SlCMINV24); c5z73A (SlCWINV11, SlChlINV12, SlCWINV13, SlChlINV14, SlChlINV22); c3ugfB (SlVaINV9, SlCyINV10); and c2ac1A (SlCWINV5, SlCWINV6, SlCWINV7, SlCWINV8, SlChlINV15, SlCWINV16, SlCWINV17, SlCWINV18, SlChlINV19, SlChlINV20, SlChlINV21, SlCMINV23) were employed in the modelling. In the neutral/alkaline subfamily, the secondary structures of modelled invertase proteins were primarily β-strands (21–45 %) with the α-helices occurring at only 5–13 %. Conversely, α-helices constituted the secondary structures of modelled invertase proteins (43–55 %) in the acidic and vacuolar subfamilies, whereas β-strands occurred at only 6–18 % (Figure S4). To further decipher the functions of the invertases, we performed GO annotation and enrichment analyses (Table S5). The GO enrichment indicates that, the invertase genes play vital roles in regulation of biological, cellular and molecular processes in tomato. Overall, 18 invertase proteins were involved in diverse interactions and positioned on the peripheries of a relational network constructed. The complex nature of the network of invertase proteins interaction with other sugar/sucrose proteins suggest their co-integration in various mechanisms of stress response (Figure S5).
Discussion
In plants, invertases (EC 3.2.1.26, INV) catalyse the unalterable hydrolysis of sucrose into glucose and fructose for the regulation of sucrose metabolism (Leong et al. 2018). Invertases are indispensable in crop growth and stress responses (Li et al. 2018), carbon partitioning (Wang et al. 2017), phloem unloading and source/sink regulation (Chen et al. 2015a, b), compositional adjustment of sugars in sink tissues (Leong et al. 2018), organ formation, and sugar transport (Zhang et al. 2015). The gene structure of acid invertases (AINVs) is highly conserved with 6–8 exons. And in almost all acid invertase genes, ‘the second exon codes for only three amino acids, DPN, a conserved NDPNG motif of the catalytic domain as the smallest ever known functional motif in plant cellular biology’ (Fotopoulos 2005).
We identified nine cell wall (CWINV), 2 cell membrane (CMINV), 11 chloroplast (ChlINV), 1 cytosol (CyINV) and one vacuolar (VaINV) invertase genes in the tomato genome distributed on 8 of the 12 chromosomes. Phylogenetic analysis clustered the invertase genes into two clades. Segmental duplication contributed to the neutral/alkaline sub-family expansion. Tomato invertase genes were differentially expressed in roots, stems, leaves, flower buds and respond differentially to light, cold, drought, salinity and phytohormones (ABA, IAA, GA) (Fig. 5). Specifically, our phylogenetic analysis of invertase genes from 9 plant species clustered them into six subcellular-specific clades (chloroplast (Chl-I & II), cell wall (CW-I, II & II), and vacuole (Va) (Zhang et al. 2019a, b) (Fig. 3). Clade I encompasses genes from four monocotyledonous and five dicotyledonous species with fewer invertase members (1–5), and clades II, III, IV and V were predominantly monocotyledonous species; while Clade V and VI were mainly dicotyledonous species (Solanaceae), respectively. Genes in Clade VI were vacuolarly localised with a potential ‘orphan’ gene from B. distachyon (a monot species) and a potential pseudogene from tomato (SlCMINV24) localised in the cell membrane. Similar dichotomies between cell-wall invertases from monocots and dicots have been reported (Huang et al. 2013; Wang et al. 2019a, b; Shen et al. 2018).
All vacuolar invertases (VaINVs) from both dicotyledonous and monocotyledonous species were clustered in clade VI, which were markedly detached from the cell wall invertase (CWINV; Fig. 2), signifying potential origin of the VaINVs from CWINVs prior to the last common ancestor (LCA) of dicotyledons and monocotyledons (Wang et al. 2017). This contradicts earlier reports that, the origin of VaINVs from CWINVs preceded the LCA of rice and Arabidopsis (Marchler-Bauer et al. 2017). Again, the possibility of parallel evolution of cell-wall invertases has been reported previously by Fridman and Zamir (Fridman et al. 2000) when a similar trend was identified between cell-wall invertases of Arabidopsis and tomato with an estimated divergence time of 112 MYA (Carther et al. 2019; Table S4). This was evidenced from the species-specific clustering of the tomato invertase genes and thus, further corroborating potential parallel evolution of invertase gene duplication (Canam et al. 2008).
Genetically, genome duplication is a major source of crop adaptation to environmental stresses (Cheng et al. 2017). Gene duplication through whole-genome duplication (WGD), tandem gene duplication and segmental duplication events often characterises gene evolution. This ultimately culminates in either neo-functionalisation, sub-functionalisation and/or non-functionalisation events (Blanca et al. 2015). Genome-wide analysis of genetic diversity and identification of relevant genes for breeding is useful for crop domestication and improvement (Fridman 2003; Hirakawa et al. 2013). The structural composition of invertase proteins were highly conserved as previously reported for other plants, characteristically comprising a signal peptide, a central variable domain, and a highly conserved functional peptide domain, with most exhibiting the C-terminal extension (Figure S1). The diversity of the major functional domains may have occasioned and contributed to their distinct roles and expansion of the invertase gene family. This partly indicates a recent speciation events from ancestral populations with greater diversity within the Solanum genus (Strickler et al. 2015). Analysis of conserved motifs reveals significant gene functions (Marchler-Bauer et al. 2013) and thus, confirming the progressive functional divergence among the invertase genes (Marchler-Bauer et al. 2013).
Abiotic stresses such as cold, light, drought, and salinity potentially pose a threat to crop yield including tomato. We employed the qRT-PCR to assess transcript abundance of 6 selected genes (SlCWINV5, SlChlINV6, SlVaINV9, SlCyINV10, SlChlINV15 & SlCmINV24) in different tissues and time interval under cold, drought, light, salinity stresses, and ABA, IAA and GA treatments. These results provide clue on the potential roles of the selected genes under these abiotic stress and phytohormones (ABA, IAA and GA). Our analysis of the promoter regions for identification of common conserved CAREs revealed the involvement of invertase genes in various hormonal signalling, biotic and abiotic stress response. Earlier studies copiously implicated invertase activity in response to various stress stimuli in A. thaliana (Yamada and Osakabe 2018), Populus trichocarpa (Chen et al. 2015a, b), Z. mays (Juárez-Colunga et al. 2018), O. sativa (Ji et al. 2005) and G. max (Wang et al. 2019a, b). Assorted mechanisms of biochemical and metabolic actions in response to abiotic factors and phytohormones are often adduced (Wang et al. 2019a, b). Genes SlCWINV5, SlChlINV6, SlVaINV9, SlCyINV10, SlChlINV15 & SlCmINV24 were differentially expressed in the different tissues and at varying period when tomato plants were exposed to longer durations of cold, drought, light, salinity stress; and phytohormones (ABA, IAA and GA) (Fig. 5). However, majority of the genes that were significantly expressed and up-regulated under the phytohormonal treatments were consistently down-regulated in the stem, flowerbud and roots. The expression study reveals that, most of the invertase gene expressions are induced by phytohormones. Most of the genes (whose cis-elements relate to abiotic stresses) were highly induced under drought, salinity and cold while those with none were repressed (Fig. 5a–d). Those genes with phytohormone and stress related cis-elements (Fig. 5a–d) followed similar trend. Therefore, these key cis-regulatory elements (sequences) may be targeted for CRISPR/Cas cis-engineering to improve stress tolerance (Zafar et al. 2020). A more recent study demonstrated that natural variation in the promoter of TGW2 determines grain width and weight in rice (Ruan et al. 2020). The SlChlINV4 invertase exclusively contains cis-acting regulatory elements involved in seed-specific regulation (RY-elements). This may require functional validation in future studies for exploitation in terms of seed development with desired traits.
A thorough analysis of the invertase proteins revealed autophagy sequences in six of the Solanaceae-specific invertase proteins (SlCWINV5, SlChlINV6, SlVaINV9, SlCyINV10, SlChlINV15 & SlCmINV24) (http://bioinformatics.psb.ugent.be/beg/software) (Ullah et al. 2019). Additionally, autophagy sequences were detected in 12 of the 24 invertase proteins, and thus, further lending credence to a potential species-specific evolution of tomato invertases (Figure S2). The discovery of the unique property in these invertases suggests they might have gone through a gain-of-function event during evolution (Ullah et al. 2019). The functional mediation of cell components degradation or deleterious cytoplasmic molecules in cells is autophagy-controlled. Several studies have reported a critical role of autophagy in drought/heat tolerance in plants, typically through selective elimination of heat-induced proteins (Zhou et al. 2014a, b). Functional analysis of ATG genes has separately established the crucial roles of autophagy in nutrient reprocessing, usage in plants and regulation of plant senescence as a process of nutrient reallocation. Besides, autophagy regulates plant innate immune responses (Zhou et al. 2014a, b), oxidative induction and osmotic stresses, salinity, and varying temperatures (Zhou et al. 2014a, b; Ullah et al. 2019). However, we are unable to report what the biochemical and molecular functions of the autophagy sequences in these invertases are, and their biological functions. The presence of these autophagy sequences in the tomato-specific invertases gives the proteins a unique identity. Significantly, the presence of the autophagy sequences in these invertase proteins may implicate their roles in stress response and potential biochemical or molecular functions associated with protein stability or integration with the autophagy process. Based on the gene expression pattern (Figs. 4 and 5), these tomato-specific invertases may participate in stress response, and may be involved in other biological processes in tomato.
We studied the functional network interactions using the STRING database (https://string-db.org/cgi/network) and visualised this in the Cytoscape software (https://cytoscape.org/). A compact interaction complex was formed among the invertase proteins as majority of the invertase proteins (18) were involved in multi-functional interactions with related proteins (Figure S6). There were interaction relationships among proteins within the same sub-domain (s) and also among proteins from dissimilar subfamilies. The gene balance hypothesis posits that, genes will be preferentially retained if their products are dose sensitive, interacting either with other proteins or in networks which may affects the function of the whole and regulatory mechanisms (Birchler and Veitia 2010). This is evidenced in the network interaction complex formed, as most of the proteins were either implicated in glycolysis, sucrose synthesis or pyrophosphate-fructose 6-phosphate 1 phosphotransferase activity.
Our homology models generated residues above 89 %, and thus, suggesting models were within acceptable thresholds. Primarily, the secondary structures of the invertase proteins were α-helices (21–45 %); while 6–18 % were β-strands. The superimposed alkaline/neutral sub-domain models showed 80–100 % structural coverage and the superimposed glycosyl hydrolases sub-domain models showed 53–91 % structural coverage (Figure S4). The models indicate an ancestrally high proximity (similarity) of invertase genes to each other and/or evolutionary stabilisation of divergence through an expanded domestication event resulting in the diverse protein structures and functionality (Figure S4).
Conclusion
We identified and functionally characterised 24 putative invertase genes in the tomato genome through a genome-wide study. A significant variation was recorded in the expression profiles of these genes at different stages of tomato fruit growth and tissues under abiotic stress and phytohormonal treatments. Segmental duplication contributed to the neutral/alkaline invertase gene sub-domain expansion in tomato. Phylogenetic analysis clustered the invertase genes from nine dicots and monocots into six subcellular-specific clades. Invertase genes from the Solanum genus were distributed across six clades. Our study provides the first comprehensive analysis of the invertase gene family and their differential response to abiotic stress and phytohormones in tomato. Results of the expression profile and differential stress response of these Solanaceae-specific invertases provide new insights to facilitate further study on biological processes in tomato stress response.
References
Bailey TL, Boden M, Buske FA et al (2009) MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res 37:W202–W208. https://doi.org/10.1093/nar/gkp335
Bihmidine S, Baker RF, Hoffner C, Braun DM (2015) Sucrose accumulation in sweet sorghum stems occurs by apoplasmic phloem unloading and does not involve differential Sucrose transporter expression. BMC Plant Biol 15:186
Birchler JA, Veitia RA (2010) The gene balance hypothesis: implications for gene regulation, quantitative traits and evolution. New Phytol 186:54–62. https://doi.org/10.1111/j.1469-8137.2009.03087.x
Blanca J, Montero-Pau J, Sauvage C et al (2015) Genomic variation in tomato, from wild ancestors to contemporary breeding accessions. BMC Genom. https://doi.org/10.1186/s12864-015-1444-1
Braun D (2014) Understanding and manipulating sucrose phloem loading, unloading, metabolism, and signalling to enhance crop yield and food security. J Exp Bot 66:1713–1735
Canam T, Mak SWY, Mansfield SD et al (2008) Spatial and temporal expression profiling of cell-wall invertase genes during early development in hybrid poplar. Tree Physiol. https://doi.org/10.1093/treephys/28.7.1059
Carther K, Ketehouli T, Ye N et al (2019) Comprehensive genomic analysis and expression profiling of diacylglycerol kinase (DGK) gene family in soybean (Glycine max) under abiotic stresses. Int J Mol Sci 20:1361. https://doi.org/10.3390/ijms20061361
Chen Z, Gao K, Su X et al (2015a) Genome-wide identification of the invertase gene family in populus. PLoS ONE. https://doi.org/10.1371/journal.pone.0138540
Chen Z, Gao K, Su X et al (2015b) Genome-wide identification of the invertase gene family in populus. PLoS ONE 10:e0138540. https://doi.org/10.1371/journal.pone.0138540
Chen C, Chen H, He Y, Xia R (2018) TBtools, a toolkit for biologists integrating various biological data handling tools with a user-friendly interface. bioRxiv. https://doi.org/10.1101/289660
Cheng Y, Bian W, Pang X et al (2017) Genome-wide identification and evaluation of reference genes for quantitative RT-PCR analysis during tomato fruit development. Front Plant Sci 8:1–13. https://doi.org/10.3389/fpls.2017.01440
Cho RJ, Mindrinos M, Richards DR et al (1999) Genome-wide mapping with biallelic markers in Arabidopsis thaliana. Nat Genet 23:203–207. https://doi.org/10.1038/13833
Chourey PS, Jain M, Li QB, Carlson SJ (2006) Genetic control of cell wall invertases in developing endosperm of maize. Planta 223:159–167. https://doi.org/10.1007/s00425-005-0039-5
Ding X, Zhang L, Hao Y et al (2018) Genome-wide identification and expression analyses of the calmodulin and calmodulin-like proteins reveal their involvement in stress response and fruit ripening in papaya. Postharvest Biol Technol 143:13–27. https://doi.org/10.1016/j.postharvbio.2018.04.010
FAO (2017) The future of food and agriculture – Trends and challenges. The Food and Agricultural Organisation of the United Nations. Rome
Finn RD, Coggill P, Eberhardt RY et al (2016) The Pfam protein families database: towards a more sustainable future. Nucleic Acids Res 44:D279–D285. https://doi.org/10.1093/nar/gkv1344
Fotopoulos V (2005) Plant invertases: structure, function and regulation. J Biol Res 4:127–137
Fridman E (2003) Functional divergence of a syntenic invertase gene family in tomato, potato, and Arabidopsis. Plant Physiol 131:603–609. https://doi.org/10.1104/pp.014431
Fridman E, Pleban T, Zamir D (2000) A recombination hotspot delimits a wild-species quantitative trait locus for tomato sugar content to 484 bp within an invertase gene. Proc Natl Acad Sci USA 97:4718–4723. https://doi.org/10.1073/PNAS.97.9.4718
Goff SA, Ricke D, Lan T-H et al (2002) A draft sequence of the rice genome (Oryza sativa L. ssp. japonica). Science 296:92–100. https://doi.org/10.1126/science.1068275
Guo N, Wang G, Zong M et al (2018) Genome-wide identification, and phylogenetic and expression profiling analyses of CaM and CML genes in Brassica rapa and Brassica oleracea. Gene 677:232–244. https://doi.org/10.1016/j.gene.2018.07.038
Gur A, Zamir D (2004) Unused natural variation can lift yield barriers in plant breeding. PLoS Biol 2:e245. https://doi.org/10.1371/journal.pbio.0020245
Hernandez-Garcia CM, Finer JJ (2014) Identification and validation of promoters and cis-acting regulatory elements. Plant Sci 217–218:109–119. https://doi.org/10.1016/j.plantsci.2013.12.007
Hirakawa H, Shirasawa K, Ohyama A et al (2013) Genome-wide SNP genotyping to infer the effects on gene functions in tomato. DNA Res 20:221–233. https://doi.org/10.1093/dnares/dst005
Hu B, Jin J, Guo A-Y et al (2015) GSDS 2.0: an upgraded gene feature visualization server. Bioinformatics 31:1296–1297. https://doi.org/10.1093/bioinformatics/btu817
Huang Z, Van HJ, Gonzalez G et al (2013) Genome-wide identification, phylogeny and expression analysis of SUN, OFP and YABBY gene family in tomato. Mol Genet Genom 288:111–129. https://doi.org/10.1007/s00438-013-0733-0
Jain M, Chourey PS, Boote KJ, Allen LHJ (2010) Short-term high temperature growth conditions during vegetative-to-reproductive phase transition irreversibly compromise cell wall invertase-mediated sucrose catalysis and microspore meiosis in grain sorghum (Sorghum bicolor). J Plant Physiol 167:578–582
Ji X, Van den Ende W, Van Laere A et al (2005) Structure, evolution, and expression of the two invertase gene families of rice. J Mol Evol 60:615–634. https://doi.org/10.1007/s00239-004-0242-1
Jin A, Ozawa T, Tajiri K et al (2009) A rapid and efficient single-cell manipulation method for screening antigen-specific antibody–secreting cells from human peripheral blood. Nat Med 15:1088–1092. https://doi.org/10.1038/nm.1966
Juárez-Colunga S, López-González C, Morales-Elías NC et al (2018) Genome-wide analysis of the invertase gene family from maize. Plant Mol Biol 97:385–406. https://doi.org/10.1007/s11103-018-0746-5
Julius BT, Leach KA, Tran TM et al (2017) Sugar transporters in plants: New insights and discoveries. Plant and Cell Phys 58(9):1442–1460. https://doi.org/10.1093/pcp/pcx090
Kelley LA, Mezulis S, Yates CM et al (2015) The Phyre2 web portal for protein modeling, prediction and analysis. Nat Protoc 10:845–858. https://doi.org/10.1038/nprot.2015.053
Kimura S, Sinha N (2008) Tomato (Solanum lycopersicum): A model fruit-bearing crop. Emerging model organisms. Cold SpringHa Protoc. https://doi.org/10.1101/pdb.emo105
Klee HJ, Giovannoni JJ (2011) Genetics and control of tomato fruit ripening and quality attributes. Annual Rev Genet 45(1):41–59. https://doi.org/10.1146/annurev-genet-110410-132507
Kocal N, Sonnewald U, Sonnewald S (2008) Cell wall-bound invertase limits sucrose export and is involved in symptom development and inhibition of photosynthesis during compatible interaction between tomato and Xanthomonas campestris pv vesicatoria. Plant Physiol 148:1523–1536. https://doi.org/10.1104/pp.108.127977
Kumar S, Stecher G, Tamura K (2016) MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol 33:1870–1874. https://doi.org/10.1093/molbev/msw054
Larkin MA, Blackshields G, Brown NP et al (2007) Clustal W and clustal X version 2.0. Bioinformatics 23:2947–2948. https://doi.org/10.1093/bioinformatics/btm404
Le Hir R, Spinner L, Klemens PAW et al (2015) Disruption of the Sugar Transporters AtSWEET11and AtSWEET12 Affects Vascular Development and Freezing Tolerance in Arabidopsis. Mol Plant 8:1687–1690
Leong BJ, Lybrand D, Lou Y-R et al (2018) Evolution of metabolic novelty: a trichome-expressed invertase creates specialized metabolic diversity in wild tomato. bioRxiv. https://doi.org/10.1101/502971
Li JM, Zheng DM, Li LT et al (2014) Genome-wide function, evolutionary characterization and expression analysis of sugar transporter family genes in pear (Pyrus bretschneideri Rehd). Plant Cell Physiol 56:1721–1737. https://doi.org/10.1093/pcp/pcv090
Li J, Su X, Wang Y et al (2018) Genome-wide identification and expression analysis of the BTB domain-containing protein gene family in tomato. Genes Genom 40:1–15. https://doi.org/10.1007/s13258-017-0604-x
Liu W, Corresp JN, Shah F et al (2019) Genome-wide identification, phylogenetic and biochemical analysis of the APYRASE family members, and gene expression analysis in response to the abiotic and biotic stresses in bread wheat (Triticum aestivum). PeerJ Prepr. https://doi.org/10.7287/peerj.preprints.27674v1
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(-delta delta C(T)) method. Methods 25:402–408. https://doi.org/10.1006/meth.2001.1262
Lopes CT, Max F, Farzana K, Donaldson SL, Quaid MBG (2010) Cytoscape web: an interactive web-based network browser. Bioinformatics 26:2347–2348. https://doi.org/10.1093/bioinformatics/btq430
Ludewig F, Flügge UI (2013) Role of metabolite transporters in source-sink carbon allocation. Front Plant Sci 4:231. https://doi.org/10.3389/fpls.2013.00231
Marchler-Bauer A, Zheng C, Chitsaz F et al (2013) CDD: conserved domains and protein three-dimensional structure. Nucleic Acids Res 41:348–352. https://doi.org/10.1093/nar/gks1243
Marchler-Bauer A, Bo Y, Han L et al (2017) CDD/SPARCLE: functional classification of proteins via subfamily domain architectures. Nucleic Acids Res 45:D200–D203. https://doi.org/10.1093/nar/gkw1129
Nonis A, Ruperti B, Pierasco A, Canaguier A, Adam-Blondon AF, Di Gaspero G et al (2008) Neutral invertases in grapevine and comparative analysis with Arabidopsis, poplar and rice. Planta 229:129–142. https://doi.org/10.1007/s00425-008-0815-0
Ogawa D, Yamamoto E, Ohtani T et al (2018) Haplotype-based allele mining in the Japan-MAGIC rice population. Sci Rep 8:1–11. https://doi.org/10.1038/s41598-018-22657-3
Priest HD, Filichkin SA, Mockler TC (2009) Cis-regulatory elements in plant cell signaling. Curr Opin Plant Biol 12:643–649. https://doi.org/10.1016/j.pbi.2009.07.016
Qi X, Wu Z, Li J et al (2007) AtCYT-INV1, a neutral invertase, is involved in osmotic stress-induced inhibition on lateral root growth in Arabidopsis. Plant Mol Biol 64:575–587. https://doi.org/10.1007/s11103-007-9177-4
Roitsch T, González MC (2004) Function and regulation of plant invertases: sweet sensations. Trends Plant Sci 9:606–613. https://doi.org/10.1016/j.tplants.2004.10.009
Rozas J, Ferrer-Mata A, Sánchez-DelBarrio JC et al (2017) DnaSP 6: DNA sequence polymorphism analysis of large data sets. Mol Biol Evol 34:3299–3302. https://doi.org/10.1093/molbev/msx248
Ruan Y, Jin Y, Yang Y et al (2010) Sugar input, metabolism, and signaling mediated by invertase: Roles in development, yield potential, and response to drought and heat. Mol Plant 3(6):942–955. https://doi.org/10.1093/mp/ssq044
Ruan B, Shang L, Gao Z et al (2020) Natural variation in the promoter of TGW2 determines grain width and weight in rice. New Phytol 227:629–640. https://doi.org/10.1111/nph.16540
Sahu KK, Chattopadhyay D (2017) Genome-wide sequence variations between wild and cultivated tomato species revisited by whole genome sequence mapping. BMC Genomics 18:430. https://doi.org/10.1186/s12864-017-3822-3
Schaarschmidt S, Roitsch T, Hause B (2006) Arbuscular mycorrhiza induces gene expression of the apoplastic invertase LIN6 in tomato (Lycopersicon esculentum) roots. J Exp Bot 57:4015–4023
Schmiesing A, Emonet A, Gouhier-Darimont C, Reymond P (2016) Arabidopsis MYC transcription factors are the target of hormonal salicylic acid/jasmonic acid cross talk in response to Pieris brassicae egg extract. Plant Physiol 170:2432–2443. https://doi.org/10.1104/pp.16.00031
Shen L-B, Qin Y-L, Qi Z-Q et al (2018) Genome-wide analysis, expression profile, and characterization of the acid invertase gene family in pepper. Int J Mol Sci 20:15. https://doi.org/10.3390/ijms20010015
Shen L-B, Qin YL, Qi ZQ et al (2019) Genome-wide analysis, expression profile, and characterization of the acid invertase gene family in pepper. Int J Mol Sci 20:15. https://doi.org/10.3390/ijms20010015
Slugina MA, Shchennikova AV, Kochieva EZ (2018) LIN7 cell-wall invertase orthologs in cultivated and wild tomatoes (Solanum section lycopersicon). Plant Mol Biol Rep 36:195–209. https://doi.org/10.1007/s11105-018-1071-5
Strickler SR, Bombarely A, Munkvold JD et al (2015) Comparative genomics and phylogenetic discordance of cultivated tomato and close wild relatives. PeerJ 3:e793. https://doi.org/10.7717/peerj.793
Tang G-Q, Lüscher M, Sturm A (1999) Antisense repression of vacuolar and cell wall invertase in transgenic carrot alters early plant development and sucrose partitioning. Plant Cell 11:177–189
Tomato Genome Consortium (2012) The tomato genome sequence provides insights into fleshy fruit evolution. Nature 485:635–641. https://doi.org/10.1038/nature11119
Tymowska-Lalanne Z, Kreis M (1998) Expression of the Arabidopsis thaliana invertase gene family. Planta 207:259–265. https://doi.org/10.1007/s004250050481
Ullah I, Magdy M, Wang L et al (2019) Genome-wide identification and evolutionary analysis of TGA transcription factors in soybean. Sci Rep 9:11186. https://doi.org/10.1038/s41598-019-47316-z
Van Bel M, Proost S, Wischnitzki E et al (2012) Dissecting plant genomes with the PLAZA comparative genomics platform. PLANT Physiol 158:590–600. https://doi.org/10.1104/pp.111.189514
Wan H, Wu L, Yang Y et al (2018) Evolution of sucrose metabolism: the dichotomy of invertases and beyond. Trends Plant Sci 23:163–177. https://doi.org/10.1016/j.tplants.2017.11.001
Wang L, Zheng Y, Ding S et al (2017) Molecular cloning, structure, phylogeny and expression analysis of the invertase gene family in sugarcane. BMC Plant Biol 17:109. https://doi.org/10.1186/s12870-017-1052-0
Wang A, Chen D, Ma Q et al (2019a) The tomato high pigment1/damaged dna binding protein 1 gene contributes to regulation of fruit ripening. Hortic Res. https://doi.org/10.1038/s41438-018-0093-3
Wang Y, Jiang Z, Li Z et al (2019b) Genome-wide identification and expression analysis of the VQ gene family in soybean (Glycine max). PeerJ 7:e7509. https://doi.org/10.7717/peerj.7509
Yadav A, Radhakrishnan A, Bhanot G, Sinha H (2015) Differential regulation of antagonistic pleiotropy in synthetic and natural populations suggests its role in adaptation. G3 Genes Genom Genet 5:699–709. https://doi.org/10.1534/g3.115.017020
Yamada K, Osakabe Y (2018) Sugar compartmentation as an environmental stress adaptation strategy in plants. Semin Cell Dev Biol 83:106–114. https://doi.org/10.1016/j.semcdb.2017.12.015
Yao Y, Geng MT, Wu XH et al (2014) Genome-wide identification, 3D modeling, expression and enzymatic activity analysis of cell wall invertase gene family from cassava (Manihot esculenta Crantz). Int J Mol Sci 15:7313–7331. https://doi.org/10.3390/ijms15057313
Yu X, Wang X, Zhang W et al (2008) Antisense suppression of an acid invertase gene (MAI1) in muskmelon alters plant growth and fruit development. J Exp Bot 59:2969–2977. https://doi.org/10.1093/jxb/ern158
Zafar SA, Zaidi SSA, Pareek A et al (2020) Engineering abiotic stress tolerance via CRISPR/Cas-mediated genome editing. J Exp Bot 71(2):470–479. https://doi.org/10.1093/jxb/erz476
Zhang Z, Li J, Zhao X et al (2006) KaKs calculator : calculating Ka and Ks through model selection and model averaging. Genom Proteom Bioinform 4:259–263. https://doi.org/10.1016/S1672-0229(07)60007-2
Zhang J, Kapli P, Pavlidis P et al (2013) A general species delimitation method with applications to phylogenetic placements. Bioinformatics 29(22):2869–2876. https://doi.org/10.1093/bioinformatics/btt499
Zhang N, Jiang J, Yang Y, Wang Z (2015) Functional characterization of an invertase inhibitor gene involved in sucrose metabolism in tomato fruit. J Zhejiang Univ B 16:845–856. https://doi.org/10.1631/jzus.b1400319
Zhang W, Wang S, Yu F et al (2019a) Genome-wide identification and expression profiling of sugar transporter protein (STP) family genes in cabbage (Brassica oleracea var. capitata L.) reveals their involvement in clubroot disease responses. Genes (basel) 10:71. https://doi.org/10.3390/genes10010071
Zhang W, Wang S, Yu F et al (2019b) Genome-wide characterization and expression profiling of SWEET genes in cabbage (Brassica oleracea var. capitata L.) reveal their roles in chilling and clubroot disease responses. BMC Genom 20:93. https://doi.org/10.1186/s12864-019-5454-2
Zhou J, Wang J, Yu J-Q, Chen Z (2014a) Role and regulation of autophagy in heat stress responses of tomato plants. Front Plant Sci 5:174. https://doi.org/10.3389/fpls.2014.00174
Zhou J, Yu J-Q, Chen Z (2014b) The perplexing role of autophagy in plant innate immune responses. Mol Plant Pathol 15:637–645. https://doi.org/10.1111/mpp.12118
Acknowledgements
We gratefully acknowledge Huiyang Yu, Ai Guo and Zhang Dedi from Huazhong Agricultural University for their inputs and assistance. This work was supported by grants from the National Key Research and Development Program of China (2018YFD1000800), National Natural Science Foundation of China (31991182 and 31972426), Wuhan Frontier Projects for Applied Foundation (2019020701011492), and the Fundamental Research Funds for the Central Universities (2662018PY073).
Author information
Authors and Affiliations
Contributions
YZ conceived and designed the experiments, JKA, BK & MM finetuned the methodology, JKA, MM, SM & BK sourced the softwares and validated the data. JKA, MAM, SAT, GL, WC & YW performed the experiments, MM, BK, SM & JKA analysed the data, JKA wrote the manuscript, JK, BK, MM, SM and YZ revised the manuscript, BK and YZ contributed extensively to its final form, YZ sourced for funding and supervised the work. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that, the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Additional information
Handling Editor: Rhonda Peavy.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Ahiakpa, J.K., Magdy, M., Karikari, B. et al. Genome-Wide Identification and Expression Profiling of Tomato Invertase Genes Indicate Their Response to Stress and Phytohormones. J Plant Growth Regul 41, 1481–1498 (2022). https://doi.org/10.1007/s00344-021-10384-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00344-021-10384-5