Abstract
Pigments produced by micro-organisms could contribute to their pathogenesis and resistance. The investigation into the red pigment of R. mucilaginosa and its ability to survive and resist has not yet been explored. This study aimed to investigate the survival and resistance of the R. mucilaginosa CQMU1 strain following inhibition of pigment production by naftifine and its underlying mechanism. The red-pigmented Rhodotorula mucilaginosa CQMU1 yeast was isolated from an infected toenail of a patient with onychomycosis. Cultivation of R. mucilaginosa in liquid and solid medium showed the effect of naftifine after treatment. Then, analysis of phagocytosis and tolerance to heat or chemicals of R. mucilaginosa was used to evaluate the survival and resistance of yeast to different treatments. Naftifine reversibly inhibited the pigmentation of R. mucilaginosa CQMU1 in solid and liquid media. Depigmented R. mucilaginosa CQMU1 showed increased susceptibility toward murine macrophage cells RAW264.7 and reduced resistance toward different types of chemicals, such as 1.5-M NaCl and 0.5% Congo red. Inhibition of pigment production by naftifine affected the survival and growth of R. mucilaginosa and its resistance to heat and certain chemicals. The results obtained could further elucidate the target of new mycosis treatment.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The genus Rhodotorula includes a heterogeneous group of fungi with diverse beneficial traits, such as resistance to heavy metals and oxidative stress, and the production of enzymes and carotenoids [1]. Traditionally, they are common nonvirulent environmental inhabitants. Recently, studies reported the relationship of Rhodotorula fungi to meningitis, endocarditis, ventriculitis, and skin infections. Thus, Rhodotorula species could refer as emerging opportunistic pathogens [2, 3]. Moreover, several cases of mixed infection caused by Rhodotorula mucilaginosa were reported due to importance of antifungal drugs studies [4, 5]. Remarkably, microbial pigments reportedly demonstrate vital biological functions, such as exertion of antioxidant effects, enhancement of immune responses, and participation in preventing the development of cardiovascular diseases [6, 7]. However, certain bacterial pigments can impair neutrophil killing and promotes virulence and pathogenesis [8, 9].
Rhodotorula mucilaginosa is a basidiomycetous yeast that produces a red carotenoid pigment [7]. The levels of the red pigments of R. mucilaginosa could be changeable in response to adverse conditions, such as UV exposure [10]. However, the effects and functions of the pigment on the fungi survival and resistance toward oxidative stress or osmotic stress remain unclear. Naftifine is a synthetic allylamine antifungal agent for topical treatment of tinea infection. Recently, naftifine has been shown to block the biosynthesis of carotenoid pigment of Staphylococcus aureus through competitively inhibiting 4,4′-diapophytoene desaturase (CrtN) enzyme and then resulting in depigmentation, attenuated virulence, and increased susceptibility to innate immune clearance [9]. Whether the naftifine can reduce the pigmentation of R. mucilaginosa and how the resistance of the depigmented R. mucilaginosa changes remain largely unknown.
In 2018, we isolated a salmon/red pigment-producing yeast, the R. mucilaginosa CQMU1 strain, using samples obtained from a healthy patient presenting with a skin infection [5]. The R. mucilaginosa CQMU1 strain and squalene monooxygenase (SQLE) gene of the isolate were identified using molecular method and the sequences of the isolate were deposited on the GenBank of National Center for Biotechnology Information (NCBI). In the study, we first determined the effect of naftifine on the pigmentation of R. mucilaginosa CQMU1. Then we investigated the growth, survival, and resistance changes of the yeasts after naftifine-mediated depigmentation. The results obtained in this study is a preliminary work to enlighten the possible mechanism of R. mucilaginosa resistance and survival rate and it is a need to further explore the relationship between yeast pigment and the survival rates of R. mucilaginosa for medical purposes.
Materials and Methods
Media Culture and Strains
The R. mucilaginosa CQMU1 yeast strain was isolated using samples obtained from a patient with toenail onychomycosis at Chongqing Medical University First Affiliated Hospital, Chongqing, China [5]. Sabouraud dextrose agar (SDA) and yeast peptone dextrose (YPD) (Oxoid Ltd., Hampshire, UK) were the cultivation media. Antifungal drugs, naftifine, butenafine, and bifonazole-treated fungal cells (Sigma-Aldrich, St. Louis, MO, USA). Chemicals used for conducting survival and resistance tests included 1% methylene blue (Oxoid Ltd.), 1.5-M NaCl and 1.5-M KCl (Sichuan Easteng Test Co., Ltd., Sichuan, China), 1 mM H2O2, and 0.5% Congo red (Oxoid Ltd.). The yeast cells were incubated in a fungal and bacterial incubator MDT-60R (Zhengzhou Instrument Co., Ltd., Zhengzhou, China). No ethical approval was required as the research in this article related to micro-organisms.
Molecular Identification of R. mucilaginosa CQMU1 Yeast Strain and Squalene Monooxygenase (SQLE) Gene
The total RNA of R. mucilaginosa CQMU1 strain which was obtained from a healthy patient presenting with a skin infection [5] was extracted using RNA extraction kit (YiShan Biotech, China) followed manufacturer’s protocol. The Internal Transcribed Spacer (ITS) and 18S rRNA of the isolated sample were amplified by PCR using universal primers of ITS1-5.8-ITS2, Small Subunit (SSU) small subunit ribosomal RNA (18S rRNA), and Large Subunit (LSU) regions as listed in supplementary Table 1. Then PCR amplification for ITS1-5.8-ITS2 and LSU regions were carried out in a thermal cycler under the following conditions: one round of denaturation at 95 °C for 120 s followed by 30 cycles of denaturation at 95 °C for 30 s, annealing at 55 °C for 30 s, and extension at 72 °C for 1 min, with a final extension step at 72 °C for 10 min. Meanwhile for 18S rRNA region, one round of denaturation at 94 °C for 5 min followed by 30 cycles of denaturation at 94 °C for 20 s, annealing at 59 °C for 20 s, and extension at 72 °C for 50 s, with a final extension step at 72 °C for 1 min. All sequences were embedded on NCBI GenBank as followed: Rhodotorula mucilaginosa CQ.1 ITS (MK080558), Rhodotorula mucilaginosa CQ.2 18 s (MK084474), and Rhodotorula mucilaginosa CQ.3 LSU (MT782268).
Moreover, the whole RNA sequence of R. mucilaginosa CQMU1 strain was sequenced using Illumina HiSeqTM2500 by Gene Denovo Biotechnology Co. The RNA-seq data were deposited in the National Center for Biotechnology Information (NCBI) Short Read Archive (SRA) database with the accession number: PRJNA590855. Sample name is Rh_ctrl and the accession number is Accession: SAMN13341477, ID: 13341477. Hence from the sequence of protein-coding marker genes such as ACT gene, it can be retrieved in RNA-seq data SRA: SRS5713385. In addition, the sequence is highly homologous (95.92%) with R. mucilaginosa strain 2LJJ1 actin (ACT) gene (MK879551.1). For beta-tubulin gene (TUB2) gene, the sequence can be retrieved from SRR10525677.28441806.1 and SRR10525677.28441806.2 and 100% homologous with sequence of R. mucilaginosa strain KR (PUHQ01000037). All PCR products were checked using 1.2% gel electrophoresis before submitted for sequencing. The cDNA strands was amplified as template using the SQLE gene primer retrieved from reference gene conserved motifs and nucleotide sequences among the SQLEs included XM_018414785 (Rhodotorula graminis) and the primer is listed in supplementary Table 1.
Macrophage Cell Maintenance and Culture
The murine macrophage (RAW264.7) cell line was provided by the Basic Medical College of Chongqing Medical University and was used for conducting phagocytosis assays. The Dulbecco’s modified Eagle medium (DMEM, Gibco, Grand Island, NY, USA) supplemented with 10% heat-inactivated fetal calf serum (Gibco) was used to maintain the cell line and were incubated at 37 °C in a 0.5% CO2 incubator (Zhengzhou Instrument Co., Ltd.). The cells were seeded at a density of 1 × 105–3.0 × 105 cells/well in a volume of 100 µL in 96-well plates (triplicates) for 16 h prior to the conduction of subsequent experiments. The culture media was changed every 24 h and the cells condition was examined. Routine culture maintenance was performed for two weeks until the cells reached the almost 10% of the cell confluence.
Survival and Resistance Rate Test
Yeast Strain and Culture Conditions
The yeast cells were pre-cultivated for 24 h in SDA agar medium in a Petri dish and were then further cultivated in 200-mL Erlenmeyer flasks in 50 mL SDA liquid medium at an initial optical density of 0.1 (at 660 nm). Incubation was performed at 28 ℃. Antifungal drugs were either added directly to liquid YPD medium or added to YPD plates at concentrations 0.2, 1.2 and 5.0 mg/mL.
Macrophage Phagocytosis Assay
Experiments for pigmentation inhibition of R. mucilaginosa using the antifungal drug naftifine were conducted prior to the tests mentioned in Supplementary data 1A and B. RAW264.7 macrophage cells were seeded at a density of 1 × 105–3.0 × 105 cells/well in a volume of 100 µL in 96-well plates for 16 h prior to the conduction of subsequent experiments. The cells were supplemented with DMEM and then incubated at 37 °C in a 0.5% CO2 incubator (Zhengzhou Instrument Co., Ltd.). The macrophages survival experiments were performed in quintuplets. R. mucilaginosa was inoculated into 5-mL SDA liquid medium in a 50-mL tube and incubated at 28 °C (subjected to shaking at 150 rpm) for 7 days to achieve log-phase growth. Yeast cells were categorized into the wild-type group (WT), SQLE-inhibited group (D), and naftifine-reversible induced (R) group and were added to the media (used for conducting infection assays) on macrophage monolayers at the desired density. 10 μl of R. mucilaginosa was added into 990-μl Phosphate-Buffered Saline (PBS) (pH 7.4) and the number of yeast cells were counted using a haemocytometer. Fluorescein isothiocyanate (FITC) (Sigma-Aldrich, St. Louis, MO, USA) was dissolved in DMSO (1 mg/ml) prior usage to stain R. mucilaginosa cells during phagocytosis assay. 50 μl of FITC solution was added into the suspension of R. mucilaginosa (1 × 108 CFU) for every 1 ml. The mixture solution was kept in dark by wrapping it using tin foils and incubating at 30 °C and then shaken at 200 rpm for 4–6 h. The solution was washed with PBS for 3 times and kept at 4 °C. RAW264.7 cell was co-cultured with FITC stained R. mucilaginosa and the culture was incubated for 1.5 h. For observation under confocal laser scanning microscope, the culture medium was discarded and cell culture was gently washed using 1 × PBS for 3 times. 4% polyacetal solution was used for cell fixation for 30 min. 0.5% Triton-100 was added into in cells at room temperature for 30 min. Co-cultured cells were gently washed using 1 × PBS for 3 times and were stained with DAPI at room temperature for 15 min. RAW264.7 cell co-cultured with R. mucilaginosa were observed under confocal laser scanning microscope.
Infected monolayers were incubated at 37 °C in a 0.5% CO2 incubator for 3 h. The cells were visualized using an inverted microscope DMIRE2 inverted microscope (Leica Microsystems) at 400 × magnification and images were acquired to determine phagocytosis. The yeast cells were evaluated every hour for a duration of 24 h (yeast cells engulfed by macrophages and yeast cells not subjected to engulfment or lysis). The cells were harvested from each well to conduct plating and to calculate the colony-forming units, with the result expressed as colony-forming units per milliliter (CFU/mL). Infected macrophages remained viable for several days, and samples were analyzed approximately every 6, 12, and 24 h. Next, 20-μL lysates were transferred to a separate tube and used for conducting the CFU plating test at different temperatures (22 °C, 28 °C, and 32 °C) for thermotolerance test. Thermotolerance is the transient ability of yeast cells to survive at higher temperatures. Survival data obtained from the analysis of the number of surviving yeast cells were statistically analyzed between paired groups using the log-rank test with the PRISM version 8.0 software (GraphPad, Inc., San Diego, CA, USA) and compared with the non-phagocytosis group (P < 0.005 was considered significant).
Tenfold dilution of the lysates was prepared from the total volume of yeast medium (5 mL) obtained, and the number of fungal colonies was determined for each dilution. The dilution factor was calculated for yeast cells per total medium volume (10−4, 10−3, 10−2, and 10−1 cells/mL). The fungal cells were obtained after macrophage cell lysis and were centrifuged to remove the liquid medium. The cell lysate was subjected to washing steps by pipetting twice or thrice in sterile, fresh phosphate-buffered saline solution (Oxoid Ltd.). The lysates were transferred into new 96-well plates for conducting serial dilutions. Duplicate well contents were combined in a final volume of 200-µL serial dilutions of the lysates (1:10) were prepared in a 96-well plate by adding 25-µL lysate to 225-µL macrophage cells. Next, 10 μL of each dilution was added to YPD agar plates prepared using the standard drop-plating technique [11]. The plates were wrapped with parafilm and incubated in a humidified chamber at 22 °C, 28 °C, and 32 °C for 3 days. Yeast colonies were counted manually using plates displaying 1–100 distinct colonies. CFU calculations were performed using MS Excel. The average duplicate or triplicate dilutions that had been plated were calculated. Colonies grown on YPD agar supplemented with naftifine were compared with the group that was not treated with naftifine and the results have been presented using a graph.
Survival and Resistance Test Using Different Types of Chemicals
YPD agar media were prepared and supplemented with 1.5-M NaCl, 1.5-M KCl, 1-mM H2O2, 0.5% Congo red, or 1% methylene blue. Next, 10 mL of the R. mucilaginosa yeast suspension was obtained after lysis of macrophage cells, and 10 μL was spotted on supplemented YPD agar and subjected to incubation at 28 ℃ for 2 days until the colonies were visible; 100 viable colonies on each plate were counted. Resistance to different types of chemicals was evaluated. Quantitative data for the CFU of R. mucilaginosa and Pichia pastoris (as a control) yeast cells were statistically evaluated using the GraphPad Prism software and the complete details of the method is presented in the supplementary data. The survival of yeast cells following treatment with naftifine was assessed as per methods presented in the supplementary data.
Assessment of SQLE mRNA Expression Level by qRT-PCR
The cultured R. mucilaginosa wild-type (WT), SQLE-inhibited (D), and naftifine-reversible-induced (R) group cells were lysed by subjecting cell suspensions to liquid nitrogen treatments using a mortar and pestle to break the cell wall. Total RNA of the isolated strains was extracted using an RNA extraction kit (YiShan Biotech, Guangdong, China) as per the manufacturer’s instructions. The quantity and quality of the RNA samples were measured using the NanoDrop 2000 Spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA) and evaluated by electrophoresis on a 1.2% (w/v) agarose (XHLY Ltd., Beijing, China) 20% (v/v) formaldehyde gel, respectively. First-strand cDNA was synthesized using the PrimeScript 1st-strand cDNA synthesis kit (TaKaRa Biotechnology, Shiga, Japan). The cDNA was stored at – 80 °C until analysis. The mRNA level was evaluated by qRT-PCR using the primers listed in Supplementary Table 1. mRNA expression levels were normalized to that of GAPDH, and qRT-PCR was performed for 30 cycles using the CFX96 Real-Time PCR Detection system (Bio-Rad, Hercules, CA, USA). The following thermal conditions were used for PCR: denaturation at 95 °C for 20 s, primer annealing at 55 °C for 20 s, and extension at 95 °C for 5 s. The final cycle included extension for 10 min at 72 °C.
Protein Assessment by SDS-PAGE and Western Blotting
Protein from naftifine-treated R. mucilaginosa was used to analyze the SQLE protein expression level by SDS–polyacrylamide gel electrophoresis (SDS-PAGE) and western blotting. The protein was extracted from the yeast culture medium by centrifugation, and 70 μL of cold acetone was added to the pellet for incubation at – 20 °C for 3 h. The mixture was centrifuged, and the pellet was subjected to washing steps using phosphate-buffered saline three times; then, the washed pellet sample was mixed with 10 × loading buffer and heated at 100 °C for 2 min. The protein sample was stored at – 20 °C until use. SDS-PAGE was conducted according to the method described by Laemmli (1970). Aliquots of the lysates were subjected to boiling for 2 min, after which they were electrophoresed on 10% SDS-PAGE gels (EpiZyme, Inc., Cambridge, UK) and transferred onto a polyvinylidene fluoride membrane (Merck Millipore, Billerica, MA, USA). The membrane was blocked using nonfat dried milk diluted in Phosphate-Buffered Saline with Tween 20 (PBST) buffer at room temperature for 2 h and then subjected to probing using primary monoclonal mouse antibodies. Antibodies against SQLE and GAPDH (control) (Santa Cruz Biotechnology, Dallas, TX, USA) with 1:10,000 dilution were used for protein detection. The secondary goat-anti-mouse IgG (H + L) antibody (Santa Cruz Biotechnology, Dallas, TX, USA) was added (1:10,000 dilution) to detect the primary antibody. Signals were detected using an enhanced chemiluminescence reagent (Promega, Madison, WI, USA), followed by detection of the probed proteins using a gel imaging machine, i.e., the ChemiDoc™ MP Imaging System (170-8280) (Bio-Rad).
Statistical Analysis
The statistical significance of differences in the percentage of phagocytosis levels and CFU of R. mucilaginosa in all treatment groups was determined by performing one-way and two-way analysis of variance using the SPSS version 25.0 software (SPSS, Inc., Chicago, IL, USA) and GraphPad Prism version 8.0. All experiments were conducted triplicates and all data were analyzed using Student’s t test. n.s., non-significant; *P < 0.05; **P < 0.01; and ***P < 0.001.
Results
Reversible Inhibition of Pigmentation of R. mucilaginosa by naftifine
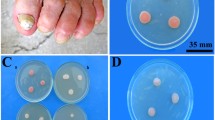
The minimum concentration necessary to induce depigmentation in R. mucilaginosa was 50 g/L (0.05 mg) in YPD liquid medium (Supplementary Fig. S1). Two classes of antifungal drugs, allylamine (naftifine and butenafine) and azole (bifonazole), were used against R. mucilaginosa. All drugs inhibited fungal growth at high concentrations (1.2 and 5.0 mg/mL); however, only naftifine inhibited the production of red pigment at the minimum concentration (0.2 mg/mL) without inhibiting yeast growth (Fig. 1A). There were no pigment color changes in the colonies grown on SDA supplemented with butenafine, and no colonies grew on bifonazole-supplemented SDA. Naftifine treatment exerted a reversible effect on carotenoid pigmentation, as transfer of depigmented cells onto new SDA restored pigmentation (Fig. 1B, a–c). Based on the above results, we used naftifine at a concentration of 0.2 mg/mL for subsequent tests.
Reversible inhibition of pigmentation of R. mucilaginosa by naftifine. Antifungal drug test for pigment inhibition. A Naftifine inhibition effect. Determination of squalene epoxidase inhibition using antifungal drugs: naftifine, butenafine, and bifonazole. (a) SDA supplemented with naftifine; (b) butenafine; and (c) bifonazole at 0.2, 1.0, and 5.0 mg/ml. B Reversible effect of naftifine on R. mucilaginosa. SDA medium; SDA supplemented with 0.2-mg/mL naftifine showed depigment; transferred depigment R. mucilaginosa recovered on SDA medium. (a) SDA medium; (b) SDA supplemented with 0.2-mg/mL naftifine showed depigment; (c) Transferred depigment R. mucilaginosa recovered on SDA medium. The figures shown are representative example of several experiments which was conducted in triplicates and repeated for three times
Survival Evaluation of R. mucilaginosa by Phagocytosis Assay
Observation of R. mucilaginosa engulfed by RAW264.7 cells under laser confocal fluorescence microscopy was made as shown in Fig. 2. There were total 17% of yeast cells of R. mucilaginosa was engulfed WT, while there were 45% yeast cells of D group and 20% of R group, which means D group was more susceptible toward macrophage cell and have less survival rate. Phagocytosis assay was conducted to evaluate the survival rate of R. mucilaginosa. The representative stage of phagocytosis is shown in Fig. 3A. The percentage rate of yeast engulfed by macrophages during phagocytosis was recorded each hour for a duration of 24 h (Fig. 3C). R. mucilaginosa uptake by macrophage cells was highest in the depigmentation group D compared to that observed in the untreated group. The number of yeast cells subjected to uptake by macrophages after 30 min of yeast infection (0 h) was the highest for group D (40%), followed by groups R (30%) and WT (20%). This finding indicated that group D was more susceptible to the phagocytosis of macrophages.
Survival evaluation of R. mucilaginosa by phagocytosis assay. Observation of R. mucilaginosa engulfed by RAW264.7 cells under laser confocal fluorescence microscopy × 1000, showing green fluorescent spots in the RAW264.7 cells after 1.5-h incubation. Red: RAW264.7 cells stained with PKH 26; green: R. mucilaginosa stained with FITC; blue: RAW264.7 cell nucleus stained with DAPI). A Wild-type (WT) R. mucilaginosa without naftifine treatment, B Depigment (D) R. mucilaginosa after treated with naftifine, C Repigment (R) R. mucilaginosa turned into red color after re-cultured the D group on new fresh agar medium (Color figure online)
A Typical field view of phagocytosis assay and the black arrows point the yeast cells. (a) control group; macrophage cells without R. mucilaginosa infected; (b) migration of phagocytes to R. mucilaginosa cells; (c) recognition of yeast cells surface; (d) engulfment of R. mucilaginosa that bound to the phagocyte cell membrane, (e) processing of engulfed cells within maturing phagosomes and digestion of the ingested particle; and (f) other macrophage recruited to the ruptured sites to ingest the released yeasts. B Comparison of yeast uptake by macrophage cells between groups: (a) untreated group named as wild-type (WT); (b) depigmentation group (D) treated by naftifine; and (c) repigmented group after naftifine treatment (R). Group D showed the highest uptake of yeast engulfed by macrophage cells and the lowest number was WT group. C Percentage of phagocytosis rate showed the number of R. mucilaginosa yeast cells engulfed by the macrophage cells in 24 h of group WT, D, and group R. Percentage of phagocytosis rate showed the number of R. mucilaginosa yeast cells engulfed by the macrophage cells in 24 h WT, D, and R. The result showed yeast uptake of depigment group is the highest at time interval 0, 1, 2, 3, 5, 7, 9, and 12–24 h. At the starting point, after 30 min of yeast infection, the WT groups show rate of yeast engulfment is more than 20%, while D group is more than 40% and R is 30%. Macrophage cells have Phagocytic clearance of R. mucilaginosa yeast cells by macrophages cells was completed after 13 h for WT and R groups while it took 14 h for D group. There are significance different between WT, D, and R groups with P value P < 0.0001. All experiments were conducted triplicates and all data were analyzed using Student’s t test on GraphPad Prism software (Color figure online)
Hence, the phagocytosis rate was rapid and vigorous in the depigmented group compared with that observed in the other groups could be due to the loss of pigmentation after naftifine treatment. Student’s t test on GraphPad Prism software revealed significant differences between the groups (P < 0.0001).
Survival and Resistance Evaluation by Determining Thermotolerance and Using Chemicals and Antifungal Drugs
Following the conduction of the phagocytosis assay, we performed survival and resistance tests by subjecting yeast cells extracted from lysed macrophages to different challenges. Evaluation of the CFU on the agar plates showed that group D had the lowest CFU count compared with the WT and R groups on YPD agar cultured at different temperatures (22 °C, 28 °C, and 32 °C) in the thermotolerance test (Fig. 4A). Furthermore, group D cells showed the lowest CFU among the three groups (Fig. 4B) after chemical treatment (Fig. 4B). Those results showed Group D was more sensitive to osmotic stress (NaCl and KCl) and a cell wall stress inducer (Congo red), but less to oxidative stress agent (H2O2). Further, we supplemented the YPD agar with different concentrations of naftifine and cultured the R. mucilaginosa cells at different temperatures. As shown in Fig. 4A, R. mucilaginosa is more suitable to grow at 28°C and the statistical data in Fig. 4C show the significant different values between the groups. The results revealed the lowest count of viable yeast cells for group D in every chemical test with P < 0.005 (0.5% Congo red: 0.0116, 1% methylene blue: 0.0200, 1.5-M NaCl: 0.0261), which indicated lowest resistance levels compared to WT and R groups after naftifine-mediated inhibition of pigment production (Fig. 4B, C). Statistical analysis was performed to determine the survival rate of R. mucilaginosa as method provided in supplementary data.
Survival and resistance evaluation by determining thermotolerance and using chemicals and antifungal drugs. A Survival and resistance test by thermotolerance and chemicals. A Morphology of colony-forming unit (CFU) of survived R. mucilaginosa on YPD agar were grown in different temperatures; 22 ℃, 28 ℃, and 32 ℃ for thermotolerance test, while B shows the morphology of CFU on different chemical tests. Chemical resistance test was conducted using 0.5% of Congo red as cell wall stressor agent, while osmotic stress response was assessed with 1.5 M of NaCl and KCl. CFU of R. mucilaginosa was counted and Group D is sensitive to osmotic, oxidative, and cell wall stress conditions. Furthermore, oxidative stress tolerance was tested with 1-mM H2O2 and 1% of methylene blue were used to test the yeast cell resistance. C shows two-way ANOVA test was performed to evaluate the significance difference between the groups. Statistical value of resistance test on different types of chemicals. CFU of R. mucilaginosa yeast was evaluated by statistical analysis using Student’s t test. Depigmented (D) group showed the lowest survival rate toward chemical and thermotolerance test. 28 ℃ is the most suitable temperature for R. mucilaginosa to grow. For test using 0.5% Congo red, there is a significant difference between WT and D groups (P = 0.0116) and D and R (P = 0.0328). For 1% methylene blue test, there is a significant difference between WT and D groups (P = 0.0200) and also significant difference for test 1.5-M NaCl for WT and D (P = 0.0261) (Color figure online)
Evaluation of SQLE Gene and Protein Expression Levels
The reported target of naftifine was squalene monooxygenase gene (SQLE). Therefore, we evaluated the changes of SQLE of the three groups (WT, D, and R). RNA-seq data showed the SQLE changed insignificantly among three groups (Supplementary data S3). The SQLE RNA level showed no significant difference among groups by qRT-PCR, although the expression levels were slightly increased after naftifine treatment (P = 0.3559) (Fig. 5A). Protein expression levels by SDS-PAGE and western blotting revealed no significant changes in expression of the 60-kDa protein upon naftifine treatment at the minimum concentration of 0.2 mg/mL (Fig. 5B and C). We further examined the protein expression of SQLE at higher drug concentrations (5, 10, 15, 20 mg/mL). The decreased expression of SQLE at higher drug concentrations indicated that the effects of naftifine were dose dependent. Further we investigated the effect of SQLE on survival and resistance by over-expressed SQLE in P. pastoris strain GS115 as showed above (supplementary data). Colonies grew on agar plates, with the SQLE-pPIC9K (SQLE-expressing) group showing the highest CFU compared to that observed in P. pastoris WT cells and P. pastoris cells without the recombinant plasmid were induced by alcohol (+ alcohol) (Fig. 5D).
Evaluation of SQLE gene and protein expression levels. Evaluation of squalene epoxidase gene expression level (qRT-PCR, SDS-PAGE, and WB results). A No significant difference of relative SQLE mRNA level: P-value = 0.3559; B No significance difference for the SQLE protein expression level (60 kDa) for SDS-PAGE and C Western Blotting result shows there is no significant difference at 0.2 mg/mL of naftifine in WT, D, and R groups but protein level was decreased when higher concentrations were supplemented at 5, 10, 15, and 20 mg/mL. D Groups named as Pichia pastoris wild type without alcohol induction (WT), P. pastoris without recombinant plasmid was induced by alcohol (+ alcohol) and SQLE-pPIC9K over-expressed (SQLE expressed). SQLE gene was over-expressed in Pichia pastoris and the survival rates was measured by counting the CFU/plates and it showed the SQLE over-expressed group has the highest CFU in comparison to other groups
Discussion
R. mucilaginosa is an emerging opportunistic pathogenic yeast that produces a red carotenoid pigment [12, 13]. However, few studies addressed the effect of pigmentation on its survival and resistance. This study presents the survival and resistance of R. mucilaginosa before and after naftifine treatment. We showed that naftifine reversibly inhibited pigmentation process, and the loss of red pigment resulted in reduced survival and resistance of R. mucilaginosa toward phagocytosis, chemical treatment, and temperature. Furthermore, over-expression of SQLE gene resulted in increased survival and resistance of the yeast cells. Those results demonstrated that pigmentation of R. mucilaginosa is important for fungi survival and resistance, which potentially points out the target of fungal therapy.
Firstly, naftifine could reduce the carotenoid pigment production of the R. mucilaginosa at a 0.02 mg/mL concentration without apparent inhibition on its growth. The depigmentation process was reversible. The above results are consistent with Mot et al.’s previous report [14]. Naftifine was first reported to block the biosynthesis of the carotenoid pigment in S. aureus through competitive inhibition of its diapophytoene desaturase (CrtN), resulting in increased sensitivity of S. aureus to ROS and decreased mortality in a murine sepsis model [9]. Further, naftifine derivatives with improved potency for CrtN inhibition and reduced S. aureus bacterial burdens during systemic infection in mice were developed [15]. The CrtN enzyme is located downstream of SQLE in the carotenogenesis pathway [9, 16]. In fungi, SQLE is involved in ergosterol biosynthesis and its inhibitor, terbinafine or naftifine, is approved for the treatment of specific fungal infections. Carotenoid pigment biosynthesis is located downstream of ergosterol biosynthesis pathway. And the previous report showed that the blockade of pigment synthesis has caused the infliction and injury of the fungal cell wall [17]. Thus, the naftifine might target the two periods of the biosynthesis pathway [9, 16], which may expand the therapy of fungi.
As for fungi, carotenoid pigments are linked to pathogenesis and resistance. Research already showed that pigment-deficient fungi demonstrated attenuated virulence because of their reduced ability to block immune recognition [18]. The green fungal conidial pigment dihydroxynaphthalene-melanin (DHN-melanin) of Aspergillus fumigatus was able to hinder phagocytosis and conidium binding to host proteins, such as fibronectin [19, 20]. In the study, we found reduced survival of R. mucilaginosa in macrophage cells and decreased resistance of R. mucilaginosa after pigment inhibition by naftifine. This reflected the decreased virulence for the depigmented R. mucilaginosa. Phagocytic clearance of fungal pathogens occurs through the recognition of cell-surface fungal-specific molecules called pathogen associated molecular patterns (PAMPs). Different species of fungi cell wall composition will be recognized specifically by the phagocytes. Meanwhile, ergosterol has been recognized as an immunologically active molecule that acts as a direct trigger of macrophage pyroptosis [21]. Thus, inhibition of SQLE could increase the susceptibility of R. mucilaginosa to be engulfed by macrophage cells. Also, this phenomenon justified the presence of pigment benefited survival and resistance rate of the fungi.
Besides, carotenogenesis has evolved as a cellular defense mechanism against oxidative damage [22]. Kejzar et al. showed that the carotenoid produced by black fungus Hortaea werneckii plays an important role in their survival and resistance at extreme conditions, such as the high NaCl concentrations [23]. For H2O2 oxidative stress test, surprisingly, existence of red pigment had no significant effect on R. mucilaginosa, which is parallel to the black fungus H. werneckii [23]. However, Schroeder et al. have ever reported the carotenoids of R. mucilaginosa against oxidative stress [22]. This could be explained by different components of carotenoids produced by R. mucilaginosa, especially in different conditions. Further study is needed to address the issue.
Moreover, we found that naftifine indeed induced the reduction of SQLE protein levels at higher concentrations. Although no significant reduction of SQLE mRNA level after naftifine treatment was detected in our study, the SQLE protein variants exhibited reduced enzymatic activities and induced SQLE expression at the transcription level [24]. The occurrence of feedback induction for SQLE expression could explain partially the effect of naftifine on SQLE, as showed in a previous study [25, 26]. The SQLE gene expression is regulated in a coordinated manner via blockade at multiple stages in the sterol biosynthesis pathway [26, 27]. Thus, effects of naftifine could reduce SQLE at both transcriptional and protein levels, resulting in decreased survival and resistance.
One limitation of the study is that the SQLE reduction is induced by naftifine rather than deletion of the gene, and SQLE is over-expressed in the P. pastoris strain GS115. We attempted to knockout the SQLE gene by performing several experiments using the CRISPR/Cas9 system or gene gun methods; however, no positive SQLE-deficient clones were obtained. SQLE catalyzes the epoxidation of squalene to yield 2,3-oxidosqualene, the key step of sterol biosynthesis pathways in fungi, and increases the content of squalene, leading to the infliction of injury to the cell wall and membrane. As an essential role played by SQLE in fungal metabolism, the fungi likely cannot grow without SQLE gene. However, despite this limitation, reduction of the R. mucilaginosa survival rate following naftifine-mediated inhibition confirms the critical role of SQLE in the process of ergosterol synthesis. Further study of the mechanisms involve in the carotenoids production and the yeast cells survival is suggested. Moreover, some studies have proven that melanin also can provide protection to extremes in temperature, both heat and cold for some fungi such as Cryptococcus neoformans [28] and Exophiala (Wangiella) dermatitidis [29]. Recent study also reported the role of melanin which acts as virulence factor for some fungi species and this knowledge is critical in understanding fungal cell structure and pathogenesis [30].
Conclusion
Thus, we found reversible inhibition of pigmentation by naftifine to R. mucilaginosa, and Depigmented R. mucilaginosa, showed decreased survival and growth to adverse conditions. Inhibition of R. mucilaginosa pigment production by naftifine affected the survival and resistance rate toward heat and certain chemicals. The results obtained could further elucidate the target of new mycosis treatment targeting on squalene monooxygenase (SQLE) gene. Besides, we may utilize R. mucilaginosa as the yeast model to study the relation of the pigment and yeast cell survival and resistance. The findings of this study have to be seen in light of some limitations. Firstly, there are lack of references of R. mucilaginosa pigment function on antifungal drug test. Besides, the key challenge in this study is to view the phagocytosis process of macrophage cells RAW264.7 cell line using limited equipment that we have. Besides, loss of function phenotype approach using gene knockout was implemented but unfortunately the method used has disrupted the genes and destroyed the yeast cells. In future, researchers can continue to explore the precise mechanism of yeast cell survival using macrophage assay.
Data Availability
The data of this study are presented in the article text and tables. Additional details are available by contacting the corresponding author upon reasonable request. Some data are provided in full in the results sections of this paper and as supplementary data.
Code Availability
Not applicable.
References
Yang Q, Li Y, Apaliya MT, Zheng X, Serwah BNA, Zhang X, Zhang H (2018) The response of Rhodotorula mucilaginosa to patulin based on lysine crotonylation. Front Microbiol 9:2025. https://doi.org/10.3389/fmicb.2018.02025
Miceli MH, Díaz JA, Lee SA (2011) Emerging opportunistic yeast infections. Lancet Infect Dis 11(2):142–151. https://doi.org/10.1016/S1473-3099(10)70218-8
Ioannou P, Vamvoukaki R, Samonis G (2019) Rhodotorula species infections in humans: a systematic review. Mycoses 62(2):90–100. https://doi.org/10.1111/myc.12856
Jarros IC, Barros ILE, Prado A, Corrêa JL, Malacrida AM, Negri M, Svidzinski TIE (2022) Rhodotorula sp. and Trichosporon sp. are more virulent after a mixed biofilm. Mycopathologia 187(1):85–93. https://doi.org/10.1007/s11046-021-00606-5
Idris NFB, Huang G, Jia Q, Yuan L, Li Y, Tu Z (2019) Mixed infection of toe nail caused by Trichosporon asahii and Rhodotorula mucilaginosa. Mycopathologia 185:373–376. https://doi.org/10.1007/s11046-019-00406-y
Merhan O (2017) Biochemistry and antioxidant properties of carotenoids. Carotenoids 5:51. https://doi.org/10.5772/67592
Milani A, Basirnejad M, Shahbazi S, Bolhassani A (2017) Carotenoids: biochemistry, pharmacology and treatment. Br J Pharmacol 174(11):1290–1324. https://doi.org/10.1111/bph.13625
Liu GY, Essex A, Buchanan JT, Datta V, Hoffman HM, Bastian JF, Fierer J, Nizet V (2005) Staphylococcus aureus golden pigment impairs neutrophil killing and promotes virulence through its antioxidant activity. J Exp Med 202(2):209–215. https://doi.org/10.1084/jem.20050846
Chen F, Di H, Wang Y, Cao Q, Xu B, Zhang X, Yang N, Liu G, Yang CG, Xu Y, Jiang H, Lian F, Zhang N, Li J, Lan L (2016) Small-molecule targeting of a diapophytoene desaturase inhibits S. aureus virulence. Nat Chem Biol 12(3):174–179. https://doi.org/10.1038/nchembio.2003
Moliné M, Flores MR, Libkind D et al (2010) Photoprotection by carotenoid pigments in the yeast Rhodotorula mucilaginosa: the role of torularhodin. Photochem Photobiol Sci 9:1145–1151. https://doi.org/10.1039/c0pp00009d
Zelver N, Hamilton M, Pitts B, Goeres D, Walker D, Sturman P, Heersink J (1999) Measuring antimicrobial effects on biofilm bacteria: from laboratory to field. Methods Enzymol 310:608–628. https://doi.org/10.1016/s0076-6879(99)10047-8
Tuon FF, Costa SF (2008) Rhodotorula infection. A systematic review of 128 cases from literature. Rev Iberoam Micol 25:135–140. https://doi.org/10.1016/s1130-1406(08)70032-9
Davoli P, Mierau V, Weber RWS (2004) Carotenoids and fatty acids in red yeasts Sporobolomyces roseus and Rhodotorula glutinis. Appl Microbiol Biotechnol 40:392–397. https://doi.org/10.1023/B:ABIM.0000033917.57177.f2
Moț AC, Pârvu M, Pârvu AE, Roşca-Casian O, Dina NE, Leopold N, Silaghi-Dumitrescu R, Mircea C (2017) Reversible naftifine-induced carotenoid depigmentation in Rhodotorula mucilaginosa (A. Jörg.) FC Harrison causing onychomycosis. Sci Rep 7(1):1–12. https://doi.org/10.1038/s41598-017-11600-7
Li B, Ni S, Chen F, Mao F, Wei H, Liu Y et al (2018) Discovery of potent benzocycloalkane derived diapophytoene desaturase inhibitors with an enhanced safety profile for the treatment of MRSA. VISA, and LRSA infections. ACS Infect Dis 4:208–217. https://doi.org/10.1016/j.ejmech.2017.12.090
Huang G, Idris NFB, Li Y, Wang Y, Tu Z (2020) Naftifine inhibits pigmentation through down-regulation on expression of phytoene desaturase gene CAR1 in Rhodotorula mucilaginosa. Afr J Microbiol Res 14(5):166–174. https://doi.org/10.5897/AJMR2020.9316
Padyana AK, Gross S, Jin L, Cianchetta G, Narayanaswamy R, Wang F, Wang R, Fang C, Lv X, Biller SA, Dang L, Mahoney CE, Nagaraja N, Pirman D, Sui Z, Popovici-Muller J, Smolen GA (2019) Structure and inhibition mechanism of the catalytic domain of human squalene epoxidase. Nat Commun 10:97. https://doi.org/10.1038/s41467-018-07928-x
Nosanchuk JD, Casadevall A (2006) Impact of melanin on microbial virulence and clinical resistance to antimicrobial compounds. Antimicrob Agents Chemother 50:3519–3528. https://doi.org/10.1128/AAC.00545-06
Jahn B et al (1997) Isolation and characterization of a pigmentless-conidium mutant of Aspergillus fumigatus with altered conidial surface and reduced virulence. Infect Immun 12:5110–5117. https://doi.org/10.1128/iai.65.12.5110-5117.1997
Pihet M et al (2009) Melanin is an essential component for the integrity of the cell wall of Aspergillus fumigatus conidia. BMC Microbiol 9:177. https://doi.org/10.1186/1471-2180-9-177
Koselny K, Mutlu N, Minard AY, Kumar A, Krysan DJ, Wellington M (2018) A genome-wide screen of deletion mutants in the filamentous Saccharomyces cerevisiae background identifies ergosterol as a direct trigger of macrophage pyroptosis. MBio 9(4):e01204-e1218. https://doi.org/10.1128/mBio.01204-18
Schroeder WA, Johnson EA (1993) Antioxidant role of carotenoids in Phaffia rhodozyma. J Ind Microbiol Biotechnol 139:907–912. https://doi.org/10.1099/00221287-139-5-907
Kejžar A, Gobec S, Plemenitaš A, Lenassi M (2013) Melanin is crucial for growth of the black yeast Hortaea werneckii in its natural hypersaline environment. Fungal Biol 117(5):368–379. https://doi.org/10.1016/j.funbio.2013.03.006
Leber R, Landl K, Zinser E, Ahorn H, Spok A, Kohlwein SD, Turnowsky F, Daum G (1998) Dual localization of squalene epoxidase, SQLE protein, in yeast reflects a relationship between the endoplasmic reticulum and lipid particles. Mol Biol Cell 9:375–386. https://doi.org/10.1091/mbc.9.2.375
Wentzinger LF, Bach TJ, Hartmann MA (2002) Inhibition of squalene synthase and squalene epoxidase in tobacco cells triggers an up-regulation of 3-hydroxy-3-methylglutaryl coenzyme a reductase. Plant Physiol 130:334–346. https://doi.org/10.1104/pp.004655
Gill S, Stevenson J, Kristiana I, Brown AJ (2011) Cholesterol-dependent degradation of squalene monooxygenase, a control point in cholesterol synthesis beyond HMG-CoA reductase. Cell Metab 13(3):260–273. https://doi.org/10.1016/j.cmet.2011.01.015
Germann M, Gallo C, Donahue T, Shirzadi R et al (2005) Characterizing sterol defect suppressors uncovers a novel transcriptional signaling pathway regulating zymosterol biosynthesis. J Biol Chem 280:35904–35913. https://doi.org/10.1074/jbc.M504978200
Rosas AL, Casadevall A (1997) Melanization affects susceptibility of Cryptococcus neoformans to heat and cold. FEMS Microbiol Lett 153:265–272
Paolo WF, Dadachova E, Mandal P et al (2006) Effects of disrupting the polyketide synthase gene WdPKS1 in Wangiella [Exophiala] dermatitidis on melanin production and resistance to killing by antifungal compounds, enzymatic degradation, and extremes in temperature. BMC Microbiol 6:55. https://doi.org/10.1186/1471-2180-6-55
Agustinho DP, and Nosanchuk J (2017). Functions of Fungal Melanins. In book Reference Module in Life Science. https://doi.org/10.1016/B978-0-12-809633-8.12091-6.
Acknowledgements
We are grateful to The first Affiliated Hospital of Chongqing Medical University for providing the clinical isolates and laboratory equipment.
Funding
This research was funded by Chongqing Research Program of Basic Research and Frontier Technology [Grant No. cstc2021jcyj-msxmX0158], and Scientific and Technological Research Program of Chongqing Municipal Education Commission [Grant No. KJQN202113201]. The funding bodies had no role in the design of the study and collection, analysis, and interpretation of data and in writing the manuscript.
Author information
Authors and Affiliations
Contributions
NFBI performed the experiments (lead), analyzed the data (lead), and wrote the original draft of the manuscript (lead). QJ (supporting) and HL (supporting) conducted sampling and sample identification, and YG (supporting) and WY provided assistance in the purchase of chemicals (lead) and culture media preparation (supporting). H provided assistance with the generation of graphical data (supporting) and manuscript editing. TZ designed the study (lead) and supervised the work (lead) and was responsible for conceptualization (lead), data curation (lead), funding acquisition (lead), and methodology formulation (lead). All authors have read and agreed to the published version of the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The author(s) declare no conflicts of interest.
Ethical Approval
The Ethics Committee of Chongqing Medical University has approved this study as reference number 2020211 and the approval was given on 10th October 2020.
Informed Consent
Informed consent was obtained from all individual participants, authors, and patients, included in the study.
Consent to Participations
The patient of Chongqing Medical University has given the consent to participate in this study as approved by the Ethics Committee of Chongqing Medical University given on 10th October 2020 with reference number 2020011.
Consent for Publications
The Ethics Committee of Chongqing Medical University has approved this study as reference number 2020211 and the data obtained can be published in this journal.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Idris, N.F.B., Jia, Q., Lu, H. et al. Reduced Survival and Resistance of Rhodotorula mucilaginosa Following Inhibition of Pigment Production by Naftifine. Curr Microbiol 80, 285 (2023). https://doi.org/10.1007/s00284-023-03388-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00284-023-03388-9