Abstract
Objectives
To evaluate the quality of radiomics studies using cardiac magnetic resonance imaging (CMR) according to the radiomics quality score (RQS), Transparent Reporting of a multivariable prediction model for Individual Prognosis Or Diagnosis (TRIPOD) guidelines, and the standards defined by the Image Biomarker Standardization Initiative (IBSI) and identify areas needing improvement.
Materials and methods
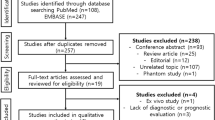
PubMed and Embase were searched to identify radiomics studies using CMR until March 10, 2021. Of the 259 identified articles, 32 relevant original research articles were included. Studies were scored according to the RQS, TRIPOD guidelines, and IBSI standards by two cardiac radiologists.
Results
The mean RQS was 14.3% of the maximum (5.16 out of 36). RQS were low for the demonstration of validation (−60.6%), calibration statistics (1.6%), potential clinical utility (3.1%), and open science (3.1%) items. No study conducted a phantom study or cost-effectiveness analysis. The adherence to TRIPOD guidelines was 55.9%. Studies were deficient in reporting title (3.1%), stating objective in abstract and introduction (6.3% and 9.4%), missing data (0%), discrimination/calibration (3.1%), and how to use the prediction model (3.1%). According to the IBSI standards, non-uniformity correction, image interpolation, grey-level discretization, and signal intensity normalization were performed in two (6.3%), four (12.5%), six (18.8%), and twelve (37.5%) studies, respectively.
Conclusion
The quality of radiomics studies using CMR is suboptimal. Improvements are needed in the areas of validation, calibration, clinical utility, and open science. Complete reporting of study objectives, missing data, discrimination/calibration, how to use the prediction model, and preprocessing steps are necessary.
Key Points
• The quality of science in radiomics studies using CMR is currently inadequate.
• RQS were low for validation, calibration, clinical utility, and open science; no study conducted a phantom study or cost-effectiveness analysis.
• In stating the study objective, missing data, discrimination/calibration, how to use the prediction model, and preprocessing steps, improvements are needed.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Cardiac magnetic resonance imaging (CMR) comprehensively evaluates cardiac structure and function and characterizes myocardial tissue. It is considered the gold standard to assess ventricular function and volume [1]. The ability of CMR to distinguish heart tissue characteristics facilitates assessing myocardial viability, determining a differential diagnosis, and predicting prognosis in several cardiomyopathies [2, 3]. In addition to qualitatively interpreting CMR images by visual inspection, the use of quantitative parameters, including parametric mapping techniques, is growing [4]. However, it still has limitations with broad overlap between health and various diseases [5], necessitating a method that utilizes the information of CMR images to the maximum.
Radiomics is an emerging field of research that converts digital medical images into mineable data to extract quantitative features, expanding the utility of images into decision-making and personalized healthcare management [6]. Traditionally, radiomics studies have been mainly conducted in oncology [7], while it has not been extensively conducted in cardiology. However, its application to cardiac imaging has been gradually increasing, especially with CMR [8, 9].
Early studies of echocardiography conducted quantitative texture analysis for differential diagnosis [10, 11], but the experience of radiomics in echocardiography is limited due to low reproducibility. In cardiac CT, several studies reported promising results of radiomics for characterization of coronary plaque [12, 13], perivascular adipose tissue [14, 15], myocardium [16, 17], and cardiac mass [18, 19]. In CMR, several studies have demonstrated feasibility and potential clinical utility of radiomics analysis for diagnostic or prognostic purposes [8]. Since radiomics analysis provides additional objective data from existing images, it has potential to be added to the routine clinical workflow [9].
Despite promising results, CMR radiomics is rarely applied in clinical practice. An important reason for this gap is that standardization of the methodology and reproducibility and robustness of the features are not sufficiently verified [20]. Therefore, generating radiomics data using high-quality science and reporting practices is essential to its clinical application [21].
The radiomics quality score (RQS) was developed to assess the methodology and analysis of a radiomics study [22] and has been applied to oncology or dementia studies [23,24,25,26,27,28,29,30]. Transparent Reporting of a multivariable prediction model for Individual Prognosis Or Diagnosis (TRIPOD) guidelines are used to evaluate studies that apply diagnostic or prognostic prediction models [31]. Transparent reporting reduces the risk of bias and enhances the clinical utility of a prediction model. Standards defined by the Image Biomarkers Standardization Initiative (IBSI) provide uniformity for image biomarker nomenclature and definitions, image processing workflows, tools for verifying radiomics software implementations, and reporting of radiomic studies [32].
To the best of our knowledge, no previous study has comprehensively evaluated CMR radiomics studies using these three sets of guidelines. Therefore, the purpose of this study was to assess the quality of radiomics studies using CMR with RQS, TRIPOD guidelines, and IBSI standards to identify areas in need of improvement.
Materials and methods
Article search strategy
Two cardiac radiologists with 6 and 9 years of experience, respectively, designed the search strategy. Each radiologist independently performed systematic searches of PubMed and EMBASE on March 10, 2021, to identify relevant original research articles reporting CMR radiomics studies. The search terms are listed in Supplementary Materials.
Study selection
Two radiologists independently reviewed the search results. Articles were initially screened based on titles and abstracts of articles meeting the inclusion criteria. A study was selected for inclusion if it analyzed radiomics using CMR images, involved human participants, collected data from in vivo studies, was written in English, and had available full text. Full-text articles were then reviewed for eligibility. A study was excluded if it was test-retest study without investigating diagnostic or prognostic utility, or did not evaluate the diagnostic or prognostic performance (Fig. 1).
Data extraction
Two radiologists independently extracted data, and any disagreements were resolved by consensus. The extracted parameters included (a) article information: authors, year of publication, journal type (clinical, imaging, or computer science journal), diseases, study topic, the intended use of radiomics features (diagnostic or prognostic), and the sequence(s) used for feature extraction; (b) enrollee characteristics: number of participants and primary diagnosis.
Method quality based on RQS
The RQS consists of 16 items and is divided into six domains [22]. The details of RQS are presented in Supplementary Materials. Two radiologists independently assessed the RQS of each article. If disagreement occurred between the two reviewers, a final decision was made by a consensus through discussion. The topics which were subject to further discussions until a consensus was reached included feature reduction, discrimination statistics, non-radiomics features, biological correlates, and comparison to “gold standard”.
Reporting completeness based on TRIPOD guidelines
The TRIPOD checklist consists of 22 main criteria assessed by a total of 37 items. Items 21 and 22 were excluded from this study because they refer to supplementary and funding information. Therefore, each article was evaluated with 35 parameters to assess reporting completeness [31]. Furthermore, the type of radiomics model in each article was categorized as development only (type 1a), development and validation using resampling (type 1b), random split-sample validation (type 2a), non-random split-sample validation (type 2b), validation using separate data (type 3), or validation only (type 4). The details of the TRIPOD checklist and data extraction method are presented in Supplementary Materials.
Image preprocessing quality and radiomics feature extraction based on IBSI standards
Two radiologists evaluated whether the preprocessing and processing items were detailed in the methods based on the IBSI guidelines (https://ibsi.readthedocs.io/en/latest/): non-uniformity correction, image interpolation, grey-level discretization, signal intensity normalization, radiomics feature extraction software, and segmentation method.
Statistical analysis
The RQS score was calculated for each article (score range, −8 to 36), and the RQS for all articles was expressed as the mean ± standard deviation, and median with interquartile range. For the six domains in the RQS (protocol quality and segmentation, feature selection and validation, biologic/clinical validation and utility, model performance index, high level of evidence, and open science and data), basic adherence was assigned when a score of at least 1 point was obtained without minus points. The basic adherence to RQS criteria (range, 0–16) and each item scored for the TRIPOD guidelines (range, 0–35) were counted and calculated as proportions (%). The TRIPOD item 5c (“if done” item) and validation items 10c, 10e, 12, 13c, 17, and 19a were excluded from the numerator and denominator when calculating overall adherence. The six IBSI standards were scored, and the adherence to each standard was calculated as a proportion (%). Subgroup analyses were performed by categorizing articles based on the year of publication (publication before January 1, 2019 [n = 12], or after January 1, 2019 [n = 20]) and journal impact factor (4.0 or higher [n = 16] or lower than 4.0 [n = 16]), according to the 2020 Journal Citation Reports. The impact factor 4.0 was chosen based on the median value of impact factors of included articles. The non-parametric Mann-Whitney U test was used to compare RQS scores within each group. Fisher’s exact test was used to compare proportions of RQS, TRIPOD guidelines, and IBSI standards for small sample sizes. All statistical analyses were performed using SPSS version 24.0 (IBM), and a p-value < 0.05 was considered statistically significant.
Results
Characteristics of radiomics studies using CMR
The characteristics of the 32 included radiomics studies [33,34,35,36,37,38,39,40,41,42,43,44,45,46,47,48,49,50,51,52,53,54,55,56,57,58,59,60,61,62,63,64] are summarized in Table 1 and Fig. 2. The journal, impact factor, primary diagnosis, study topic, biomarker, number of participants, and model type are summarized in Supplementary Table 1. The mean number of patients per study was 244.5 (standard deviation, 850.9; range, 23–4891).
The proportions of studies by journal type are as follows: imaging journals (59.4%), clinical journals (28.1%), and computer science journals (12.5%). Radiomics analyses were conducted for diagnostic purposes (50%) and to identify prognostic biomarkers (50%). The two most frequent primary diagnoses were non-ischemic cardiomyopathy (50%) and ischemic cardiomyopathy (31.3%). The study purposes included differential diagnosis (46.9%), adverse event prediction (34.4%), late gadolinium enhancement (LGE) prediction (9.4%), functional recovery prediction (6.3%), and genomic classification (3.1%). Mapping was the most studied sequence (40.6%), followed by LGE (31.3%), cine (18.8%), T1-weighted image (3.1%), T2-weighted image (3.1%), and two or more sequences (6.3%). Most studies performed manual segmentation (90.6%), whereas three (9.4%) performed automatic segmentation. Studies were more frequently conducted at 1.5T (62.5%) than 3T (25%). Three studies (9.4%) included both 1.5T and 3T scans, and one study did not report the magnetic field strength. Three studies were published in 2010–2015, nine in 2016–2018, and twenty in 2019–2021. Four of the 32 studies were published in journals ranked in the top 10% in the field by impact factor [44, 46, 50, 52].
RQS according to the six key domains
The RQS evaluation is summarized in Table 2. The RQS of the 32 studies combined expressed as a percentage of the ideal score according to the six key domains is shown in Fig. 3. The mean overall RQS is 5.16 ± 5.85 (range, −5 to 21), or 14.3 ± 16.3% of the ideal score of 36. Key domain 2 (feature selection and validation) has the lowest percentage (−21.5%) of the ideal score, followed by domain 6 (open science and data, 3.1%) and domain 5 (high level of evidence, 19.1%). Domain 4 has the highest percentage of the ideal score (34.9%).
Basic adherence to the RQS according to the six key domains
The basic adherence to the 16 RQS criteria is documented in Table 2. The overall basic adherence to the RQS is 35.5%.
Thirty studies (93.8%) followed the domain 1 criterion of having a well-documented image protocol, and one study used a public protocol [60]. Only one study used a test-retest approach at different times to evaluate robustness to temporal variability [43], and no study performed a phantom study. Multiple segmentations for the same region were performed in 19 studies (59.4%) [33,34,35,36,37,38,39, 41, 42, 44,45,46, 48,49,50,51,52, 59, 63].
Twenty-three studies (71.9%) performed the domain 2 criteria of feature reduction or adjustment for multiple testing [33,34,35,36,37,38,39, 41, 42, 44, 46,47,48,49,50,51,52, 54, 55, 59,60,61, 63]. Nine studies (28.1%) performed validation from the same institute [35, 37, 39, 41, 44, 47, 56, 57, 64]. No study performed external validation.
Twelve studies (37.5%) performed the domain 3 criterion of cutoff analysis. Twenty-nine studies (90.6%) applied discrimination statistics using receiver operating characteristics curve and/or area under the curve, and only one study (3.1%) adopted calibration statistics without a resampling method [35].
Twenty-two studies (68.8%) performed the domain 4 criterion of discussed biological correlation of the radiomics features, and eighteen studies (56.3%) compared the radiomics features to the gold standard methods. Seven studies (21.9%) performed multivariable analysis with non-radiomics features [36, 40, 45, 48, 49, 58, 63], and one study (3.1%) derived the potential clinical utility [35].
Seven articles (21.9%) reported performing the domain 5 criterion of a prospective study [36, 41, 45, 46, 52, 57, 63]. No study conducted a cost-effectiveness analysis. Three studies (9.4%) performed the domain 6 criterion of using open-source scans or code [38, 41, 60].
Reporting completeness of radiomics-based multivariable prediction models using TRIPOD
The mean number of TRIPOD items reported is 16.5 ± 4.0 (standard deviation; range, 9–25) out of the 35 items considered. The adherence rate for TRIPOD is 55.9% ± 12.2% (standard deviation; range, 32.1%–78.6%) when “if relevant” and “if done” items are excluded from the numerator and denominator. The reporting completeness of individual TRIPOD items is shown in Table 3.
Quality of image preprocessing and radiomics feature extraction according to IBSI
Most studies did not report preprocessing steps in detail (Table 4). Two studies (6.3%) performed non-uniformity correction, four studies (12.5%) performed image interpolation, six studies (18.8%) performed grey-level discretization, and twelve studies (37.5%) performed signal intensity normalization. The software packages used for radiomics feature extraction were MaZda (43.8%), MATLAB (34.4%), TexRAD (12.5%), PyRadiomics (3.1%), and AVIEW (3.1%). One study did not mention the software used.
Subgroup analysis
The results of the subgroup analysis are shown in Table 5. The total RQS is higher for the twenty studies published since 2019 than the twelve studies published before 2019, but the difference is not statistically significant (mean 6.4 ± 6.4 vs. 3.08 ± 4.25; p = 0.114). The studies published since 2019 score higher for domain 1 (p = 0.02) and domain 4 (p = 0.005) items. The most significant difference is observed for the “comparison to gold standard” parameter (p = 0.007). The more recently published studies also have higher adherence rates for the TRIPOD criterion of “describing eligibility criteria” (p = 0.049). No significant differences are observed between the two groups for ISBI standards.
The total RQS is higher for studies published in the journals with higher ( ≥ 4.0) impact factor than those published in journals with lower ( < 4.0) impact factor (mean 7.56 ± 4.29 vs. 2.75 ± 6.32; p = 0.007). The studies published in higher impact factor journals score higher for domain 5 (p = 0.035). The studies published in higher impact factor journals also have higher adherence rates for TRIPOD criterion of “key dates” (p = 0.043), “modeling” (p = 0.037), and “unadjusted association” (p = 0.033). No statistically significant differences are observed between the two groups for IBSI standards.
Discussion
Our systematic review evaluated the quality of science in CMR radiomics studies using the RQS, TRIPOD guidelines, and IBSI standards. Radiomics studies scored poorly in science quality and reporting completeness, with 14.3% of the ideal RQS and 55.9% adherence to the TRIPOD guidelines. No study performed a phantom study or cost-effective analysis, and approximately half of the TRIPOD items were not adequately recorded. Most studies did not report preprocessing steps according to the IBSI standards. These results imply that CMR radiomics studies need significant improvement in scientific quality.
The number of radiomics studies performed using CMR is relatively small. However, the studies published to date demonstrate promising results for differential diagnosis and prognosis. Because radiomic features extracted from CMR images can be affected by the equipment, protocol, or personnel, it is vital to establish reproducibility and reduce variability for clinical application [9]. Most CMR radiomics studies focused on the left ventricle for myocardial tissue characterization in non-ischemic and ischemic cardiomyopathies, and mapping was the most frequently used sequence for feature extraction.
The overall mean RQS for CMR studies was 5.16 ± 5.85 (14.3 ± 16.3%), which is lower than the RQS reported for some oncology studies (mean score 9.4–11) [23, 24], but similar to that reported in other radiomics studies on various tumors and dementia (mean score 3.6–6.9) [25,26,27,28,29,30]. The basic adherence rate was low for domain 5 (10.9%) and domain 6 (9.4%); such rates are similar to oncology studies [23,24,25, 27], suggesting that there are limitations to pursuing higher levels of evidence and open science. No phantom study or cost-effective analysis was performed in the included CMR studies, which is consistent with non-cardiac studies [23, 24, 26, 27] and indicates low-level evidence. The basic adherence rate of the calibration statistics (3.1%) was lower than that of oncology studies (range 9.9–29.9%) [23,24,25, 27], suggesting that improvement is needed. The low basic adherence rates for test-retest (3.1%) and potential clinical utility (3.1%) were consistent with other studies [24, 26]. The basic adherence rate of validation (28.1%) was lower than studies on tumors or dementia (range 46.2–70.1%) [23, 24, 26, 27] but was similar to one sarcoma study (26.9%) [25]. The nine CMR studies that performed validation used datasets from the same institute, and no study performed external validation, resulting in a low RQS score. Because radiomic features extracted from CMR images can be largely affected by protocol, standardization, compensation for multicenter effects, and external validation are necessary and crucial for the clinical application of radiomics.
Our results are in line with the recently published study that assessed RQS in cardiac MRI and CT radiomics studies [65]. The median total RQS in the previous study was 7, which was higher than in our study (mean 5.16, median 4.5). The difference could be due to the different range of included articles and interobserver variability for RQS assessment. For example, our study included fewer articles with internal validation (28.1% vs. 45%), and more articles with prospective design (21.9% vs. 4%). The greatest difference was found in potential clinical utility item (3.1% vs. 94%). In our study, an article earned points if the clinical utility was objectively assessed, such as decision curve analysis to demonstrate net improvement. On the other hand, discussing the possible utility of radiomics without an adequate analysis did not provide points. Despite some differences, both studies revealed that improvement is needed in validation, calibration, cost-effectiveness, and open science items.
The overall adherence to the TRIPOD guidelines was 55.9%, consistent with previous radiomics studies [23, 27]. Specifically, the rates of adherence to TRIPOD guidelines were low for reporting title (3.1%), missing data (0%), discrimination/calibration (3.1%), and how to use the prediction model (3.1%). Most articles did not explicitly describe development or validation in the title (item 1), abstract (item 2), or introduction (item 3b). Only one study [35] conducted calibration statistics (item 10d) and constructed a nomogram for clinical use (item 15b), resulting in low adherence rates for these items. Improving the adherence of research conduct and reporting according to the TRIPOD guidelines could improve the quality of radiomics models.
The image preprocessing steps performed before extracting radiomics features affect the repeatability and reproducibility of results [66, 67]. However, most CMR radiomics studies fail to describe the preprocessing steps according to the IBSI standards of non-uniformity correction, image interpolation, grey-level discretization, and signal intensity normalization (6.3%, 12.5%, 18.8%, and 37.5%, respectively), and the adherence rates are lower than those of radiomics studies in cancer patients [25, 27]. Making more concerted efforts to conduct image preprocessing steps according to IBSI standards could improve the reproducibility of CMR radiomics studies.
The subgroup analysis revealed that the total RQS was higher in studies published since 2019 than studies published before 2019; however, the differences did not reach statistical significance. More recent studies have been conducted with better protocol quality and stability (domain 1) and biologic/clinical validation and utility (domain 4). The adherence rates for most TRIPOD guidelines also tended to be higher in the more recent studies. However, for IBSI standards, even the more recent studies exhibited inadequate adherence.
When grouped according to impact factor, studies published in journals with a higher (≥4.0) impact factor had higher RQS scores than studies published in journals with a lower (<4.0) impact factor. The studies published in higher impact factor journals had a higher level of evidence (domain 5), because they included more prospective studies. In TRIPOD items, they more clearly defined the study dates, type of model, and unadjusted association between candidate predictor and outcome. However, no significant difference was observed for IBSI standards between articles grouped by journal impact factor.
Our results suggest that the RQS, TRIPOD guidelines, and IBSI standards were not adequately followed even in recently published studies or articles published in more highly ranked journals. We conclude that additional effort is needed to improve the scientific quality and reporting completeness in CMR radiomics studies.
There are several limitations to our study. First, the number of included CMR radiomics studies was small. Second, some items of RQS and TRIPOD may be too idealistic for most studies to meet. For example, it is not easy to perform multiple CMR scans in clinical practice. In addition, it is challenging to meet all the TRIPOD guidelines because most CMR radiomics studies are retrospective. However, it is necessary to be as transparent as possible about protocol parameters and data extraction methods. Lastly, we did not assess the inter-reader reproducibility for scores due to the consensus approach.
In conclusion, the overall scientific quality and reporting completeness of CMR radiomics studies are inadequate. Improvements are needed in the areas of validation, calibration, clinical utility, and open science. Complete reporting of study objectives, missing data, discrimination/calibration, and how to use the prediction model is necessary, and image preprocessing steps must be reported in detail.
Abbreviations
- CMR:
-
Cardiac magnetic resonance imaging
- IBSI:
-
Image Biomarker Standardization Initiative
- LGE:
-
Late gadolinium enhancement
- RQS:
-
Radiomics quality score
- TRIPOD:
-
Transparent Reporting of a multivariable prediction model for Individual Prognosis Or Diagnosis
References
West AM, Kramer CM (2009) Comprehensive cardiac magnetic resonance imaging. J Invasive Cardiol 21:339–345
Lee E, Ibrahim EH, Parwani P, Bhave N, Stojanovska J (2020) Practical guide to evaluating myocardial disease by cardiac MRI. AJR Am J Roentgenol 214:546–556
Patel AR, Kramer CM (2017) Role of cardiac magnetic resonance in the diagnosis and prognosis of nonischemic cardiomyopathy. JACC Cardiovasc Imaging 10:1180–1193
Messroghli DR, Moon JC, Ferreira VM et al (2017) Clinical recommendations for cardiovascular magnetic resonance mapping of T1, T2, T2* and extracellular volume: a consensus statement by the Society for Cardiovascular Magnetic Resonance (SCMR) endorsed by the European Association for Cardiovascular Imaging (EACVI). J Cardiovasc Magn Reson 19:75
Thongsongsang R, Songsangjinda T, Tanapibunpon P, Krittayaphong R (2021) Native T1 mapping and extracellular volume fraction for differentiation of myocardial diseases from normal CMR controls in routine clinical practice. BMC Cardiovasc Disord 21:270
Gillies RJ, Kinahan PE, Hricak H (2016) Radiomics: images are more than pictures, they are data. Radiology 278:563–577
Liu Z, Wang S, Dong D et al (2019) The applications of radiomics in precision diagnosis and treatment of oncology: opportunities and challenges. Theranostics 9:1303–1322
Raisi-Estabragh Z, Izquierdo C, Campello VM et al (2020) Cardiac magnetic resonance radiomics: basic principles and clinical perspectives. Eur Heart J Cardiovasc Imaging 21:349–356
Hassani C, Saremi F, Varghese BA, Duddalwar V (2020) Myocardial radiomics in cardiac MRI. AJR Am J Roentgenol 214:536–545
Pinamonti B, Picano E, Ferdeghini EM et al (1989) Quantitative texture analysis in two-dimensional echocardiography: application to the diagnosis of myocardial amyloidosis. J Am Coll Cardiol 14:666–671
Lattanzi F, Bellotti P, Picano E et al (1996) Quantitative texture analysis in two-dimensional echocardiography: application to the diagnosis of myocardial hemochromatosis. Echocardiography 13:9–20
Kolossvary M, Karady J, Kikuchi Y et al (2019) Radiomics versus visual and histogram-based assessment to identify atheromatous lesions at coronary CT angiography: an ex vivo study. Radiology 293:89–96
Kolossvary M, Karady J, Szilveszter B et al (2017) Radiomic features are superior to conventional quantitative computed tomographic metrics to identify coronary plaques with napkin-ring sign. Circ Cardiovasc Imaging:10
Oikonomou EK, Williams MC, Kotanidis CP et al (2019) A novel machine learning-derived radiotranscriptomic signature of perivascular fat improves cardiac risk prediction using coronary CT angiography. Eur Heart J 40:3529–3543
Lin A, Kolossvary M, Yuvaraj J et al (2020) Myocardial infarction associates with a distinct pericoronary adipose tissue radiomic phenotype: a prospective case-control study. JACC Cardiovasc Imaging 13:2371–2383
Kay FU, Abbara S, Joshi PH, Garg S, Khera A, Peshock RM (2020) Identification of high-risk left ventricular hypertrophy on calcium scoring cardiac computed tomography scans: validation in the DHS. Circ Cardiovasc Imaging 13:e009678
Mannil M, von Spiczak J, Manka R, Alkadhi H (2018) Texture analysis and machine learning for detecting myocardial infarction in noncontrast low-dose computed tomography: unveiling the invisible. Invest Radiol 53:338–343
Nam K, Suh YJ, Han K, Park SJ, Kim YJ, Choi BW (2019) Value of computed tomography radiomic features for differentiation of periprosthetic mass in patients with suspected prosthetic valve obstruction. Circ Cardiovasc Imaging 12:e009496
Chun SH, Suh YJ, Han K et al (2021) Differentiation of left atrial appendage thrombus from circulatory stasis using cardiac CT radiomics in patients with valvular heart disease. Eur Radiol 31:1130–1139
Jang J, Ngo LH, Mancio J et al (2020) Reproducibility of segmentation-based myocardial radiomic features with cardiac MRI. Radiol Cardiothorac Imaging 2:e190216
O'Connor JP, Aboagye EO, Adams JE et al (2017) Imaging biomarker roadmap for cancer studies. Nat Rev Clin Oncol 14:169–186
Lambin P, Leijenaar RTH, Deist TM et al (2017) Radiomics: the bridge between medical imaging and personalized medicine. Nat Rev Clin Oncol 14:749–762
Park JE, Kim D, Kim HS et al (2020) Quality of science and reporting of radiomics in oncologic studies: room for improvement according to radiomics quality score and TRIPOD statement. Eur Radiol 30:523–536
Park JE, Kim HS, Kim D et al (2020) A systematic review reporting quality of radiomics research in neuro-oncology: toward clinical utility and quality improvement using high-dimensional imaging features. BMC Cancer 20:29
Crombe A, Fadli D, Italiano A, Saut O, Buy X, Kind M (2020) Systematic review of sarcomas radiomics studies: bridging the gap between concepts and clinical applications? Eur J Radiol 132:109283
Won SY, Park YW, Park M, Ahn SS, Kim J, Lee SK (2020) Quality reporting of radiomics analysis in mild cognitive impairment and Alzheimer's disease: a roadmap for moving forward. Korean J Radiol 21:1345–1354
Won SY, Park YW, Ahn SS et al (2021) Quality assessment of meningioma radiomics studies: bridging the gap between exploratory research and clinical applications. Eur J Radiol 138:109673
Wang H, Zhou Y, Li L, Hou W, Ma X, Tian R (2020) Current status and quality of radiomics studies in lymphoma: a systematic review. Eur Radiol 30:6228–6240
Ursprung S, Beer L, Bruining A et al (2020) Radiomics of computed tomography and magnetic resonance imaging in renal cell carcinoma-a systematic review and meta-analysis. Eur Radiol 30:3558–3566
Zhong J, Hu Y, Si L et al (2021) A systematic review of radiomics in osteosarcoma: utilizing radiomics quality score as a tool promoting clinical translation. Eur Radiol 31:1526–1535
Moons KG, Altman DG, Reitsma JB et al (2015) Transparent Reporting of a multivariable prediction model for Individual Prognosis or Diagnosis (TRIPOD): explanation and elaboration. Ann Intern Med 162:W1–W73
Zwanenburg A, Vallieres M, Abdalah MA et al (2020) The Image Biomarker Standardization Initiative: standardized quantitative radiomics for high-throughput image-based phenotyping. Radiology 295:328–338
Son J, Hong YJ, Kim S et al (2021) Radiomics feature analysis using native T1 mapping for discriminating between cardiac tumors and thrombi. Acad Radiol 5:S1076-6332(20)30702-9
Shi RY, Wu R, An DL et al (2021) Texture analysis applied in T1 maps and extracellular volume obtained using cardiac MRI in the diagnosis of hypertrophic cardiomyopathy and hypertensive heart disease compared with normal controls. Clin Radiol 76:236 e239–236 e219
Ma Q, Ma Y, Wang X et al (2021) A radiomic nomogram for prediction of major adverse cardiac events in ST-segment elevation myocardial infarction. Eur Radiol 31:1140–1150
Chen BH, An DA, He J et al (2021) Myocardial extracellular volume fraction radiomics analysis for differentiation of reversible versus irreversible myocardial damage and prediction of left ventricular adverse remodeling after ST-elevation myocardial infarction. Eur Radiol 31:504–514
Neisius U, El-Rewaidy H, Kucukseymen S et al (2020) Texture signatures of native myocardial T1 as novel imaging markers for identification of hypertrophic cardiomyopathy patients without scar. J Magn Reson Imaging 52:906–919
Muthulakshmi M, Kavitha G (2020) An integrated multi-objective whale optimized support vector machine and local texture feature model for severity prediction in subjects with cardiovascular disorder. Int J Comput Assist Radiol Surg 15:601–615
Ma Q, Ma Y, Yu T, Sun Z, Hou Y (2021) Radiomics of non-contrast-enhanced T1 mapping: diagnostic and predictive performance for myocardial injury in acute ST-segment-elevation myocardial infarction. Korean J Radiol 22:535–546
Gould J, Porter B, Sidhu BS et al (2020) High mean entropy calculated from cardiac MRI texture analysis is associated with antitachycardia pacing failure. Pacing Clin Electrophysiol 43:737–745
El-Rewaidy H, Neisius U, Nakamori S et al (2020) Characterization of interstitial diffuse fibrosis patterns using texture analysis of myocardial native T1 mapping. PLoS One 15:e0233694
Alis D, Guler A, Yergin M, Asmakutlu O (2020) Assessment of ventricular tachyarrhythmia in patients with hypertrophic cardiomyopathy with machine learning-based texture analysis of late gadolinium enhancement cardiac MRI. Diagn Interv Imaging 101:137–146
Schofield R, Ganeshan B, Fontana M et al (2019) Texture analysis of cardiovascular magnetic resonance cine images differentiates aetiologies of left ventricular hypertrophy. Clin Radiol 74:140–149
Neisius U, El-Rewaidy H, Nakamori S, Rodriguez J, Manning WJ, Nezafat R (2019) Radiomic analysis of myocardial native T1 imaging discriminates between hypertensive heart disease and hypertrophic cardiomyopathy. JACC Cardiovasc Imaging 12:1946–1954
Gould J, Porter B, Claridge S et al (2019) Mean entropy predicts implantable cardioverter-defibrillator therapy using cardiac magnetic resonance texture analysis of scar heterogeneity. Heart Rhythm 16:1242–1250
Baessler B, Luecke C, Lurz J et al (2019) Cardiac MRI and texture analysis of myocardial T1 and T2 maps in myocarditis with acute versus chronic symptoms of heart failure. Radiology 292:608–617
Larroza A, Lopez-Lereu MP, Monmeneu JV et al (2018) Texture analysis of cardiac cine magnetic resonance imaging to detect nonviable segments in patients with chronic myocardial infarction. Med Phys 45:1471–1480
Gibbs T, Villa ADM, Sammut E et al (2018) Quantitative assessment of myocardial scar heterogeneity using cardiovascular magnetic resonance texture analysis to risk stratify patients post-myocardial infarction. Clin Radiol 73:1059e1017–1059e1026
Cheng S, Fang M, Cui C et al (2018) LGE-CMR-derived texture features reflect poor prognosis in hypertrophic cardiomyopathy patients with systolic dysfunction: preliminary results. Eur Radiol 28:4615–4624
Baessler B, Mannil M, Oebel S, Maintz D, Alkadhi H, Manka R (2018) Subacute and chronic left ventricular myocardial scar: accuracy of texture analysis on nonenhanced cine MR images. Radiology 286:103–112
Baessler B, Mannil M, Maintz D, Alkadhi H, Manka R (2018) Texture analysis and machine learning of non-contrast T1-weighted MR images in patients with hypertrophic cardiomyopathy-preliminary results. Eur J Radiol 102:61–67
Baessler B, Luecke C, Lurz J et al (2018) Cardiac MRI texture analysis of T1 and T2 maps in patients with infarctlike acute myocarditis. Radiology 289:357–365
Amano Y, Suzuki Y, Yanagisawa F, Omori Y, Matsumoto N (2018) Relationship between extension or texture features of late gadolinium enhancement and ventricular tachyarrhythmias in hypertrophic cardiomyopathy. Biomed Res Int 2018:4092469
Larroza A, Materka A, Lopez-Lereu MP, Monmeneu JV, Bodi V, Moratal D (2017) Differentiation between acute and chronic myocardial infarction by means of texture analysis of late gadolinium enhancement and cine cardiac magnetic resonance imaging. Eur J Radiol 92:78–83
Kotu LP, Engan K, Borhani R et al (2015) Cardiac magnetic resonance image-based classification of the risk of arrhythmias in post-myocardial infarction patients. Artif Intell Med 64:205–215
Kotu LP, Engan K, Skretting K et al (2013) Probability mapping of scarred myocardium using texture and intensity features in CMR images. Biomed Eng Online 12:91
Wang J, Yang F, Liu W et al (2020) Radiomic analysis of native T1 mapping images discriminates between MYH7 and MYBPC3-related hypertrophic cardiomyopathy. J Magn Reson Imaging 52:1714–1721
Pham VT, Lin C, Tran TT et al (2020) Predicting ventricular tachyarrhythmia in patients with systolic heart failure based on texture features of the gray zone from contrast-enhanced magnetic resonance imaging. J Cardiol 76:601–609
Mannil M, Kato K, Manka R et al (2020) Prognostic value of texture analysis from cardiac magnetic resonance imaging in patients with Takotsubo syndrome: a machine learning based proof-of-principle approach. Sci Rep 10:20537
Cetin I, Raisi-Estabragh Z, Petersen SE et al (2020) Radiomics signatures of cardiovascular risk factors in cardiac MRI: results from the UK Biobank. Front Cardiovasc Med 7:591368
Engan K, Eftestol T, Orn S, Kvaloy JT, Woie L (2010) Exploratory data analysis of image texture and statistical features on myocardium and infarction areas in cardiac magnetic resonance images. Annu Int Conf IEEE Eng Med Biol Soc 2010:5728–5731
Amano Y, Yanagisawa F, Omori Y et al (2020) Detection of myocardial tissue alterations in hypertrophic cardiomyopathy using texture analysis of T2-weighted short inversion time inversion recovery magnetic resonance imaging. J Comput Assist Tomogr 44:341–345
Zhou H, An DA, Ni Z et al (2021) Texture analysis of native T1 images as a novel method for noninvasive assessment of uremic cardiomyopathy. J Magn Reson Imaging 54:290–300
Shao XN, Sun YJ, Xiao KT et al (2018) Texture analysis of magnetic resonance T1 mapping with dilated cardiomyopathy: a machine learning approach. Medicine (Baltimore) 97:e12246
Ponsiglione A, Stanzione A, Cuocolo R et al (2021) Cardiac CT and MRI radiomics: systematic review of the literature and radiomics quality score assessment. Eur Radiol. https://doi.org/10.1007/s00330-021-08375-x
Traverso A, Wee L, Dekker A, Gillies R (2018) Repeatability and reproducibility of radiomic features: a systematic review. Int J Radiat Oncol Biol Phys 102:1143–1158
Moradmand H, Aghamiri SMR, Ghaderi R (2020) Impact of image preprocessing methods on reproducibility of radiomic features in multimodal magnetic resonance imaging in glioblastoma. J Appl Clin Med Phys 21:179–190
Funding
This study has received funding from the National Research Foundation of Korea (NRF) grant funded by the Korean government (MSIT) (No.2021R1A2C4002195).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Guarantor
The scientific guarantor of this publication is Young Joo Suh.
Conflict of Interest
The authors of this manuscript declare no relationships with any companies, whose products or services may be related to the subject matter of the article.
Statistics and Biometry
One of the authors has significant statistical expertise.
Informed Consent
Written informed consent was not required because of the nature of our study, which was a study based on research articles.
Ethical Approval
Institutional Review Board approval was not required because of the nature of our study, which was a study based on research articles.
Methodology
• retrospective
• cross-sectional study
• performed at one institution
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
ESM 1
(DOCX 29 kb)
Rights and permissions
About this article
Cite this article
Chang, S., Han, K., Suh, Y.J. et al. Quality of science and reporting for radiomics in cardiac magnetic resonance imaging studies: a systematic review. Eur Radiol 32, 4361–4373 (2022). https://doi.org/10.1007/s00330-022-08587-9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00330-022-08587-9