Abstract
Purpose
To retrospectively evaluate the performance of different manual segmentation methods of placenta MR images for predicting Placenta Accreta Spectrum (PAS) disorders in patients with placenta previa (PP) using a Machine Learning (ML) Radiomics analysis.
Methods
64 patients (n=41 with PAS and n= 23 without PAS) with PP who underwent MRI examination for suspicion of PAS were retrospectively selected. All MRI examinations were acquired on a 1.5 T using T2-weighted (T2w) sequences on axial, sagittal and coronal planes. Ten different manual segmentation methods were performed on sagittal placental T2-weighted images obtaining five sets of 2D regions of interest (ROIs) and five sets of 3D volumes of interest (VOIs) from each patient. In detail, ROIs and VOIs were positioned on the following areas: placental tissue, retroplacental myometrium, cervix, placenta with underneath myometrium, placenta with underneath myometrium and cervix. For feature stability testing, the same process was repeated on 30 randomly selected placental MRI examinations by two additional radiologists, working independently and blinded to the original segmentation. Radiomic features were extracted from all available ROIs and VOIs. 100 iterations of 5-fold cross-validation with nested feature selection, based on recursive feature elimination, were subsequently run on each ROI/VOI to identify the best-performing method to classify instances correctly.
Results
Among the segmentation methods, the best performance in predicting PAS was obtained by the VOIs covering the retroplacental myometrium (Mean validation score: 0.761, standard deviation: 0.116).
Conclusion
Our preliminary results show that the VOI including the retroplacental myometrium using T2w images seems to be the best method when segmenting images for the development of ML radiomics predictive models to identify PAS in patients with PP.
Graphical abstract

Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Placenta accreta spectrum (PAS) disorders cover a range of clinical conditions where the placenta is abnormally attached to the uterus and does not spontaneously separate at delivery, resulting in massive haemorrhage, which can be life-threatening and usually necessitates hysterectomy [1, 2]. Based on the different grades of clinical and histological findings, PAS has been recently classified into three categories by the International Federation of Gynecology and Obstetrics (FIGO) in order to differentiate between abnormal adherence and abnormal invasion among the spectrum of disorders [3]. The pathogenesis of most cases of PAS is thought to be placental implantation at an area of defective decidualisation caused by pre-existing damage to the endometrial-myometrial interface [4]. Indeed, the most important risk factors related to the development of PAS are the placenta previa (PP) and the number of previous caesarean deliveries [5, 6]. The antenatal detection of PAS in high-risk patients is crucial to establish the most appropriate obstetrician management and avoid the occurrence of haemorrhage during delivery [7, 8]. Current placental imaging techniques rely on ultrasound (US) and magnetic resonance imaging (MRI) [9,10,11]. Given its high contrast resolution, MRI enables the characterisation of the uterus and placental tissues, thus delineating the entire placental-myometrium interface [12, 13]. The major challenge of MR placental imaging is related to the high inter-reader variability in interpreting abnormal morphological signs of PAS disorders, which requires imaging expertise and methodology [14,15,16,17]. Radiomics is a multistep process that converts medical images into multi-dimensional data to quantify tissue heterogeneity in terms of grey-level patterns and pixel inter-relationships within the image, thus potentially overcoming the limitations of traditional qualitative image assessment [18]. Recent advances in PAS imaging have leveraged the feasibility of the radiomics approach combined with machine learning algorithms in developing high-performance computational systems for the extraction and analysis of quantitative features from medical imaging data to support diagnosis and prognosis tasks [19, 20]. An increasing number of radiomic studies exploring the use of placental MRI radiomics for the detection and prognosis of PAS disorders have shown encouraging results [21,22,23]. Nevertheless, the most critical issues limiting the clinical translation of such approaches concern the reproducibility and generalizability of the various steps that compose the radiomics pipeline, in particular, a major methodological drawback is represented by the lack of features robustness testing for multiple segmentations across delineation techniques and regional anatomy variabilities [24, 25].
In this work, we aimed to retrospectively evaluate the performance of different manual segmentation methods of placenta MR images for predicting PAS disorders in patients with PP using a radiomics analysis empowered with ML technique.
Materials and methods
Patient selection
The local Institutional Review Board approved this retrospective study, and the requirement for informed consent was waived.
Consecutive pregnant patients at high risk for PAS who underwent MR examinations at our institution between January 2018 to January 2020 were retrieved. The standard of reference was based on the clinical diagnostic criteria and confirmed histopathological diagnosis, according to the recent FIGO classification [3]. The following clinical data were collected from medical records for each patient: maternal and gestational age as well as number of previous caesarean deliveries. Inclusion criteria were: >18-year-old patients with PP; patients with available clinical intraoperative findings and histological proof of PAS after CS or total hysterectomy. We excluded: 1) patients for whom MR images were incomplete for the retrospective evaluation; 2) MR examinations significantly affected by mother/fetal motion artifacts; and 3) patients with incomplete clinical and/or histological data.
MR acquisition protocol
Placental MRI was performed using a 1.5 T scanner (Gyroscan, Intera, Philips, Best, The Netherlands) with a phased-array body coil. The following MR sequences were acquired: Single-shot Turbo-Spin-Echo (TSE) T2-weighted (T2w) sequence (FOV 405 × 321 mm, matrix: 232 × 164, slice thickness 5–6 mm, number of slice 40, Flip angle: 90°, GAP 1, TR/TE = 381/80 ms) on axial, sagittal and coronal planes; breath-holding was requested to minimize respiratory motion artifact; and Thrive Spectral Attenuated Inversion Recovery (SPAIR) T1-weighted sequence (FOV: 395 × 280x340 mm, matrix: 192 × 192, slice thickness 4 mm, number of slice 60, Flip angle: 10°, GAP 2, TR/TE = 3.6/1.7 ms). Contrast agent was not administered. Total MR examination duration time was around 20 min.
Segmentation methods
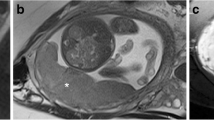
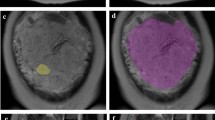
MR images were reviewed and analysed by an experienced radiologist (>10 years) who ensured the recognition of the different uterine, placental and cervical tissues. The best delineation of entire placental–myometrial interface occurs on sagittal T2w planes. Thus, ten different manual segmentations methods were performed on sagittal placental T2w images obtaining the following sets of 2D regions of interest (ROIs) and 3D volumes of interest (VOIs) from each patient. In detail, ROIs and VOIs were positioned in the following anatomical areas:
-
placental tissue
-
retroplacental myometrium
-
cervix
-
placenta with underneath myometrium
-
placenta with underneath myometrium and cervix.
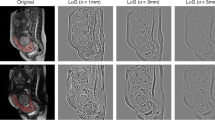
On sagittal T2w images, VOIs were manually delineated slice-by-slice on the entire tissue of interest whereas ROIs segmentation was performed in the midline of the uterus where the placental tissue, retroplacental myometrium and cervix were better represented. The segmentation process was performed using a freely available segmentation software (ITKSnap v3.8.0). Figure 1 shows the ROIs positioned on the different tested anatomical structures. Figure 2 reports an example of retroplacental myometrium delineation using 2D and 3D approaches. The same process was repeated for feature stability on 30 randomly selected placental MRI examinations by two additional radiologists, working independently and blinded to the original segmentations testing. These additional segmentations were exclusively employed to conduct feature reproducibility analysis. The segmentations used to build the classification model were all performed by the first reader.
Radiomics feature extraction and selection
Image pre-processing and feature extraction were conducted according to the Image Biomarker Standardisation Initiative (IBSI) guidelines using an open-source Python radiomics software package (PyRadiomics, v3.0.1) [26].
The following pre-processing steps were applied. First, all images and corresponding segmentation masks were resampled to a 3 × 3 mm resolution for ROI sets and 3 x 3 x 3 mm resolution in VOI sets, to ensure rotational invariance of texture features. Grey-level intensity values were normalized by subtracting the mean intensity and dividing by the standard deviation. Then, scaling by a factor of 100 and an array shift of +300 were performed, resulting in an expected [0, 600] final intensity range. Discretization was performed using a fixed bin count method (bin number = 7). To take into account the effect on radiomic feature robustness of image filtering, wavelet decomposition with all possible combinations of high- and low-pass filtering were applied as well as Laplacian of Gaussian edge enhancement (sigma = 3, 4 and 5), thus generating additional image sets for feature extraction. All available radiomic features for 2D and 3D masks were extracted from both original and filtered images, subdivided into the following classes: first-order (histogram analysis), 2D or 3D shape-based (for ROIs and VOIs, respectively), Gray Level Co-occurrence Matrix, Gray Level Size Zone Matrix, Gray Level Run Length Matrix, Neighbouring Gray Tone Difference Matrix and Gray Level Dependence Matrix (as defined in https://pyradiomics.readthedocs.io/en/latest/features.html).
Subsequently, a multistep selection process was carried to reduce the dimensionality of the dataset. First, non-reproducible features were excluded through feature stability ICC analysis, applied on data extracted from the multi-reader annotations. Features were considered stable if the ICC 95% confidence interval lower bound was ≥ 0.75. Then, low variance (< 0.1) and high pairwise Pearson correlation (≥ 0.80) were removed from the dataset. The final step of the feature selection process was based on data class labels and therefore nested within the ML pipeline described in the following section.
Machine learning analysis
Given the sample size available, a ML pipeline based on a nested cross-validation approach was employed. In particular, the following steps were iteratively performed within a 5-fold cross validation process: 1) scaling with a MinMax scaler (range = 0-1), 2) class balancing through Synthetic Minority Oversampling Technique, 3) recursive feature elimination with a nested 5-fold cross-validation, 4) hyperparameter tuning through random search with a nested 5-fold cross-validation. Given the tabular nature of the data and sample size, an ExtraTrees ensemble classifier was selected. Regarding the last step, the search grid parameters were as follows:
-
Number of trees = 100–1000
-
Criterion = entropy, Gini
-
Maximum tree depth = 1–10
-
Maximum tree features = 1–5
-
Bootstrap = True, false
-
Class weight = None, balanced
-
Maximum samples = 0–100%.
The ML pipeline was repeated for 100 iterations on each segmentation dataset. The classifier performance was estimated using the mean validation score with related standard deviation values. The analysis was performed by using the “numpy”, “pandas”, “imbalanced-learn” and “scikit-learn” Python packages [27].
Results
Patient population
Seventy-seven consecutive MRI examinations of high-risk patients with for PAS who underwent Caesarean section in our institution were retrieved. Thirteen patients were excluded because MRI scans were early interrupted due to claustrophobia (n = 5) or affected by fetal/mother motion artifact (n = 5) and for the lack of complete clinical/histological data (n = 3). Therefore, a final population of sixty-four patients was enrolled having a mean age 34.4 ± 4.9 years and mean gestational age 34.6 ± 2.3 weeks. Based on the FIGO classification criteria, 41 patients were confirmed to have PAS.
Feature selection and machine learning analysis
A total of 737 and 1106 radiomics features were extracted respectively from overall 2D and 3D delineation datasets. Stable features selected at ICC analysis for each segmentation methodology are reported in Table1. The mean validation scoring obtained by the best ML model in all segmentation approaches is summarized in Table 2. Among the tested segmentation approaches, the best performance in predicting PAS in patients with PP was obtained by the VOIs covering the retroplacental myometrium achieving a mean validation score of 0.761 (Standard Deviation = 0.116). The Supplementary Table include the final selected features from the 3D retroplacental myometrium segmentation method after the dimensionality reduction process.
Discussion
In this retrospective methodological study, we tested ten different segmentations methods built on MRI-based radiomic ML analysis demonstrating that 3D delineation of retroplacental myometrium obtained the best performance for the prediction of PAS in patients with PP.
One of the major challenges for reliable radiomics is the lack of feature reproducibility testing to variations in the segmentation methodology, especially for manual techniques, which might lead to poorly generalizable radiomics models hindering their introduction in clinical practice [24, 28, 29].
Efforts towards a radiomics harmonization framework have been embraced by exploratory works in oncological imaging which investigated the influence of segmentation variability on each step of the radiomics feature workflow in terms of boundaries variations [30, 31], tumour site [32], the adoption of 2D or 3D annotations [33], and the use of manual or (semi)automatic method [34].
In this perspective, Kocak et al. [35] systematically reviewed feature reproducibility strategies implemented in the original studies evaluating the radiomics profiling of renal lesions, pointing out that reproducibility analysis focused on segmentation variability was applied only in less than half of the revised investigations (18/44) .
In a recent systematic review [36], a quality appraisal of MRI-based radiomic studies focused on the detection and prognosis of PAS disorders revealed an overall heterogeneous and suboptimal methodological quality. Commonly encountered reproducibility concerns regarded the reporting of calibration statistics, the employment of validation strategies and the analyse of feature robustness to segmentation variabilities. Of note, different 2D or 3D approaches have been adopted in delineating uterine and placental tissue among the nine reviewed studies, without testing the feature robustness to regional anatomy variabilities or to repeated segmentations [36].
To our knowledge, our work is the first rigorously evaluating manual image segmentation in different areas of the uterine–placental environment, using the same feature selection algorithm and ML classifier, in order to identify the most robust and informative ROI/VOI delineation criteria. This might facilitate higher generalizability of radiomics features extraction in terms of regional variations and interobserver variations. In this light, the findings in our cohort suggest that VOIs delineation encompassing MRI retroplacental tissue may be the most efficient and reproducible segmentation method, possibly setting a standard for future studies in the field. As reported in Table 2, the 2D retroplacental myometrium segmentation method was slightly less performing than to the corresponding 3D sampling. In detail, among 2D segmentation approaches, 2D retroplacental myometrium (0.713) was the second-best approach shortly behind 2D placenta with underneath myometrium (0.730) and overall (2D and 3D) third in terms of average accuracy. Interestingly, the 2D approach slightly outperforming 2D retroplacental myometrium still included the retroplacental myometrium (i.e., 2D placenta with underneath myometrium). These findings can be explained by the number of pixels included in the ROIs/VOIs and thus in the analysis. Having more pixels to compute radiomics features can make them more informative and noisier; conversely, fewer pixels might not be sufficient to compute clinically valuable features but decrease the noise of non-relevant pixels. Since retroplacental myometrium is a thin structure and 2D segmentation inherently collects fewer pixels, it is likely that when working with 2D data, including additional information from the placental tissue is also beneficial. On the other hand, when working with 3D data, the number of included pixels from the more biologically relevant structure (retroplacental myometrium) is sufficient per se to provide clinically relevant information, and adding more pixels merely generates noise.
Our results are reasonably on the same line of recent evidence-based histopathologic findings arguing that rather than abnormal invasion, the main factor responsible of abnormal placentation in PAS is due to a defect in the uterine wall [37, 38]. Indeed, these emerging insights challenge the historical concept of abnormal placentation shifting the pathophysiologic paradigm of PAS from a destructive villous “invasive model” to the uterine “dehiscence model” in which myometrium disarray is induced by uterine scarification process such as caesarean section, leading to altered spatial relationship between the uterine wall and the anchoring villi implanted within and around the scar [39,40,41]. Therefore, our results identifying the retroplacental myometrium sampling as the most performing segmentation-based radiomics method are supported by these recent histopathological findings corroborating the primary role of the defective myometrium uterine and subsequent utero-placental interface abnormalities in the development of PAS; of course, additional similar studies are required to confirm this hypothesis.
Some limitations of the present study need to be acknowledged and discussed. First of all, the relatively small sample size did not allow to validate the final segmentation method performance on an external dataset. Nevertheless, a 5-fold cross-validation resampling procedure was used to estimate the ML model performance with nested feature selection based on recursive feature elimination method in order to prevent the risk of overfitting and avoid information leak between training and test folds. As a second limitation, we did not evaluate to what degree radiomic feature robustness can be influenced by the use of semi- or automatic methods. However, given the high variability in annotation approaches for uteroplacental tissue in radiomics focused literature [36], we believe that establishing a reliable delineation criteria could increase standardization regardless of segmentation methodology (e.g., automated Deep Learning networks). Finally, for PAS disorders diagnosis we considered the recent FIGO classification guidelines, which propose integrated criteria combining clinical finding at delivery and histopathologic criteria which not be applied on just delivered placenta tissue [3]; albeit the adoption of this system may lead to overestimating positive cases, it is the only internationally recognized classification thus enabling the appropriate comparison of different specialist centres and different management strategies.
In conclusion, on the basis of this preliminary experience, the VOIs including the retroplacental myometrium using T2-weighted images seems to be the best method when segmenting images for the development of reproducible ML radiomics predictive models to identify PAS in patients at high risk. In particular, our results in terms of pixel computation analysis might reflect the underlying microscopic morphologic alterations occurring in abnormal placentation which lies in the distortion of utero-placental interface induced by myometrial scarification events. Future investigations on multiple datasets are needed to confirm our findings and encourage the adoption of a common segmentation standard method to analyze MR images.
References
R.M. Silver, K.D. Barbour, Placenta Accreta Spectrum, Obstet. Gynecol. Clin. North Am. 42 (2015) 381–402. https://doi.org/https://doi.org/10.1016/j.ogc.2015.01.014.
E. Jauniaux, F. Chantraine, R.M. Silver, J. Langhoff-Roos, FIGO consensus guidelines on placenta accreta spectrum disorders: Epidemiology, Int. J. Gynecol. Obstet. 140 (2018) 265–273. https://doi.org/https://doi.org/10.1002/ijgo.12407.
E. Jauniaux, D. Ayres‐de‐Campos, J. Langhoff‐Roos, K.A. Fox, S. Collins, G. Duncombe, P. Klaritsch, F. Chantraine, J. Kingdom, L. Grønbeck, K. Rull, M. Tikkanen, L. Sentilhes, T. Asatiani, W. Leung, T. AIhaidari, D. Brennan, M. Seoud, A.M. Hussein, R. Jegasothy, K.N. Shah, D. Bomba‐Opon, C. Hubinont, P. Soma‐Pillay, N.T. Mandić, P. Lindqvist, B. Arnadottir, I. Hoesli, R. Cortez, FIGO classification for the clinical diagnosis of placenta accreta spectrum disorders, Int. J. Gynecol. Obstet. 146 (2019) 20–24, doi: https://doi.org/10.1002/ijgo.12761.
J.L. Hecht, R. Baergen, L.M. Ernst, P.J. Katzman, S.M. Jacques, E. Jauniaux, T.Y. Khong, L.A. Metlay, L. Poder, F. Qureshi, J.T. Rabban, D.J. Roberts, S. Shainker, D.S. Heller, Classification and reporting guidelines for the pathology diagnosis of placenta accreta spectrum (PAS) disorders: recommendations from an expert panel, Mod. Pathol. (2020). https://doi.org/https://doi.org/10.1038/s41379-020-0569-1.
H. Imafuku, K. Tanimura, Y. Shi, A. Uchida, M. Deguchi, Y. Terai, Clinical factors associated with a placenta accreta spectrum, Placenta. (2021). https://doi.org/https://doi.org/10.1016/j.placenta.2021.08.001.
R.M. Silver, M.B. Landon, D.J. Rouse, K.J. Leveno, C.Y. Spong, E.A. Thom, A.H. Moawad, S.N. Caritis, M. Harper, R.J. Wapner, Y. Sorokin, M. Miodovnik, M. Carpenter, A.M. Peaceman, M.J. O’Sullivan, B. Sibai, O. Langer, J.M. Thorp, S.M. Ramin, B.M. Mercer, Maternal Morbidity Associated With Multiple Repeat Cesarean Deliveries, Obstet. Gynecol. 107 (2006) 1226–1232. https://doi.org/https://doi.org/10.1097/01.AOG.0000219750.79480.84.
A. Mehrabadi, J.A. Hutcheon, S. Liu, S. Bartholomew, M.S. Kramer, R.M. Liston, K.S. Joseph, Contribution of Placenta Accreta to the Incidence of Postpartum Hemorrhage and Severe Postpartum Hemorrhage, Obstet. Gynecol. 125 (2015) 814–821. https://doi.org/https://doi.org/10.1097/AOG.0000000000000722.
L. Allen, E. Jauniaux, S. Hobson, J. Papillon-Smith, M.A. Belfort, FIGO consensus guidelines on placenta accreta spectrum disorders: Nonconservative surgical management, Int. J. Gynecol. Obstet. 140 (2018) 281–290. https://doi.org/https://doi.org/10.1002/ijgo.12409.
V. Romeo, L. Sarno, A. Volpe, M.I. Ginocchio, R. Esposito, P.P. Mainenti, M. Petretta, R. Liuzzi, M. D’Armiento, P. Martinelli, A. Brunetti, S. Maurea, US and MR imaging findings to detect placental adhesion spectrum (PAS) in patients with placenta previa: a comparative systematic study, Abdom. Radiol. 44 (2019) 3398–3407. https://doi.org/https://doi.org/10.1007/s00261-019-02185-y.
M. De Oliveira Carniello, L.G. Oliveira Brito, L.O. Sarian, J.R. Bennini, Diagnosis of placenta accreta spectrum in high‐risk women using ultrasonography or magnetic resonance imaging: systematic review and meta‐analysis, Ultrasound Obstet. Gynecol. 59 (2022) 428–436, doi: https://doi.org/10.1002/uog.24861.
S. Hong, Y. Le, K.U. Lio, T. Zhang, Y. Zhang, N. Zhang, Performance comparison of ultrasonography and magnetic resonance imaging in their diagnostic accuracy of placenta accreta spectrum disorders: a systematic review and meta-analysis, Insights Imaging. 13 (2022) 1–13. https://doi.org/https://doi.org/10.1186/S13244-022-01192-W/TABLES/2.
S. Maurea, V. Romeo, P.P. Mainenti, M.I. Ginocchio, G. Frauenfelder, F. Verde, R. Liuzzi, M. D’Armiento, L. Sarno, M. Morlando, M. Petretta, P. Martinelli, A. Brunetti, Diagnostic accuracy of magnetic resonance imaging in assessing placental adhesion disorder in patients with placenta previa: Correlation with histological findings, Eur. J. Radiol. 106 (2018) 77–84. https://doi.org/https://doi.org/10.1016/j.ejrad.2018.07.014.
H. Kapoor, M. Hanaoka, A. Dawkins, A. Khurana, Review of MRI imaging for placenta accreta spectrum: Pathophysiologic insights, imaging signs, and recent developments, Placenta. 104 (2021) 31–39. https://doi.org/https://doi.org/10.1016/j.placenta.2020.11.004.
L. Alamo, A. Anaye, J. Rey, A. Denys, G. Bongartz, S. Terraz, S. Artemisia, R. Meuli, S. Schmidt, Detection of suspected placental invasion by MRI: Do the results depend on observer’ experience?, Eur. J. Radiol. 82 (2013) e51–e57. https://doi.org/https://doi.org/10.1016/j.ejrad.2012.08.022.
C.L.A. Ghezzi, C.K. Silva, A.S. Casagrande, S.S. Westphalen, C.C. Salazar, J. Vettorazzi, Diagnostic performance of radiologists with different levels of experience in the interpretation of MRI of the placenta accreta spectrum disorder, Br. J. Radiol. (2021). https://doi.org/https://doi.org/10.1259/bjr.20210827.
S. Maurea, F. Verde, P.P. Mainenti, L. Barbuto, F. Iacobellis, V. Romeo, R. Liuzzi, G. Raia, G. De Dominicis, C. Santangelo, L. Romano, A. Brunetti, Qualitative evaluation of MR images for assessing placenta accreta spectrum disorders in patients with placenta previa: A pilot validation study, Eur. J. Radiol. (2022). https://doi.org/https://doi.org/10.1016/j.ejrad.2021.110078.
P. Jha, L. Pōder, C. Bourgioti, N. Bharwani, S. Lewis, A. Kamath, S. Nougaret, P. Soyer, M. Weston, R.P. Castillo, A. Kido, R. Forstner, G. Masselli, Society of Abdominal Radiology (SAR) and European Society of Urogenital Radiology (ESUR) joint consensus statement for MR imaging of placenta accreta spectrum disorders., Eur. Radiol. 30 (2020) 2604–2615. https://doi.org/https://doi.org/10.1007/s00330-019-06617-7.
P. Lambin, R.T.H. Leijenaar, T.M. Deist, J. Peerlings, E.E.C. de Jong, J. van Timmeren, S. Sanduleanu, R.T.H.M. Larue, A.J.G. Even, A. Jochems, Y. van Wijk, H. Woodruff, J. van Soest, T. Lustberg, E. Roelofs, W. van Elmpt, A. Dekker, F.M. Mottaghy, J.E. Wildberger, S. Walsh, Radiomics: the bridge between medical imaging and personalized medicine, Nat. Rev. Clin. Oncol. 14 (2017) 749–762. https://doi.org/https://doi.org/10.1038/nrclinonc.2017.141.
J. Guiot, A. Vaidyanathan, L. Deprez, F. Zerka, D. Danthine, A. Frix, P. Lambin, F. Bottari, N. Tsoutzidis, B. Miraglio, S. Walsh, W. Vos, R. Hustinx, M. Ferreira, P. Lovinfosse, R.T.H. Leijenaar, A review in radiomics: Making personalized medicine a reality via routine imaging, Med. Res. Rev. 42 (2022) 426–440. https://doi.org/https://doi.org/10.1002/med.21846.
V. Romeo, S. Maurea, The new era of advanced placental tissue characterization using MRI texture analysis: Clinical implications, EBioMedicine. 51 (2020) 102588. https://doi.org/10.1016/j.ebiom.2019.11.049.
V. Romeo, C. Ricciardi, R. Cuocolo, A. Stanzione, F. Verde, L. Sarno, G. Improta, P.P. Mainenti, M. D’Armiento, A. Brunetti, S. Maurea, Machine learning analysis of MRI-derived texture features to predict placenta accreta spectrum in patients with placenta previa, Magn. Reson. Imaging. 64 (2019). https://doi.org/10.1016/j.mri.2019.05.017.
H. Sun, H. Qu, L. Chen, W. Wang, Y. Liao, L. Zou, Z. Zhou, X. Wang, S. Zhou, Identification of suspicious invasive placentation based on clinical MRI data using textural features and automated machine learning., Eur. Radiol. 29 (2019) 6152–6162. https://doi.org/https://doi.org/10.1007/s00330-019-06372-9.
Q. Wu, K. Yao, Z. Liu, L. Li, X. Zhao, S. Wang, H. Shang, Y. Lin, Z. Wen, X. Zhang, J. Tian, M. Wang, Radiomics analysis of placenta on T2WI facilitates prediction of postpartum haemorrhage: A multicentre study, EBioMedicine. 50 (2019) 355–365. https://doi.org/https://doi.org/10.1016/j.ebiom.2019.11.010.
A. Stanzione, R. Cuocolo, L. Ugga, F. Verde, V. Romeo, A. Brunetti, S. Maurea, Oncologic Imaging and Radiomics: A Walkthrough Review of Methodological Challenges, Cancers (Basel). 14 (2022) 4871. https://doi.org/https://doi.org/10.3390/cancers14194871.
C. Ricciardi, R. Cuocolo, F. Verde, G. Improta, A. Stanzione, V. Romeo, S. Maurea, M. D’Armiento, L. Sarno, M. Guida, M. Cesarelli, Resolution Resampling of Ultrasound Images in Placenta Previa Patients: Influence on Radiomics Data Reliability and Usefulness for Machine Learning, in: 2021: pp. 1011–1018. https://doi.org/10.1007/978-3-030-64610-3_113.
A. Zwanenburg, M. Vallières, M.A. Abdalah, H.J.W.L. Aerts, V. Andrearczyk, A. Apte, S. Ashrafinia, S. Bakas, R.J. Beukinga, R. Boellaard, M. Bogowicz, L. Boldrini, I. Buvat, G.J.R. Cook, C. Davatzikos, A. Depeursinge, M.-C. Desseroit, N. Dinapoli, C.V. Dinh, S. Echegaray, I. El Naqa, A.Y. Fedorov, R. Gatta, R.J. Gillies, V. Goh, M. Götz, M. Guckenberger, S.M. Ha, M. Hatt, F. Isensee, P. Lambin, S. Leger, R.T.H. Leijenaar, J. Lenkowicz, F. Lippert, A. Losnegård, K.H. Maier-Hein, O. Morin, H. Müller, S. Napel, C. Nioche, F. Orlhac, S. Pati, E.A.G. Pfaehler, A. Rahmim, A.U.K. Rao, J. Scherer, M.M. Siddique, N.M. Sijtsema, J. Socarras Fernandez, E. Spezi, R.J.H.M. Steenbakkers, S. Tanadini-Lang, D. Thorwarth, E.G.C. Troost, T. Upadhaya, V. Valentini, L. V. van Dijk, J. van Griethuysen, F.H.P. van Velden, P. Whybra, C. Richter, S. Löck, The Image Biomarker Standardization Initiative: Standardized Quantitative Radiomics for High-Throughput Image-based Phenotyping, Radiology. 295 (2020) 328–338. https://doi.org/10.1148/radiol.2020191145.
F. Pedregosa, G. Varoquaux, A. Gramfort, V. Michel, B. Thirion, O. Grisel, M. Blondel, A. Müller, J. Nothman, G. Louppe, P. Prettenhofer, R. Weiss, V. Dubourg, J. Vanderplas, A. Passos, D. Cournapeau, M. Brucher, M. Perrot, É. Duchesnay, Scikit-learn: Machine Learning in Python, (2012).
B. Baeßler, K. Weiss, D. Pinto dos Santos, Robustness and Reproducibility of Radiomics in Magnetic Resonance Imaging, Invest. Radiol. 54 (2019) 221–228. https://doi.org/https://doi.org/10.1097/RLI.0000000000000530.
A. Stanzione, R. Galatola, R. Cuocolo, V. Romeo, F. Verde, P.P. Mainenti, A. Brunetti, S. Maurea, Radiomics in Cross-Sectional Adrenal Imaging: A Systematic Review and Quality Assessment Study, Diagnostics. 12 (2022) 578. https://doi.org/https://doi.org/10.3390/diagnostics12030578.
B. Kocak, E. Ates, E.S. Durmaz, M.B. Ulusan, O. Kilickesmez, Influence of segmentation margin on machine learning–based high-dimensional quantitative CT texture analysis: a reproducibility study on renal clear cell carcinomas, Eur. Radiol. (2019). https://doi.org/https://doi.org/10.1007/s00330-019-6003-8.
X. Zhang, L. Zhong, B. Zhang, L. Zhang, H. Du, L. Lu, S. Zhang, W. Yang, Q. Feng, The effects of volume of interest delineation on MRI-based radiomics analysis: evaluation with two disease groups, Cancer Imaging. 19 (2019) 89. https://doi.org/https://doi.org/10.1186/s40644-019-0276-7.
M. Pavic, M. Bogowicz, X. Würms, S. Glatz, T. Finazzi, O. Riesterer, J. Roesch, L. Rudofsky, M. Friess, P. Veit-Haibach, M. Huellner, I. Opitz, W. Weder, T. Frauenfelder, M. Guckenberger, S. Tanadini-Lang, Influence of inter-observer delineation variability on radiomics stability in different tumor sites, Acta Oncol. (Madr). 57 (2018) 1070–1074. https://doi.org/https://doi.org/10.1080/0284186X.2018.1445283.
S. Gitto, R. Cuocolo, I. Emili, L. Tofanelli, V. Chianca, D. Albano, C. Messina, M. Imbriaco, L.M. Sconfienza, Effects of Interobserver Variability on 2D and 3D CT- and MRI-Based Texture Feature Reproducibility of Cartilaginous Bone Tumors, J. Digit. Imaging. 34 (2021) 820–832. https://doi.org/https://doi.org/10.1007/s10278-021-00498-3.
Q. Qiu, J. Duan, Z. Duan, X. Meng, C. Ma, J. Zhu, J. Lu, T. Liu, Y. Yin, Reproducibility and non-redundancy of radiomic features extracted from arterial phase CT scans in hepatocellular carcinoma patients: Impact of tumor segmentation variability, Quant. Imaging Med. Surg. (2019). https://doi.org/10.21037/qims.2019.03.02.
B. Kocak, E.S. Durmaz, C. Erdim, E. Ates, O.K. Kaya, O. Kilickesmez, Radiomics of Renal Masses: Systematic Review of Reproducibility and Validation Strategies, Am. J. Roentgenol. 214 (2020) 129–136. https://doi.org/https://doi.org/10.2214/AJR.19.21709.
A. Stanzione, F. Verde, R. Cuocolo, V. Romeo, P. Paolo Mainenti, A. Brunetti, S. Maurea, Placenta Accreta Spectrum Disorders and Radiomics: Systematic review and quality appraisal, Eur. J. Radiol. 155 (2022) 110497. https://doi.org/10.1016/j.ejrad.2022.110497.
B.D. Einerson, J. Comstock, R.M. Silver, D.W. Branch, P.J. Woodward, A. Kennedy, Placenta Accreta Spectrum Disorder, Obstet. Gynecol. 135 (2020) 1104–1111. https://doi.org/https://doi.org/10.1097/AOG.0000000000003793.
E. Jauniaux, D. Jurkovic, A.M. Hussein, G.J. Burton, New insights into the etiopathology of placenta accreta spectrum, Am. J. Obstet. Gynecol. 227 (2022) 384–391. https://doi.org/https://doi.org/10.1016/j.ajog.2022.02.038.
A.M. Hussein, R.A. Elbarmelgy, R.M. Elbarmelgy, M.M. Thabet, E. Jauniaux, Prospective evaluation of impact of <scp>post‐Cesarean</scp> section uterine scarring in perinatal diagnosis of placenta accreta spectrum disorder, Ultrasound Obstet. Gynecol. 59 (2022) 474–482. https://doi.org/https://doi.org/10.1002/uog.23732.
E. Jauniaux, A.M. Hussein, N. Zosmer, R.M. Elbarmelgy, R.A. Elbarmelgy, H. Shaikh, G.J. Burton, A new methodologic approach for clinico-pathologic correlations in invasive placenta previa accreta, Am. J. Obstet. Gynecol. 222 (2020) 379.e1-379.e11. https://doi.org/https://doi.org/10.1016/j.ajog.2019.11.1246.
E. Jauniaux, A.M. Hussein, R.M. Elbarmelgy, R.A. Elbarmelgy, G.J. Burton, Failure of placental detachment in accreta placentation is associated with excessive fibrinoid deposition at the utero-placental interface, Am. J. Obstet. Gynecol. (2022). https://doi.org/https://doi.org/10.1016/j.ajog.2021.08.026.
Author information
Authors and Affiliations
Contributions
I confirm that all the authors have contributed significantly to this manuscript, have seen and approved the final manuscript, and have agreed to its submission to the Abdominal Radiology.
Corresponding author
Ethics declarations
Conflict of interest
All authors declare that they have no conflicts of interest.
Ethical approval
IRB approved.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Verde, F., Stanzione, A., Cuocolo, R. et al. Segmentation methods applied to MRI-derived radiomic analysis for the prediction of placenta accreta spectrum in patients with placenta previa. Abdom Radiol 48, 3207–3215 (2023). https://doi.org/10.1007/s00261-023-03963-5
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00261-023-03963-5