Abstract
Objective
The aim of this study was to investigate whether intraplacental texture features from routine placental MRI can objectively and accurately predict invasive placentation.
Material and methods
This retrospective study includes 99 pregnant women with pathologically confirmed placental invasion and 56 pregnant women with simple placenta previa. All participants underwent magnetic resonance imaging after 24 gestational weeks. The placenta was segmented in sagittal images from both turbo spin echo (TSE) and balanced turbo field echo (bTFE) sequences. Textural features were extracted from the both original and Laplacian of Gaussian (LoG)-filtered MRI images. An automated machine learning algorithm was applied to the extracted feature sets to obtain the optimal preprocessing steps, classification algorithm, and corresponding hyper-parameters.
Results
A gradient boosting classifier using all textual features from original and LoG-filtered TSE images and bTFE images identified by the automated machine learning algorithm achieved the optimal performance with sensitivity, specificity, accuracy, and area under ROC curve (AUC) of 100%, 88.5%, 95.2%, and 0.98 in the prediction of placental invasion. In addition, textural features that contributed to the prediction of placental invasion differ from the features significantly affected by normal placenta maturation.
Conclusions
Quantifying intraplacental heterogeneity using LoG filtration and texture analysis highlights the different heterogeneous appearance caused by abnormal placentation relative to normal maturation. The predictive model derived from automated machine learning yielded good performance, indicating the proposed radiomic analysis pipeline can accurately predict placental invasion and facilitate clinical decision-making for pregnant women with suspicious placental invasion.
Key Points
• The intraplacental texture features have high efficiency in prediction of invasive placentation after 24 gestational weeks.
• The features with dominated predictive power did not overlap with the features significantly affected by gestational age.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Placental invasion represents a spectrum of placental adhesive disorders that occur mostly in patients with placenta previa or prior cesarean section, with common complications including catastrophic perinatal hemorrhage and bladder, bowel, and urethral injuries [1]. Early diagnosis of placental invasion before delivery is critical for appropriate treatment planning [2]. Although ultrasound is the mainstay in the imaging of placental invasion [3], MRI is especially recommended for cases of posterior placenta and cases whose ultrasound results are equivocal and/or clinical suspicion is high [4, 5].
According to a recent meta-analysis of prenatal diagnosis of invasive placentation, the sensitivity of MRI for diagnosing placental invasion ranged from 81.3 to 95.1% (mean 90.2%) with a specificity ranging from 76.7 to 94.4% (mean 88.2%) [6]. It worth note that one significant factor that affects diagnostic performance is observer experience. A recent study showed that more experienced radiologists performed significantly better than junior radiologists (sensitivity 90.9% and specificity 75% for senior attending physicians, 81.8% and 61.8% for junior attending physicians) [7]. Some scoring systems for the diagnostic placental invasion were proposed but have not been widely tested [8].
Marked placental heterogeneity and irregular thick intraplacental T2 dark bands are well-established MR imaging makers of placental invasion [9]. However, visual characterization of placental heterogeneity and identification of abnormal T2 dark bands can be difficult and highly subjective. Thus, the use of quantitative image analysis has been an attractive field to overcome the subjectivity of visual interpretation and improve diagnostic accuracy.
The fast evolution of medical imaging has fostered a comprehensive analysis method for medical images called radiomics [10]. Radiomics generally refers to the extraction and analysis of large amounts of quantitative features from medical images. In comparison with the traditional visual inspection, radiomics aims to transform medical images into minable data [11]. Using sophisticated machine learning tools, researchers have developed radiomic models that can potentially improve diagnostic, prognostic, and predictive accuracy [12].
Texture features in radiomics represent the spatial distribution and arrangement of the pixels in a region of interest. It offers a means of capturing localized image variations arising from tissue characteristics not readily quantified by the human eye [13]. For this reason, we hypothesized that texture features may have the potential to improve the diagnostic performance over conventional visual based diagnosis by providing detailed information about intraplacental heterogeneity and T2 hypointense striped pattern. Thus, the purpose of this study was to leverage machine learning algorithms to develop a MR texture-based radiomic model to differentiate between normal and invasive placentation for decision support and treatment planning.
Material and methods
Participant
Our institutional review board approved this retrospective study and waived the requirement to obtain informed consent. The privacy of the patients was protected by de-identifying images prior analysis. To accrue our cohort, we performed a database search on MRIs performed on pregnant women from July 2015 to March 2017. Two hundred forty-eight consecutive pregnant women with suspected abnormal placentation due to the presence of placenta previa on routine second trimester ultrasound examination were initially selected. In all, 93 patients were excluded for various reasons including multiple pregnancy (n = 5), those who underwent MRI scan before 24 gestational weeks (n = 27) [14], presence of significant fetal/maternal anomalies (n = 18), severe motion or other type of artifact on either imaging sequence (n = 12), and deliveries performed outside of our institution (resulting in lack of final diagnosis) (n = 31). The final cohort included 155 patients scanned because of suspicion of placental invasion.
MR imaging parameters
All MRI examinations were performed on a 1.5-T clinical scanner (Achieva; Philips Medical Systems) with a body array coil. T2-weighted images through the uterus and placenta were obtained with a TSE sequence (TR 465 ms, effective TE 80 ms, 432 × 432 matrix over a field of view of 405 × 405 mm, 5 mm slice thickness, SENSE factor 2) for full placenta coverage in the sagittal plane and T2-/T1-weighted images were also obtained with bTFE sequence (TR 3 ms, TE 1.5 ms, 384 × 384 matrix over a field of view of 375 × 375 mm, 5 mm slice thickness, no parallel imaging) with the same slice position as the TSE sequence.
Placenta delineation
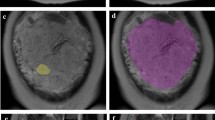
The placenta was segmented using an online learning-based semi-automated algorithm implemented in Slic-Seg [15] by providing scribbles to define the object and background. The masks of segmented placenta were reviewed and manually adjusted by a radiologist experienced in obstetric imaging using the ITK-SNAP software [16]. A detailed illustration of steps on semi-automatic placenta segmentation is provided in supplementary Table S1.
Radiomic feature extraction
Radiomic feature calculations were performed by using the Pyradiomics package (version 2.0.0) [17]. For a consistent calculation of 3D textural features, intensities of all images were normalized as described in Appendix A. Image intensities were then further discretized before calculation of texture features [18]. Discretization is a preprocessing step, which reduce overall gray level of original image by setting the voxels within bin width to the same value, to make calculation of texture features tractable. Discretization was performed by using a fixed bin width = 5. Textural features from the following matrixes were extracted from pre-delineated masks:
-
Gray-level co-occurrence matrix (GLCM) (22 features)
-
Gray-level size zone matrix (GLSZM) (16 features)
-
Gray-level run length matrix (GLRLM) (16 features)
-
Neighboring gray tone difference matrix (NGTDM) (5 features)
-
Gray-level dependence matrix (GLDM) (14 features)
The above features were also calculated on the normalized images after LoG filtering with kernel size (σ) of 1 mm (fine-scale filtration), 3 mm (medium-scale filtration), and 5 mm (coarse-scale filtration) (Fig. 1).
In total, 624 radiomic features (78 textural features × 2 imaging sequences × (1 original image + 3 filtered images)) were generated for each subject. A detailed list of extracted features is provided in supplementary Table S2.
Correction for gestational age effects
After 24 weeks gestation, the placenta can become slightly lobulated, which may lead to increased signal heterogeneity with increasing gestational age in MRI appearance [19]. To remove gestational age induced textural changes while retaining disease-associated textural variation, a linear de-trending strategy was used [20]. Multiple univariate linear regression models were fitted between each MRI-derived textural feature and corresponding gestational age in the simple placenta previa group only. This procedure models gestational age-related textural changes as a linear drift. Then, the regression coefficients with statistical significance were used to remove the gestational age-related changes for all individuals to obtain corrected values.
Automated machine learning
A genetic algorithm-based automated machine learning algorithm implemented in the Tree-based Pipeline Optimization Tool (TPOT) [21] was used to construct the optimal classification model.
To investigate the diagnostic value of two imaging sequences and the scale at which the disease-associated features exhibited, extracted features were categorized into 14 feature sets and fed into TPOT as follows:
-
1.
Textural features from original TSE image (73 features)
-
2.
Textural features from LoG-filtered (σ = 1 mm) TSE image (73 features)
-
3.
Textural features from LoG-filtered (σ = 3 mm) TSE image (73 features)
-
4.
Textural features from LoG-filtered (σ = 5 mm) TSE image (73 features)
-
5.
Textural features from LoG-filtered (σ = 1, 3, and 5 mm) TSE image (219 features)
-
6.
Textural features from original + LoG-filtered (σ = 1, 3, and 5 mm) TSE image (292 features)
-
7.
Textural features from original bTFE image (73 features)
-
8.
Textural features from LoG-filtered (σ = 1 mm) bTFE image (73 features)
-
9.
Textural features from LoG-filtered (σ = 3 mm) bTFE image (73 features)
-
10.
Textural features from LoG-filtered (σ = 5 mm) bTFE image (73 features)
-
11.
Textural features from LoG-filtered (σ = 1, 3, and 5 mm) bTFE image (219 features)
-
12.
Textural features from original + LoG-filtered (σ = 1, 3, and 5 mm) bTFE image (292 features)
-
13.
Textural features from original TSE and bTFE image (146 features)
-
14.
Textural features from original + LoG-filtered (σ = 1, 3, and 5 mm) TSE and bTFE image (584 features)
A hold-out scheme was used to evaluate each generated model. Sixty percent of all involved subjects were randomly selected according to their diagnosis to form the training cohort; the rest were treated as a testing cohort. Metrics including sensitivity, specificity, accuracy, kappa score [22], and AUC were used to score each model. The full workflow is depicted in Fig. 2.
To compare the diagnostic performance of the radiomic model with visual assessment, MR images in the testing cohort were reviewed by the same radiologist that completed image segmentation. A definitive diagnosis was made for each case to indicate the presence of placental invasion or not. The sensitivity, specificity, and overall accuracy from human reader were calculated and compared with the best-performed radiomic model.
Results
Patient characteristics
One hundred fifty-five pregnant women who meet inclusion criteria were enrolled in the current study. Of which 99 had histologically confirmed placental invasion (PI) (65 accreta, 31 increta, and 3 percreta) and 56 had simple placenta previa (SPP). The mean age for the PI group (30.86 ± 3.49 years, range from 21 to 38) was significantly higher than the SPP group (28.84 ± 3.34 years, range from 19 to 35) (p < 0.001, univariate two-sample t test). The mean gestational age for the PI group was 32.90 ± 2.61 weeks (range from 24.71 to 36.0), while for the SPP group it was 31.82 ± 3.35 weeks (range from 24.14 to 35.71). No significant difference was observed in gestational age (p = 0.22, univariate two-sample t test) between the two groups.
Effect of gestational age on placental textural features
Twenty features were found to be significantly affected (with Bonferroni-adjusted p < 0.05) by gestational age in the SPP group (Table 1). Most identified features showed intensity non-uniformity in different imaging sequences, feature matrixes, and filter sizes. These 20 features were linear de-trended using the fitted coefficients in both SPP and PI groups before classification analysis.
Classification model derived from automated machine learning
Sixty-three pregnant women with PI and 30 pregnant women with SPP were randomly selected from the study cohort to form the training set, with the remaining 36 PI (28 accreta, 6 increta, and 2 percreta) and 26 SPP cases treated as a testing set. No significant difference was observed in either age or gestational age between training and testing set for either PI or SPP group.
The TPOT identified the optimal machine learning models for each feature set in the training set. The performance of each identified model was evaluated using the independent testing set. The identified models and performances are listed in Table 2. ROC curves were plotted to illustrate diagnostic performance of each model (Fig. 3).
The best overall performance was achieved by the gradient boosting classifier [23] with all 584 features (highest scores in all evaluated metrics). Of the 26 cases of simple placenta previa in the testing set, 3 cases were misclassified as having invasion by this model and the remaining 23 cases were correctly classified as no invasion. In addition, 28 cases of accreta, 6 cases of increta, and 2 cases of percreta in the testing set were all correctly classified as having invasion by this model. Moreover, the models built on all TSE features (model 6) and all LoG-filtered TSE features (model 5) also obtained the highest specificity. Diagnostic performance from the senior radiologist achieved the sensitivity of 88.9% (32 of 36), specificity of 88.5% (23 of 26), and the overall accuracy of 88.7% (55 of 62).
Significant features contributing to classification
According to the optimal model (gradient boosting classifier with all 584 features), the importance of each input feature can be measured during model training. Figure 4a lists the top 10 features with the most predictive power. The distributions of top 10 features in both groups were box plotted in Fig. 4c. The separability of the two groups was visualized by t-distributed stochastic neighbor embedding (t-SNE) with the top 10 features presented in Fig. 4b. Representative images from subtypes of placental invasion with corresponding top discriminative textural features are shown in Fig. 5.
Discussion
In this study, we developed a radiomic analysis pipeline with integration of textural features and automated machine learning for pre-delivery prediction of invasive placentation after 24 gestational weeks. In general, the gradient boosting classifier constructed on textural features from original and LoG-filtered TSE and bTFE images, which achieved the predictive performance with sensitivity, specificity, accuracy, and AUC of 100%, 88.5%, 95.2%, and 0.98 respectively in the external evaluation, was automatically identified as the optimal prediction model. This radiomic model showed comparable specificity and slightly better sensitivity and over-accuracy than an experienced radiologist, which indicate quantitative textural analysis is more sensitive than the perception of even an experienced radiologist on the threshold of multiple MRI signatures. Moreover, the gradient boosting classifiers built on LoG-filtered TSE images and built on original and LoG-filtered TSE images also achieved the highest specificity of 88.5%, which indicate it is possible to obtain comparable prediction performance using only TSE images. Our findings show a relatively high accuracy of invasive placentation prediction and then suggest the proposed radiomic approach is feasible and promising for MRI-based pre-delivery prediction.
Machine learning in radiology aims at training mathematical models to recognize patterns in image features and to assist diagnosis by linking these patterns to prediction targets such as diagnosis and outcome [24]. Most existing radiomic studies selected a machine learning pipeline according to the preference or the popularity of the algorithm. It has been proven that the choice of modeling technique can affect prediction performance in radiomics [25]. However, identification of an optimal machine learning pipeline for a given problem often requires expert knowledge of the algorithm as well as the target problem, because there are dozens of possible choices to make in the steps of data preprocessing, model selection, and hyperparameter tuning [24]. As the complexity of these tasks is often beyond non-ML-experts, automated machine learning (autoML) was recently proposed to automatically determine a well-performing machine learning pipeline under certain optimization strategy to solve the ever-growing challenge of applying machine learning in radiomics and other fields.
Target segmentation is also a crucial step in radiomic studies, which can significantly affect subsequent feature extraction [26]. For target tissue with irregular shape or fuzzy boundary, manual segmentation is demanded. However, manual segmentation is labor intensive and time consuming, making it impractical in clinical practice. In the current study, we exploited an interactive, learning-based method for the segmentation of the placenta, which only requires a few user-provided scribbles to roughly indicate target tissue and background in a few slices. This semi-automatic interactive segmentation adds to the practicality of the proposed workflow, allowing it to be integrated into routine clinical diagnostics.
Of note, the predictive power of textural features is related to the imaging sequence and whether or not filtering is used. Features from the unfiltered bTFE images (model 7) were found to be more predictive than features from unfiltered TSE images (model 1). However, when LoG filtering was used, diagnostic performance of models built on TSE images (model 6) outperformed models built on bTFE images (model 12), especially in specificity. The bTFE sequence belongs to gradient echo technique and thus sensitive to magnetic field inhomogeneity and off resonance effects [27]. The noise arising from local field inhomogeneity may interfere with prediction and thus cause poorer specificity. In addition, the current result also proved that features from LoG-filtered images are more predictive than features from unfiltered images. The LoG filter serves as a generic differential operator that responds to local image variations [28]. The response is strongest when the kernel size σ matches the spatial extent of local textural patterns in the image. A small σ emphasizes on fine textures (change over a short distance), whereas a large σ emphasizes coarse textures (gray-level change over a large distance). The filtered volumes encode properties of tissue heterogeneity and local structure that may be connected to the invasive placentation.
There is already considerable evidence in the literature of the ability of intraplacental heterogeneity to predict invasive placentation. Disorganized hypertrophied abnormalities of intraplacental vascularity and fibrin deposition can cause intraplacental heterogeneity [29]. However, intensity heterogeneity is subjective and difficult to quantify for human readers and normally progresses with advancing gestational age after the second trimester. These factors complicate the visual assessment of placental invasion. The current results demonstrate that the features predict placental invasion did not overlap with the features change along with gestational age, suggesting the intensity inhomogeneity caused by maturation or disease have different pattern and scale in the view of computerized textural analysis. The features that change along with gestational age mostly belong to run length and dependence matrix family in coarse scale. The run length matrix provides a way of charactering voxels having the same gray-level value occurring in a line across certain direction [30]. The run length non-uniformity of GLRLM measures the distribution of lines of voxels with same intensity. This metric is low when lines are equally distributed in length and increases when a few outliers of line length dominate the histogram. The dependence matrix measures the number of connected voxels within a certain distance that are dependent on the central voxel. Similarly, gray-level non-uniformity of GLDM increases when gray-level outliers dominate the histogram. These features are indicators of a stripe-like pattern in the region, which may reflect the formation of cytoledon and septum during normal placenta maturation [31].
The top-ranked features contributing to prediction were inferred from the best predictive model (model 14). Most of them were derived from the GLCM and GLSZM in both fine and coarse scale. The feature with dominative predictive power was the correlation metric based on GLCM from TSE images with fine filtration. The co-occurrence matrix describes the pair-wise arrangement of voxels with the same discretized gray level at a given offset [32]. The metric of correlations measures the joint probability occurrence of the specified voxel pairs. The higher correlation in fine-filtered TSE images of patients with placenta invasion indicates certain fine structures related to abnormal placentation, which may be a sign of subtle fibrin deposition or abnormal microvessel patterns. The GLSZM counts the number of zones of voxels with identical discretized gray level. Within GLSZM, the zone percentage measures the fraction of the number of realized zones and the maximum number of potential zones, with higher values indicating the targeted region consists of small zones, while the zone entropy measures the uncertainty/randomness in the distribution of zone sizes and gray levels. A higher value indicates more heterogeneity in the texture patterns. The differences in these features between the two groups in the present study indicate a unique heterogeneous textural pattern, which is not related to gestational age, which appeared in the placenta with invasion.
We need to point out certain limitations of the current study. Due to retrospective nature of this study, some bias may inevitably exist or may have affected the analysis. The sample size (n = 155) was relatively small, but with the advantage that all data were collected at a single site using a single scanner. To some extent, the use of the same MRI equipment and acquisition protocol in our data not only strengthens the study but also limits generalization of the results. The results obtained in this study require further external validation in data from different vendors and with different acquisition parameters.
In spite of these limitations, the results reported here are encouraging as they provide pilot evidence of the potential of image filtration and measures of quantitative intraplacental texture for facilitating clinical decision-making for pregnant women with suspected placental invasion. The results also provide support for the initiation of a new prospective study to systematically evaluate the diagnostic capability of the combination of imaging features and clinical parameters for improving diagnosis and patient care.
Abbreviations
- AUC:
-
Area under the ROC curve
- bTFE:
-
Balanced turbo field echo
- LoG:
-
Laplacian of Gaussian
- PI:
-
Placental invasion
- SPP:
-
Simple placenta previa
- TSE:
-
Turbo spin echo
References
Oyelese Y, Smulian JC (2006) Placenta previa, placenta accreta, and vasa previa. Obstet Gynecol 107:927–941. https://doi.org/10.1097/01.AOG.0000207559.15715.98
Garmi G, Salim R (2012) Epidemiology, etiology, diagnosis, and management of placenta accreta. Obstet Gynecol Int 2012:1–7. https://doi.org/10.1155/2012/873929
Baughman WC, Corteville JE, Shah RR (2008) Placenta accreta: spectrum of US and MR imaging findings. Radiographics 28:1905–1916. https://doi.org/10.1148/rg.287085060
Kocher MR, Sheafor DH, Bruner E et al (2017) Diagnosis of abnormally invasive posterior placentation: the role of MR imaging. Radiol Case Rep 12:295–299. https://doi.org/10.1016/j.radcr.2017.01.014
Delli Pizzi A, Tavoletta A, Narciso R et al (2019) Prenatal planning of placenta previa: diagnostic accuracy of a novel MRI-based prediction model for placenta accreta spectrum (PAS) and clinical outcome. Abdom Radiol 44:1873–1882. https://doi.org/10.1007/s00261-018-1882-8
D’Antonio F, Iacovella C, Palacios-Jaraquemada J et al (2014) Prenatal identification of invasive placentation using magnetic resonance imaging: systematic review and meta-analysis. Ultrasound Obstet Gynecol 44:8–16. https://doi.org/10.1002/uog.13327
Alamo L, Anaye A, Rey J et al (2013) Detection of suspected placental invasion by MRI: do the results depend on observer’ experience? Eur J Radiol 82:e51–e57. https://doi.org/10.1016/j.ejrad.2012.08.022
Ueno Y, Kitajima K, Kawakami F et al (2014) Novel MRI finding for diagnosis of invasive placenta praevia: evaluation of findings for 65 patients using clinical and histopathological correlations. Eur Radiol 24:881–888. https://doi.org/10.1007/s00330-013-3076-7
Lax A, Prince MR, Mennitt KW et al (2007) The value of specific MRI features in the evaluation of suspected placental invasion. Magn Reson Imaging 25:87–93. https://doi.org/10.1016/j.mri.2006.10.007
Lambin P, Rios-Velazquez E, Leijenaar R et al (2012) Radiomics: extracting more information from medical images using advanced feature analysis. Eur J Cancer 48:441–446. https://doi.org/10.1016/j.ejca.2011.11.036
Gillies RJ, Kinahan PE, Hricak H (2016) Radiomics: images are more than pictures, they are data. Radiology 278:563–577. https://doi.org/10.1148/radiol.2015151169
Langs G, Röhrich S, Hofmanninger J et al (2018) Machine learning: from radiomics to discovery and routine. Radiologe 58:1–6. https://doi.org/10.1007/s00117-018-0407-3
Castellano G, Bonilha L, Li LM, Cendes F (2004) Texture analysis of medical images. Clin Radiol 59:1061–1069. https://doi.org/10.1016/j.crad.2004.07.008
Horowitz JM, Berggruen S, McCarthy RJ et al (2015) When timing is everything: are placental MRI examinations performed before 24 weeks’ gestational age reliable? Am J Roentgenol 205:685–692. https://doi.org/10.2214/AJR.14.14134
Wang G, Zuluaga MA, Pratt R et al (2016) Slic-Seg: a minimally interactive segmentation of the placenta from sparse and motion-corrupted fetal MRI in multiple views. Med Image Anal 34:137–147. https://doi.org/10.1016/j.media.2016.04.009
Yushkevich PA, Piven J, Hazlett HC et al (2006) User-guided 3D active contour segmentation of anatomical structures: significantly improved efficiency and reliability. Neuroimage 31:1116–1128. https://doi.org/10.1016/j.neuroimage.2006.01.015
Van Griethuysen JJM, Fedorov A, Parmar C et al (2017) Computational radiomics system to decode the radiographic phenotype. Cancer Res 77:e104–e107. https://doi.org/10.1158/0008-5472.CAN-17-0339
Yip SSF, Aerts HJWL (2016) Applications and limitations of radiomics. Phys Med Biol 61:R150–R166. https://doi.org/10.1088/0031-9155/61/13/R150
Fadl S, Moshiri M, Fligner CL et al (2017) Placental imaging: normal appearance with review of pathologic findings. Radiographics 37:979–998. https://doi.org/10.1148/rg.2017160155
Dukart J, Schroeter ML, Mueller K (2011) Age correction in dementia – matching to a healthy brain. PLoS One 6:e22193. https://doi.org/10.1371/journal.pone.0022193
Olson RS, Bartley N, Urbanowicz RJ, Moore JH (2016) Evaluation of a Tree-based Pipeline Optimization Tool for automating data science. Proc 2016 Genet Evol Comput Conf - GECCO ‘16 485–492. https://doi.org/10.1145/2908812.2908918
Cohen J (1960) A coefficient of agreement for nominal scales. Educ Psychol Meas 20:37–46. https://doi.org/10.1177/001316446002000104
Natekin A, Knoll A (2013) Gradient boosting machines, a tutorial. Front Neurorobot 7. https://doi.org/10.3389/fnbot.2013.00021
Wang S, Summers RM (2012) Machine learning and radiology. Med Image Anal 16:933–951. https://doi.org/10.1016/j.media.2012.02.005
Parmar C, Grossmann P, Bussink J et al (2015) Machine learning methods for quantitative radiomic biomarkers. Sci Rep 5:13087. https://doi.org/10.1038/srep13087
Larue RTHMHM, Defraene G, De Ruysscher D et al (2017) Quantitative radiomics studies for tissue characterization: a review of technology and methodological procedures. Br J Radiol 90:20160665. https://doi.org/10.1259/bjr.20160665
Scheffler K, Lehnhardt S (2003) Principles and applications of balanced SSFP techniques. Eur Radiol 13:2409–2418. https://doi.org/10.1007/s00330-003-1957-x
Neycenssac F (1993) Contrast enhancement using the Laplacian-of-a-Gaussian filter. CVGIP Graph Model Image Process 55:447–463. https://doi.org/10.1006/cgip.1993.1034
Rahaim NSA, Whitby EH (2015) The MRI features of placental adhesion disorder and their diagnostic significance: systematic review. Clin Radiol 70:917–925. https://doi.org/10.1016/j.crad.2015.04.010
Tang X (1998) Texture information in run-length matrices. IEEE Trans Image Process 7:1602–1609. https://doi.org/10.1109/83.725367
Blaicher W, Brugger PC, Mittermayer C et al (2006) Magnetic resonance imaging of the normal placenta. Eur J Radiol 57:256–260. https://doi.org/10.1016/j.ejrad.2005.11.025
Haralick RM, Shanmugam K, Dinstein I (1973) Textural features for image classification. IEEE Trans Syst Man Cybern SMC-3:610–621. https://doi.org/10.1109/TSMC.1973.4309314
Funding
This study was supported by the National Natural Science Foundation of China (81601458), the Ministry of Science and Technology of China (2016YFC0100803, 2018YFC1004603), the Health and Family Planning Commission of Sichuan Province (16ZD017), and Bureau of Science and Technology of Chengdu City (2014-HM01-00314-SF).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Guarantor
The scientific guarantor of this publication is S. Zhou.
Conflict of interest
The authors declare that they have no conflict of interest.
Statistics and biometry
One of the authors (H. Sun) has significant statistical expertise.
Informed consent
Written informed consent was waived by the Institutional Review Board.
Ethical approval
Institutional Review Board approval was obtained.
Methodology
• retrospective
• cross-sectional study
• performed at one institution
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
ESM 1
(DOCX 31312 kb)
Rights and permissions
About this article
Cite this article
Sun, H., Qu, H., Chen, L. et al. Identification of suspicious invasive placentation based on clinical MRI data using textural features and automated machine learning. Eur Radiol 29, 6152–6162 (2019). https://doi.org/10.1007/s00330-019-06372-9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00330-019-06372-9