Abstract
Objectives
To develop a proof-of-concept “interpretable” deep learning prototype that justifies aspects of its predictions from a pre-trained hepatic lesion classifier.
Methods
A convolutional neural network (CNN) was engineered and trained to classify six hepatic tumor entities using 494 lesions on multi-phasic MRI, described in Part 1. A subset of each lesion class was labeled with up to four key imaging features per lesion. A post hoc algorithm inferred the presence of these features in a test set of 60 lesions by analyzing activation patterns of the pre-trained CNN model. Feature maps were generated that highlight regions in the original image that correspond to particular features. Additionally, relevance scores were assigned to each identified feature, denoting the relative contribution of a feature to the predicted lesion classification.
Results
The interpretable deep learning system achieved 76.5% positive predictive value and 82.9% sensitivity in identifying the correct radiological features present in each test lesion. The model misclassified 12% of lesions. Incorrect features were found more often in misclassified lesions than correctly identified lesions (60.4% vs. 85.6%). Feature maps were consistent with original image voxels contributing to each imaging feature. Feature relevance scores tended to reflect the most prominent imaging criteria for each class.
Conclusions
This interpretable deep learning system demonstrates proof of principle for illuminating portions of a pre-trained deep neural network’s decision-making, by analyzing inner layers and automatically describing features contributing to predictions.
Key Points
• An interpretable deep learning system prototype can explain aspects of its decision-making by identifying relevant imaging features and showing where these features are found on an image, facilitating clinical translation.
• By providing feedback on the importance of various radiological features in performing differential diagnosis, interpretable deep learning systems have the potential to interface with standardized reporting systems such as LI-RADS, validating ancillary features and improving clinical practicality.
• An interpretable deep learning system could potentially add quantitative data to radiologic reports and serve radiologists with evidence-based decision support.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Deep learning (DL) systems based on convolutional neural networks (CNNs) have shown potential to revolutionize the process of radiological diagnosis [1,2,3]. Unlike other artificial intelligence techniques, CNNs do not need to be taught specific radiological features to learn how to interpret images [4]. A synergistic workflow that combines the experience of radiologists and the computational power of artificial intelligence systems may substantially improve the efficiency and quality of clinical care. Part I of this article series demonstrated a proof-of-concept 3D CNN for the classification of liver lesions on multi-phasic MRI [5]. Although CNNs have demonstrated high performance in diagnostic classification tasks, their “black box” design limits their clinical adoption [6,6,8]. Despite recent advances in interpretable machine learning [9], deep learning models still do not provide information about the factors used in decision-making in a manner that can be understood by radiologists and other physicians, which prevents them from incorporating their results into an informed decision-making process. The inability to explain their reasoning also leads to a lack of safeguards and accountability when they fail. DL systems that demonstrate high accuracy in a more transparent manner are more likely to gain clinical acceptance.
This is especially applicable when incorporating DL into standardized reporting systems such as the Liver Imaging Reporting and Data System (LI-RADS). While LI-RADS has changed the diagnostic workflow of malignant lesions and contributed to higher quality diagnosis and reporting [10,10,12], most studies have demonstrated moderate inter-observer agreement for LI-RADS categories [13,13,14,15,16,17,19]. Recent studies also highlighted issues regarding the application of LI-RADS ancillary features, which are primarily based on a combination of biological plausibility, single-center retrospective studies, and expert opinion with somewhat low level of evidence [20, 21]. For example, the application of such features resulted in an increased number of misclassifications [10, 14, 22] and ancillary features were not seen as a useful tool for assigning definite LR classes [13]. Moreover, the application of a number of ancillary features may be inefficient, as they affected the final diagnosis in at most 10% of cases [13, 19]. The American College of Radiology has called for novel systems to increase the efficiency and accuracy of LI-RADS and to make it more feasible for daily radiology practice [21]. Interpretable DL systems could help to address this gap by automating the validation, detection, and standardized reporting of diagnostic imaging features, providing a way for radiologists to efficiently interact with such tools in a shared decision-making paradigm.
This study investigates an integrative interpretable DL approach for DL systems used in clinical radiology, using techniques for identifying, localizing, and scoring imaging features. In addition to developing a liver lesion classifier for multi-phasic MRI (Part I), the aim of Part II was to develop a proof-of-concept interpretable system that justifies aspects of its decisions through internal analysis of relevant radiologic features.
Materials and methods
Deep learning system and model-agnostic interpretability
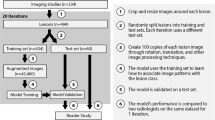
This single-center retrospective study is based on an institutional review board–approved protocol, and the requirement for written consent was waived. The specific methods for patient selection, lesion reference standard, MRI technique, image processing techniques, and DL model are described in Part I of this study [5]. Briefly, a CNN was utilized with three convolutional layers and two fully connected layers, which was capable of differentiating benign cysts, cavernous hemangiomas, focal nodular hyperplasias (FNHs), HCCs, intrahepatic cholangiocarcinomas (ICCs), and colorectal carcinoma (CRC) metastases after being trained on 434 hepatic lesions from these classes. This study was integrated into the Part I DL workflow so that the system could be trained to classify lesion types before incorporating techniques to identify, localize, and score their radiological features (Fig. 1). Specifically, the current study utilized the DL model from Part I which has been trained on a large dataset including 494 lesions. Additionally, custom algorithms were applied to analyze specific hidden layers of this pre-trained neural network in a model-agonistic approach. This method is also known as post hoc analysis (not to be confused with the post hoc analysis in statistics) and is generalizable to various pre-trained machine learning neural networks [23, 24]. Under the taxonomy of established interpretability methods, these algorithms fall under the general category of feature summary statistic. In terms of scope, the methods used describe local interpretability where the focus is on individual predictions, as opposed to global scope where the entire model behaviour is analysed. These selected techniques are especially useful for the purposes of communicating feature information to radiologists. These algorithms are described in detail below.
Radiological feature selection
Fourteen radiological features were selected comprising lesion imaging characteristics that are observable on multi-phasic MRI and are commonly utilized in day-to-day radiological practice for differentiating between various lesion types [25, 26] (Table 1). This includes LI-RADS features for HCC classification, including arterial phase hyperenhancement, washout, and pseudocapsule. Up to 20 hepatic lesions in the training set that best exemplified each feature were selected (Fig. 2). From this sample, ten were randomly selected in each repetition of this study. Imaging features with similar appearances were grouped. A test set of 60 lesions was labeled with the most prominent imaging features in each image (1–4 features per lesion). This test set was the same as that used to conduct the reader study in Part I.
Feature identification with probabilistic inference
For each radiological feature, a subset of ten sample lesions with that feature was passed through the CNN, and the intermediate outputs of the 100 neurons in the fully connected layer were inspected. By analyzing these neuronal outputs among the ten samples, each radiological feature was associated with specific patterns in these neurons. The test image was passed through the CNN to obtain its intermediate outputs, which were compared to the outputs associated with each feature. When the intermediate outputs of a test image are similar to the outputs observed for lesions with a particular feature, then the feature is likely to be present in the test image (see Fig. 3). The intermediate outputs were modeled as a 100-dimensional random variable and the training dataset was used to obtain its empirical distribution (refer to “marginal distributions” and “conditional distributions” in [27]. Using kernel density estimation, the features present in each test image were probabilistically inferred. The neuronal outputs of augmented versions of all images were used to provide more robust estimates of the probability distributions. As described in Part I, image augmentation creates copies of images with stochastic distortions.
CNN model architecture used to infer the lesion entity and radiological features based on the input image, shown for an example of intrahepatic cholangiocarcinoma. Patterns in the convolutional layers are mapped back to the input image to establish feature maps for each identified feature. As well, relevance scores are assigned to the features based on the correspondence between patterns in the convolutional layers, the lesion classification, and the identified features
The CNN system’s performance was assessed by its ability to correctly identify the radiological features in the test set of 60 labeled lesions. Performance was evaluated in 20 iterations with separately trained models using different choices of the ten sample lesions. Positive predictive value (PPV) and sensitivity (Sn) were measured for the entire population (averaged over the total number of features across all lesions). This was performed for each feature individually and for each lesion class.
Feature mapping with weighted activations
After identifying the radiological features observed in an input lesion image, 3D feature maps were derived from the CNN’s layer activations to show where features are observed within each image. For this analysis, the post-activation neuronal outputs of the final convolutional layer were used, which has 128 channels. The original images have 24 × 24 × 12 resolution and pass through padded convolutions and a 2 × 2 × 2 max pooling layer before reaching this layer at 12 × 12 × 6 spatial dimensions. The feature map was constructed for a test image by obtaining this layer’s output and applying a weighted average over the 128 channels using different weights for each of the 1–4 radiological features identified within the image. The resulting 12 × 12 × 6 feature maps were upsampled using trilinear interpolation to correspond to the 24 × 24 × 12 resolution of the original image. The mapping to the three MRI phases cannot be readily traced. The channel weights used for each feature were determined by correlating the channel with at most one of the features based on the channel outputs observed in the sample lesions labeled with the feature.
Feature scoring with influence functions
Among the radiological features identified in an image, some features may be more important for classifying the lesion than others. The contribution of each identified feature to the CNN’s decision was analyzed by impairing the CNN’s ability to learn the specific feature and examining how this impacts the quality of the CNN’s classification. If the feature is not important for classifying the lesion, then the CNN should still make the correct decision, even if it can no longer identify the feature. The CNN’s ability to learn a particular feature can be hampered by removing examples of that feature from its training set. Although repeatedly removing examples and retraining the model is prohibitively time-consuming, Koh et al. developed an approximation of this process that calculates an “influence function” [28]. The influence function of a feature with respect to a particular image estimates how much the probability of the correct lesion classification deteriorates for that image as examples of the feature are removed from the CNN’s training set. Thus, the radiological feature that is most influential for classifying a particular lesion is the feature with the largest influence function for that image. Scores were obtained for each feature by measuring their respective influence functions, then dividing each by the sum of the influences. No ground truth was used for the optimal weighting of radiological features for diagnosing a given image, since a CNN does not “reason” about radiological features in the same way as a radiologist. The definition and further interpretation of the influence function are provided in Supplement 1.
Results
Characteristics of the 296 patients included in this study are described in Part I of this article series. CNN model classification performance is also described in detail in Part I.
Feature identification with probabilistic inference
A total of 224 annotated images were used across the 14 radiological features, and some images were labeled with multiple features. After being presented with a randomly selected subset of 140 out of 224 sample lesions, the model obtained a PPV of 76.5 ± 2.2% and Sn of 82.9 ± 2.6% in identifying the 1–4 correct radiological features for the 60 manually labeled test lesions over 20 iterations (see Table 2).
Among individual features, the model was most successful at identifying relatively simple enhancement patterns. With a mean number of 2.6 labeled features per lesion, the model achieved a precision of 76.5 ± 2.2% with a recall of 82.9 ± 2.6% (see Table 3). It achieved the best performance at identifying arterial phase hyperenhancement (PPV = 91.2%, Sn = 90.3%), hyperenhancing mass on delayed phase (PPV = 93.0%, Sn = 100%), and thin-walled mass (PPV = 86.5%, Sn = 100%). In contrast, the model performed relatively poorly on more complex features, struggling to identify nodularity (PPV = 62.9%, Sn = 60.8%) and infiltrative appearance (PPV = 33.0%, Sn = 45.0%). The CNN also overestimated the frequency of central scars (PPV = 32.0%, Sn = 80.0%), which only appeared once among the 60 test lesions.
The model misclassified lesions with higher frequency when the radiological features were also misclassified. For the 12% of lesions that the model misclassified over 20 iterations, its PPV and Sn were reduced to 56.6% and 63.8%, respectively. Furthermore, the feature that the model predicted with the highest likelihood was only correct in 60.4% of cases—by comparison, the feature that the model predicts with the greatest likelihood in correctly classified lesions was correct 85.6% of the time.
This effect was also observed when the feature identification metrics are grouped by lesion classes, as the model generally identified features most accurately for classes in which the lesion entity itself was classified with high accuracy. The model obtained the highest PPV for benign cyst features at 100% and lowest for CRC metastasis features at 61.2%. The model attained the highest sensitivity for hemangioma features at 96.1% and lowest for HCC features at 64.2%. The lesion classifier performed better on both cysts (Sn = 99.5%, Sp = 99.9%) and hemangiomas (Sn = 93.5%, Sp = 99.9%) relative to HCCs (Sn = 82.0%, Sp = 96.5%) and CRC metastases (Sn = 94.0%, Sp = 95.9%).
Feature mapping with weighted activations
The feature maps (Fig. 4) were consistent with radiological features related to borders: enhancing rim and capsule/pseudocapsule, and a thin wall yield feature maps that trace these structures. Additionally, the model’s feature maps for hypoenhancing and hyperenhancing masses were well localized and consistent with their location in the original image: hypoenhancing core/mass and nodularity had fairly well-defined bounds, as did arterial phase hyperenhancement and hyperenhancing mass in delayed phase. Iso-intensity in venous/delayed phase was also well defined, capable of excluding the hyperenhancing vessels in its map. In contrast, features describing enhancement patterns over time were more diffuse and poorly localized. There was slight misregistration between phases included in the hemangioma example, contributing to artifacts seen in the feature map for nodular peripheral hyperenhancement.
2D slices of the feature maps and relevance scores for examples of lesions from each class with correctly identified features. The color and ordering of the feature maps correspond to the ranking of the feature relevance scores, with the most relevant feature’s map in red. The feature maps are created based on the entire MRI sequence, and do not correspond directly to a single phase. These results are taken from a single iteration
Feature scoring with influence functions
The most relevant radiological feature for cavernous hemangiomas was progressive centripetal filling, with a score of 48.6% compared with 34.0% for hyperenhancing mass on delayed phase and 21.6% for nodular/discontinuous peripheral hyperenhancement. Thin-walled mass was a more relevant feature for classifying benign cysts than hypoenhancing mass (67.1% vs. 46.6%; Table 4). The most relevant feature for correctly classifying FNHs was iso-intensity on venous/delayed phase (79.4%), followed by arterial phase hyperenhancement (65.8%) and central scar (37.4%). The relevance scores for HCC imaging features were 49.5% for capsule/pseudo-capsule, 48.5% for heterogeneous lesion, 40.3% for washout, and 38.4% for arterial phase hyperenhancement. The relevance scores for ICC imaging features were 58.2% for progressive hyperenhancement, 47.3% for heterogeneous lesion, 43.8% for infiltrative appearance, and 37.2% for nodularity. The most relevant imaging feature for correctly classifying CRC metastases was progressive hyperenhancement (67.2%), followed by hypoenhancing core (52.0%) and enhancing rim (46.9%).
Discussion
This study demonstrates the development of a proof-of-concept prototype for the automatic identification, mapping, and scoring of radiological features within a DL system, enabling radiologists to interpret elements of decision-making behind classification decisions. While DL algorithms have the opportunity to markedly enhance the clinical workflow of diagnosis, prognosis, and treatment, transparency is a vital component. Indeed, it is unlikely that clinicians would accept automated diagnostic decision support without some measure of “evidence” to justify predictions. The method of identifying and scoring radiological features allows the algorithm to communicate factors used in making predictions. Radiologists can then quickly validate these features by using feature maps or similar interpretability techniques to check whether the system has accurately identified the lesion’s features in the correct locations.
The CNN was able to identify most radiological features fairly consistently despite being provided with a small sample of lesions per class, in addition to being trained to perform an entirely different task (classifying the lesion entity in Part I). For many simple imaging features such as hyperenhancing or hypoenhancing masses, the model was able to accurately and reliably determine its presence, location, and contribution to the lesion classification. However, it had greater difficulty identifying or localizing features that consist of patterns over multiple phases than patterns that are visible from a single phase or constant across all phases. It struggled in particular on more complex features that may appear quite variable across different lesions such as infiltrative appearance, suggesting that these features are not well understood by the CNN or that more examples of these features need to be provided. By highlighting which radiological features the CNN fails to recognize, this system may provide engineers with a path to identify possible failure modes and fine-tune the model, for example, by training it on more samples with these features.
A general relationship was observed between the model’s misclassification of a lesion entity and its misidentification of radiological features, which could provide researchers and clinicians with the transparency to identify when and how a CNN model fails. If the model predicts non-existent imaging features, clinicians will be aware that the model has likely made a mistake. Moreover, this gives developers an example of a potential failure mode in the model. An interpretable DL system can be utilized as a tool for validation of imaging guidelines, particularly for entities which are uncommon or have evolving imaging criteria, such as bi-phenotypic tumors and ICCs [12, 29, 30]. As shown in the results on feature scoring, the model tends to put greater weight on imaging features that have greater uniqueness and differential diagnostic power in the respective lesion class. An interpretable CNN could be initially presented with a large set of candidate imaging features. Then by selecting the imaging features with the highest relevance score output by the model, one could determine which features are most relevant to members of a given lesion class. This approach also addresses the need for more quantitative evidence-based data in radiology reports.
An interpretable DL system could help to address the large number of ancillary imaging features that are part of the LI-RADS guidelines and similar systems by providing feedback on the importance of various radiological features in performing differential diagnosis. With further refinements, the presented concepts could potentially be used to validate newly proposed ancillary features in terms of frequency of occurrence, by applying it to a large cohort and analyzing the CNN’s predictions. Features that are predicted with low frequency or relevance could be considered for exclusion from LI-RADS guidelines. This could be a first step towards providing a more efficient and clinically practical protocol [13, 19]. An interpretable DL model could also enable the automated implementation of such complex reporting systems as LI-RADS, by determining and reporting standardized descriptions of the radiological features present. By enabling such systems to become widely adopted, there is potential for the reporting burden on radiologists to be alleviated, data quality to improve, and the quality and consistency of patient diagnosis to increase.
Since the present study is designed as a proof-of-concept development, there are multiple limitations that future studies will address. As a single-institution study with limited data availability, a relatively small number of sample lesions was included for each lesion type. This will be remedied by eventually utilizing larger multi-institutional datasets. In addition, while feature extraction could be easily validated with ground truth confirmation by radiological readers, there is intrinsically no existing ground truth criteria for validating feature maps and relevance scores. As a result, more formal validation of these elements will require an aggregate of forthcoming studies that demonstrate reproducibility under different DL models and datasets. Such a system would also need to demonstrate similar functionality using different choices of radiological features and lesion types. Future work will demonstrate this technique on LI-RADS ancillary features, which will require incorporating a more complex CNN model capable of analyzing other types of MRI sequences.
In summary, this study demonstrates a proof-of-concept interpretable deep learning system for clinical radiology. This provides a technique for interrogating relevant portions of an existing CNN, offering rationale for classifications through internal analysis of relevant imaging features. With further refinement and validation, such methods have the potential to eventually provide a cooperative approach for radiologists to interact with deep learning systems, facilitating clinical translation into radiology workflows. Transparency and comprehensibility are key barriers towards the practical integration of deep learning into clinical practice [31]. An interpretable approach can serve as a model for addressing these issues as the medical community works to translate useful aspects of deep learning into clinical practice.
Abbreviations
- CNN:
-
Convolutional neural network
- CRC:
-
Colorectal carcinoma
- DL:
-
Deep learning
- FNH:
-
Focal nodular hyperplasia
- HCC:
-
Hepatocellular carcinoma
- ICC:
-
Intrahepatic cholangiocarcinoma
- LI-RADS:
-
Liver Imaging Reporting and Data System
- PPV:
-
Positive predictive value
- Sn:
-
Sensitivity
References
Rajpurkar P, Irvin J, Zhu K et al (2018) Deep learning for chest radiograph diagnosis: a retrospective comparison of the CheXNeXt algorithm to practicing radiologists. PLoS Med 15:e1002686. https://doi.org/10.1371/journal.pmed.1002686
Chartrand G, Cheng PM, Vorontsov E et al (2017) Deep learning: a primer for radiologists. Radiographics 37:2113–2131
Anthimopoulos M, Christodoulidis S, Ebner L, Christe A, Mougiakakou S (2016) Lung pattern classification for interstitial lung diseases using a deep convolutional neural network. IEEE Trans Med Imaging 35:1207–1216
Greenspan H, Van Ginneken B, Summers RM (2016) Guest editorial deep learning in medical imaging: overview and future promise of an exciting new technique. IEEE Trans Med Imaging 35:1153–1159
Hamm CA, Wang CJ, Savic LJ et al (2019) Deep learning for liver tumor diagnosis part I: development of a convolutional neural network classifier for multi-phasic MRI. Eur Radiol. https://doi.org/10.1007/s00330-019-06205-9
Olden JD, Jackson DA (2002) Illuminating the “black box”: a randomization approach for understanding variable contributions in artificial neural networks. Ecol Model 154:135–150
Kiczales G (1996) Beyond the black box: open implementation. IEEE Softw 13(8):10–11
Dayhoff JE, DeLeo JM (2001) Artificial neural networks: opening the black box. Cancer 91:1615–1635
Olah C, Satyanarayan A, Johnson I et al (2018) The building blocks of interpretability. Distill 3:e10. https://doi.org/10.23915/distill.00010
Corwin MT, Lee AY, Fananapazir G, Loehfelm TW, Sarkar S, Sirlin CB (2018) Nonstandardized terminology to describe focal liver lesions in patients at risk for hepatocellular carcinoma: implications regarding clinical communication. AJR Am J Roentgenol 210:85–90
Mitchell DG, Bruix J, Sherman M, Sirlin CB (2015) LI-RADS (Liver Imaging Reporting and Data System): summary, discussion, and consensus of the LI-RADS Management Working Group and future directions. Hepatology 61:1056–1065
Mitchell DG, Bashir MR, Sirlin CB (2018) Management implications and outcomes of LI-RADS-2, -3, -4, and -M category observations. Abdom Radiol (NY) 43:143–148
Barth B, Donati O, Fischer M et al (2016) Reliability, validity, and reader acceptance of LI-RADS-an in-depth analysis. Acad Radiol 23:1145
Davenport MS, Khalatbari S, Liu PS et al (2014) Repeatability of diagnostic features and scoring systems for hepatocellular carcinoma by using MR imaging. Radiology 272:132
Ehman EC, Behr SC, Umetsu SE et al (2016) Rate of observation and inter-observer agreement for LI-RADS major features at CT and MRI in 184 pathology proven hepatocellular carcinomas. Abdom Radiol (NY) 41:963–969
Zhang YD, Zhu FP, Xu X et al (2016) Classifying CT/MR findings in patients with suspicion of hepatocellular carcinoma: comparison of liver imaging reporting and data system and criteria-free Likert scale reporting models. J Magn Reson Imaging 43:373–383
Bashir M, Huang R, Mayes N et al (2015) Concordance of hypervascular liver nodule characterization between the organ procurement and transplant network and liver imaging reporting and data system classifications. J Magn Reson Imaging 42:305
Liu W, Qin J, Guo R et al (2017) Accuracy of the diagnostic evaluation of hepatocellular carcinoma with LI-RADS. Acta Radiol. https://doi.org/10.1177/0284185117716700:284185117716700
Fowler KJ, Tang A, Santillan C et al (2018) Interreader reliability of LI-RADS version 2014 algorithm and imaging features for diagnosis of hepatocellular carcinoma: a large international multireader study. Radiology 286:173–185
Cruite I, Santillan C, Mamidipalli A, Shah A, Tang A, Sirlin CB (2016) Liver imaging reporting and data system: review of ancillary imaging features. Semin Roentgenol 51:301–307. https://doi.org/10.1053/j.ro.2016.05.004
Sirlin CB, Kielar AZ, Tang A, Bashir MR (2018) LI-RADS: a glimpse into the future. Abdom Radiol (NY) 43:231–236
Kim YY, An C, Kim S, Kim MJ (2017) Diagnostic accuracy of prospective application of the Liver Imaging Reporting and Data System (LI-RADS) in gadoxetate-enhanced MRI. Eur Radiol. https://doi.org/10.1007/s00330-017-5188-y
Molnar C (2019) Interpretable machine learning. A guide for making black box models explainable. https://christophm.github.io/interpretable-ml-book/
Fisher A, Rudin C, Dominici F (2018) Model class reliance: variable importance measures for any machine learning model class, from the “Rashomon” perspective. arXiv preprint arXiv:180101489
Federle MP, Jeffrey RB, Woodward PJ, Borhani A (2009) Diagnostic imaging: abdomen. Published by Amirsys. Lippincott Williams & Wilkins
Victoria C, Sirlin CB, Cui J et al (2018) LI-RADS v2018 CT/MRI Manual. Available via https://www.acr.org/-/media/ACR/Files/Clinical-Resources/LIRADS/Chapter-16-Imaging-features.pdf?la=en
Everitt BS (2002) The Cambridge dictionary of statistics. Cambridge University Press
Koh PW, Liang P (2017) Understanding black-box predictions via influence functions. AarXiv preprint arXiv:170304730
Narsinh KH, Cui J, Papadatos D, Sirlin CB, Santillan CS (2018) Hepatocarcinogenesis and LI-RADS. Abdom Radiol (NY) 43:158–168
Tang A, Bashir MR, Corwin MT et al (2018) Evidence supporting LI-RADS major features for CT- and MR imaging-based diagnosis of hepatocellular carcinoma: a systematic review. Radiology 286:29–48
Holzinger A, Biemann C, Pattichis CS, Kell DB (2017) What do we need to build explainable AI systems for the medical domain? arXiv preprint arXiv:171209923
Funding
BL and CW received funding from the Radiological Society of North America (RSNA Research Resident Grant No. RR1731). JD, JC, ML, and CW received funding from the National Institutes of Health (NIH/NCI R01 CA206180).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Guarantor
The scientific guarantor of this publication is Julius Chapiro.
Conflict of interest
The authors of this manuscript declare relationships with the following companies: JW: Bracco Diagnostics, Siemens AG; ML: Pro Medicus Limited; JC: Koninklijke Philips, Guerbet SA, Eisai Co.
Statistics and biometry
One of the authors has significant statistical expertise.
Informed consent
Written informed consent was waived by the Institutional Review Board.
Ethical approval
Institutional Review Board approval was obtained.
Methodology
• retrospective
• experimental
• performed at one institution
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
ESM 1
(PDF 28.6 kb)
Rights and permissions
About this article
Cite this article
Wang, C.J., Hamm, C.A., Savic, L.J. et al. Deep learning for liver tumor diagnosis part II: convolutional neural network interpretation using radiologic imaging features. Eur Radiol 29, 3348–3357 (2019). https://doi.org/10.1007/s00330-019-06214-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00330-019-06214-8