Abstract
Radiomics refers to the process of conversion of conventional medical images into quantifiable data (“features”) which can be further mined to reveal complex patterns and relationships between the voxels in the image. These high throughput features can potentially reflect the histology of biologic tissues at macroscopic and microscopic levels. Several studies have investigated radiomics of hepatocellular carcinoma (HCC) before and after treatment. HCC is a heterogeneous disease with diverse phenotypical and genotypical landscape. Due to this inherent heterogeneity, HCC lesions can manifest variable aggressiveness with different response to treatment options, including the newer targeted therapies. Hence, radiomics can be used as a potential tool to enable patient selection for therapies and to predict response to treatments and outcome. Additionally, radiomics may serve as a tool for earlier and more efficient assessment of response to treatment. Radiomics, radiogenomics, and radio-immunoprofiling and their potential roles in management of patients with HCC will be discussed and critically reviewed in this article.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
The recent decade has witnessed an exponential growth in field of radiomics with numerous published studies exploring the potential applications of radiomics in patient-level tumor characterization and prognostication. Radiomics refers to the process of converting medical images into quantitative imaging data (“features”) that can be mined in order to reveal complex patterns reflective of biology at the macro- and microscopic levels [1]. This task can be achieved by semantic analysis or by more complex voxel-level computational analysis. Semantic analysis is performed using a qualitative or semi-quantitative analysis of imaging studies using lesion descriptors and/or standardized reading scores by a radiologist. Examples of semantic analysis include description of the tumor enhancement pattern, appearance of the border, presence of intra-lesional arteries, presence of vascular invasion as well as the Liver Imaging Reporting and Data System v2018 (LI-RADS®) lexicon and scoring scheme. Quantitative image descriptors that are defined mathematically to describe imaging phenotypes are known as agnostic features, which can potentially remedy many of the limitations of semantic analysis such as subjectivity and inter- and intraobserver variabilities. Examples include texture features, histogram-derived parameters, and quantitative indices of tumor heterogeneity.
Given the inherent intra- and inter-tumoral heterogeneity of hepatocellular carcinoma (HCC) and selective success of newer targeted systemic agents, there has been a significant shift in the research priorities with an urgent need to perform patient-level HCC characterization to enable proper patient selection for available therapies. Advanced molecular and immunoprofiling of HCC have potential to achieve these objectives as certain gene signatures and immune phenotypes obtained from these analyses are associated with aggressive tumor behavior and poor patient outcomes [2,3,4,5]. However, these techniques require invasive tissue sampling (which is infrequently performed in daily practice) and specialized equipment and are costly. Radiogenomics and radio-immunoprofiling are emerging areas of study that focus on identifying relationships between imaging traits, genomic markers, and immune profiles of tumors and assess tumor heterogeneity to inform on patient specific tumor biology. The role of radiomics for predicting the pathology of HCC including tumor grade, presence of microvascular invasion, and certain immunohistochemical markers has been described in recently published reviews [6, 7]. Potential and promising applications of radiomics in non-invasive characterization of HCC genetic and immune profile and in prediction as well as assessment of treatment response will be the focus of review in this article.
Techniques and workflow
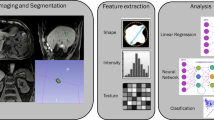
Image analysis, radiomics data curation, and data interpretation should ideally follow a meticulous and systematic workflow in order to ensure accuracy and reproducibility of the results (Fig. 1). Different methods and different software, including the proprietary software, have been used for each of these steps, which are discussed here. Some of the commonly used software include pyRadiomics [8], IBEX [9], TexRad, CERR [10], LifeX [11], and RaCaT [12].
Flowchart of the radiomics workflow demonstrating each step of the pipeline: imaging acquisition and post-processing, tumor segmentation, image pre-processing, feature extraction, selection, and modeling steps. ROI: region of interest; VOI: volume of interest; GLCM: gray level co-occurrence matrix; GLRM: gray level run length matrix; GLSZM: gray level size zone matrix; NGTDM: neighborhood gray tone difference matrix
Segmentation and image pre-processing
Segmentation is the process of delineating a region of interest (ROI) or a volume of interest (VOI), which may include part of, or the entire, tumor. Manual segmentation performed by an experienced radiologist, using computer software to outline the boundaries and to add the ROIs or VOIs in a slice-by-slice manner, is considered the gold standard. Several semi-automated and automated segmentation tools have been designed in past decade, which still have limited application in the clinical settings. State-of-the-art deep learning algorithms are promising and have demonstrated high capability in fast and accurate segmentation of medical images [13]. Despite their accuracy and reproducibility in segmentation of measurable tumors comparable to a gold standard, these algorithms can occasionally fail and require manual corrections.
Image pre-processing aims at facilitating a reproducible and reliable radiomic analysis. Inherent differences between the scanners and variations in acquisition parameters and scanning protocols unavoidably add systematic variation into the images. Removing or minimizing the effect of such variability is an essential part of the image pre-processing. Voxel size is one of the most important imaging parameters that varies significantly depending on the modality, vendor, and imaging protocols. Various radiomic features demonstrate dependence on the voxel size and the number of voxels within the ROI [14,15,16]. Voxel size resampling is required as a pre-processing step for datasets acquired with variable voxel sizes. Most common interpolation algorithms for voxel resampling utilize linear, polynomial, or spline interpolation techniques [17]. A number of other techniques have also been proposed to reduce the dependence on number of voxels within the ROI such as normalizing size-dependent features by number of voxels [14, 15].
Radiomics features are calculated from matrices that describe various spatial relationship between signal intensities of the voxels. In many cases, intensity values vary in a very wide range. The intensity discretization is a process of reducing the number of intensity values by stratifying them and using intensity levels instead. Each intensity level represents a range of the signal intensities that fall into the “bin”. Signal intensity discretization helps to reduce the number of intensity values as well as to minimize and suppress the voxel-level noise. The commonly used approaches are fixed bin size and fixed bin number [18]. In the fixed bin size method, the bin has a constant width and the number of bins varies depending on the dynamic range of signal intensities. This approach keeps the direct relationship with the original intensity scale and preserves the contrast differences. The second method is based on sorting of voxels between a fixed number of bins with the bin width decided based on the range of signal intensities. This approach adjusts the contrast between the two images and is more applicable when the imaging modality is not well calibrated.
Feature extraction
Different radiomic features are introduced to capture unique characteristics of the tissue through the quantification of various image-texture properties. Feature extraction is performed on imaging datasets using data processing software that often provides hundreds of radiomic features per segmentation. The radiomic features can be categorized into several groups including: 1) morphological features; 2) intensity-based statistical features; 3) intensity histogram features; 4) second-order statistical features; and 5) higher-order features [19]. Morphological features quantify the size and shape of the target object, based on two or three-dimensional segmentation. These features are independent from the signal intensity distribution within the ROI. They include diameter, cross-section area, volume, surface area, and many measures of shape asymmetry such as compactness, flatness, sphericity as well as minor and major axis length. Intensity-based features are computed using statistical moments of the signal intensity histogram. They include minimum and maximum intensity, range, mean, median, standard deviation, interquartile range, skewness and kurtosis to name a few. The intensity histogram is generated by discretizing the original intensity distribution into intensity bins and then computing statistical moments of the histogram. Many of these quantities are commonly used in everyday radiology practice and are familiar to all radiologists. Second-order statistical features quantify the spatial arrangement of the voxel-intensity levels within the ROI and include gray-level co-occurrence matrix (GLCM), gray level run length matrix (GLRLM), gray level size zone matrix (GLSZM), neighboring gray tone difference matrix (NGTDM), and gray level dependence matrix (GLDM) features [20, 21] (Fig. 2). The second-order texture features can also be extracted from transformed images by applying Laplacian of Gaussian (LoG) wavelet and other filters [19, 22]. Higher-order features include Fourier spectral analysis, wavelet transform-based multiscale features, and fractal analysis [23, 24]. The wavelet features quantify the local structure of the tumor corresponding to spatial scales, localization, and orientation. The power spectrum of the Fourier transform defines the relationships between the domain intensity and the domain size, whereas the fractal analysis (fractal dimensionality) quantifies the special distribution of different domains within the tumor [22, 25].
a Contrast-enhanced CT of 50-year-old male with hepatocellular carcinoma (HCC). The yellow line defines tumor boundary. b The CT texture heterogeneity map demonstrating the presence of two main domains; the first one (red–orange colors) with high average attenuation and the second one (blue-gray colors) with low average attenuation. c The intensity histogram computed from the region of interest above. The histogram can be fitted to a sum of two Gaussian curves with corresponding mean 44.3 HU and 79.9 HU and the standard deviation 16.9 HU and 14.9 HU, correlating with texture heterogeneity map. d The GLCM, GLRLM and GLZLM matrices computed from ROI after the intensity discretization to 32 Gy levels. GLCM: gray level co-occurrence matrix; GLRLM: gray level run length matrix; GLZLM: gray level zone length matrix
“Delta-radiomics” is defined as interval changes in radiomic features between baseline and follow-up study and is often used in longitudinal studies to assess individual response to treatment (Fig. 3). Two most common approaches for calculation of “delta-radiomic features” include: (1) assessing the ratio of changes relative to the baseline (delta-radiomics = [follow-up radiomics − baseline radiomics]/baseline radiomics); and (2) assessing velocity of changes over time (delta-radiomics = [follow-up radiomics − baseline radiomics]/time interval). Several studies have utilized delta-radiomics to assess response to treatment or to predict clinical outcome [26].
Baseline (a and b) and 1-month follow-up (c and d) CT scan of 64-year-old male with HCC obtained during the late arterial phase. A 2D low-pass (LL) wavelet transform of the scans highlights the region of response to sorafenib chemotherapy. Tumor boundaries (ROI) are marked with a black contour line. Mean wavelet intensity (± standard deviation) within the ROI are 133.3 ± 35.8 at the baseline scan and 91.1 ± 34.5 at the follow-up scan, respectively. In addition, a number of radiomics features (such as GLCM Energy and GLRLM LGRE) demonstrate a significant interval change between the baseline and the follow-up studies. GLCM: gray level co-occurrence matrix; GLRLM: gray level run length matrix; LGRE: low gray-level run emphasis
Reproducibility and feature selection
Higher-level radiomic feature values are affected by the feature extraction algorithms and software. Validation and reproduction of the prior results is challenging given the inconsistencies in the definition and computation of radiomic features by different software. Recently, the Image Biomarker Standardization Initiative consisting of 25 global research teams provided a consensus-based standardization for the definition of radiomic features and for the workflow of radiomic image processing [27]. Their study showed good to excellent reproducibility of the majority of radiomic features (169 features) across different modalities after standardization of definitions and techniques [27].
Features computed from the same matrix (e.g., GLCM) are highly correlated and dependent to each other. Inclusion of redundant and clustered data into a radiomic model can lead to an increased risk of overfitting and causing false-positive associations. An over-fitted model is not generalizable and leads to poor performance on other datasets. To avoid the risk of overfitting and to increase efficiency of radiomic data, only robust features with high reproducibility should be selected for model development. Feature selection aims to reduce redundancy, eliminate irrelevant features, and identify most significant features with highest prognostic value. It also helps to simplify the models without compromising their accuracy. Feature selection can be accomplished through a variety of statistical methods [28]. For instance, the Pearson correlation coefficient and the Lin’s concordance correlation coefficient (CCC) are methods used to evaluate association between different radiomic features and to perform test–retest robustness analysis. The feature selection could be performed using one of several regression methods, such as the least absolute shrinkage and selection operator (LASSO) [29]. There are also several metrics for evaluation of image segmentation performance and reproducibility, such as the Dice similarity coefficient (DSC).
Model building and performance evaluation
The goal of radiomic analysis is to construct a statistical model for prediction of a clinical occurrence such as diagnosis, prognosis, or response assessment. A radiomic model can be constructed using one of many multivariable models. Random Forest (a decision tree classifier) methodology is one of the most powerful statistical techniques for building classification models. Regularized regression techniques (e.g., using LASSO or Elastic Nets) are other popular methods for the development of machine learning classifiers from radiomics features.
The radiomic data used to build a model is ideally partitioned between three datasets. The training dataset is used during the learning process to fit the data and to assess model parameters and weights. Validation dataset provides an unbiased evaluation of the model parameters and accuracy and allows for adjustment of the model hyperparameters and classifiers. The model hyperparameters are variables that define the model configuration and cannot be estimated from the data, for example the “learning rate” for training a neural network or the k in k-fold cross-validation. In contrary, the model classifiers are variables that are estimated from the data, e.g., linear regression coefficients or weights associated with each neural network connection. Lastly, a test dataset is used to assess the final model fit. Model performance evaluation aims to assess its generalizability and accuracy in data that were not present in the original dataset. Preferably, generalizability of the models should be assessed using an external validation cohort. Open-access public domains such as Cancer Imaging Archive (NCI TCIA; funded by the Cancer Imaging Program of the National Cancer Institute) have been made available for this purpose [30]. Preliminary studies may not have sufficient data for external validation and may instead use internal validation techniques (such as random sampling, k-fold, and bootstrap cross-validation) using the original dataset [27]. Frequently, model performance is evaluated by comparing validation and test dataset outcomes. Classifier performance is usually assessed by the area under the receiver operating characteristics curve (AUC-ROC) including metrics such as sensitivity, specificity and accuracy. Metrics such as mean absolute error, mean squared errors or R2 are utilized to assess performance of regression problems. AUC-ROC metrics depend on the balance between the outcome classes in the data and should be used and interpreted with caution. Balanced accuracy and Matthews’s correlation coefficient should be considered as alternatives [18].
The reproducibility of radiomics models depends on reproducibility of radiomic features and accuracy of the statistical model. Harmonization of imaging data are essential to overcome experimental variation across multiple scanners and protocols. This is of more importance in case of multi-center and retrospective studies [27].
Radiogenomics of HCC
A considerable amount of work has been conducted over the past two decades to characterize the molecular classifications of HCC [31]. Genome-wide molecular profiling of HCC tumors has identified heterogeneous molecular aberrations across lesions, indicating significant inter-tumoral heterogeneity [32]. A variety of classification schemes have been used for genomic landscapes in HCC. Two main molecular classes of HCC are described: (1) proliferation class, typically associated with hepatitis B virus infection, has poor tumor differentiation and elevated serum alpha fetoprotein (AFP) level, and is associated with worsened outcomes; and (2) non-proliferation class, more commonly seen with hepatitis C virus infection or alcohol-related HCC, has moderate/well tumor differentiation, lower AFP values, and better outcomes [31]. In addition to the biological variability across HCC tumors, significant intratumoral heterogeneity has also been observed at histopathological and molecular genomic levels [33, 34]. Biopsies generally sample only a small portion of the tumor, precluding analysis of intra-tumoral heterogeneity within the entire lesion.
Semantic imaging traits and genomics
The majority of the radiogenomic studies to date have been based on semantic features obtained from CT or MRI. The commonly used semantic features include description of the tumor enhancement pattern, appearance of the border, presence of intra-tumoral arteries, peritumoral halo, vascular invasion or necrosis as well as the American College of Radiology Liver Reporting and Data System (ACR LIRADS ®) scoring. A greater number of radiogenomics studies have used CT compared to MRI, which may be due to the greater challenges in standardizing MR data compared with CT data. Approximately 78% of HCC global gene expression profiles could be reconstructed using combinations of 28 qualitative traits at contrast-enhanced CT [35]. In a retrospective study of 39 patients, qualitative imaging traits on CT and MRI (including infiltrative pattern, mosaic appearance, presence of macrovascular invasion, and large size) were shown to have significant associations with gene signatures of aggressive HCC phenotype (G3-Boyault, Proliferation-Chiang profiles, CK19-Villanueva, S1/S2-Hoshida) with odds ratios ranging from 4.44–12.73 (p < 0.045) [36]. These particular genes were associated with increased cellular proliferation, vascular invasion, distant metastases, and poor prognosis. In a separate study, mosaic appearance of tumor was shown to correlate with expression level of POSTN gene (r = − 0.47, p = 0.036), which is marker of tumor aggressiveness [37]. In another study, tumor size showed significant association (p = 0.005) with fractional allelic imbalance (FAI) rate index, which is a molecular marker associated with early tumor recurrence in liver transplant patients, while LI-RADS classification showed non-significant weak positive correlation (r = 0.264) [38]. The tumor margins on arterial phase were qualitatively assessed in a separate study which showed correlation with a doxorubicin response gene expression profile (p < 0.05), which may aid treatment planning for patients receiving trans-arterial chemoembolization (TACE) [39]. While semantic analysis is commonly performed in routine clinical practice, these assessments are prone to limited inter-/intra-observer reproducibility, which makes data analysis more challenging to standardize and to evaluate on a large scale [40].
Agnostic imaging traits and genomics
Agnostic features have recently shown value for prediction of gene signatures or potentially actionable targets with prognostic and therapeutic implications. In a retrospective study of 38 patients with contrast-enhanced CT, texture features correlated with prognostic gene modules that were associated with overall survival [41]. In a separate retrospective study of 48 patients, minimum ADC values obtained from histogram analysis enabled distinction between aggressive molecular subtypes (S1/S2) versus less aggressive subtype (S3) using the transcriptomic classification described by Hoshida et al. (odds ratio = 2.00, p = 0.036, AUC = 0.68) [37]. In the same study, radiomics features (10 Haralick texture features, 1 other quantitative feature) showed correlation with 14 gene expression levels (r = − 0.61–0.56, p < 0.043) (Fig. 4) [37]. The majority of correlations were found with molecular subclass signature genes, the strongest of which was between early arterial phase texture and expression of binding protein TARBP1 (r = − 0.61, p = 0.013). Regarding the therapeutic targets, correlations were demonstrated between late arterial phase texture features and immunotherapy targets PD-1 and CTLA4 mRNA expression level (r = − 0.48–0.47, p < 0.037) (Fig. 5) [37]. Associations between histogram parameters obtained from functional multiparametric MRI (DWI, blood-oxygenation level-dependent (BOLD), tissue-oxygenation level-dependent (TOLD), and dynamic contrast-enhanced MRI) and both molecular and immune characteristics of HCC were evaluated in a prospective study [42]. Central tendency parameters (mean and median) significantly correlated with gene expression of Wnt target GLUL, pharmacological target FGFR4, stemness markers EPCAM and KRT19, and immune checkpoint PDCD1 [42].
Clustergram of significant correlations, expressed as the Spearman correlation coefficient r, between radiomics features and gene expression levels. Only radiomics features and genes for which a significant association (FDR-adjusted p < 0.05) was observed are shown. Correlations are colored according to the color bar shown on the left. Nonsignificant correlations are displayed with a checkerboard pattern. Radiomics features, in particular texture features, showed significant association with gene expression levels of HCC markers and therapeutic targets. ADC apparent diffusion coefficient, EAP early arterial phase, ER enhancement ratio, HBP hepatobiliary phase, LAP late arterial phase, PVP portal venous phase. Adapted with permission from Hectors et al. [37]
Energy texture maps overlaid on T1-weighted post-contrast image obtained during the late arterial phase image. Left: 59-year-old male HBV patient with a 3.7-cm HCC lesion in right hepatic lobe. Energy texture map overlaid on T1-weighted post-contrast image obtained during the late arterial phase image. Texture feature Energy with 16 bins was 0.025 in the lesion, with corresponding molecular expression of immunotherapy target CTLA4 of 0.44. Right: 51-year-old male HBV patient with a 3.3-cm HCC lesion in right hepatic lobe. Texture feature energy with 16 bins was 0.061 in the lesion, with corresponding molecular expression of immunotherapy target CTLA4 of 0.68. The texture feature maps are colored according to the scale bar on the right with a range of [0–0.1]. Adapted with permission from Hectors et al. [37]
Radio-immunoprofiling
The success of targeted immunotherapeutic agents is determined by the immune status of the HCC tumors [43]. Gene expression analysis in 956 patients with HCC has identified a distinct immune class, characterized by expression of inflammatory response markers, in 25% of patients [44]. This “Immune activated” phenotype is characterized by dense functional CD8 cell infiltration, increased interferon-γ signaling, expression of cell checkpoint markers (such as PD-L1), and a high mutational burden [45]. HCCs with this immune profile may best respond to targeted immunotherapeutic agents [44, 46]. Furthermore, the “immunoscore”, calculated based on analysis of the type, functional orientation, density, and spatial location of tumor-infiltrating lymphocytes, was significantly associated with tumor recurrence rate and recurrence-free survival [47,48,49,50]. Therefore, immunoprofiling of HCCs is highly relevant for predicting the response to immunotherapy [2]. Multiplexed immunohistochemistry is a promising advanced technique for evaluation of immune cell distribution and localization in tumors [3]. This technique however requires invasive tissue sampling, equipment, and expertise.
Preliminary works have shown value in radiomics quantification (using both semantic and agnostic features) for HCC immune profiling. In a large-scale study of 207 HCC patients, radiomics data obtained from HCC tumors and the peritumoral regions extracted from gadoxetic-acid-enhanced MRI accurately predicted HCC immunoscore, specifically evaluating CD3 and CD8 T-cells, with AUC of 0.904 [51]. In the study by Hectors et al., central tendency parameters from functional mpMRI significantly correlated with the count of endothelial cells (CD31 monoclonal antibody), macrophages (CD68), and T-cells (CD3) (p < 0.05) [42]. In a separate study, despite showing no associations with histopathologic grade or microvascular invasion, radiomics measurements (texture features and quantitative enhancement ratios) were associated with expression of immune markers CD3, CD31, CD68, and PD-L1. The greatest number of correlations was found with CD68 (macrophage) expression, including several texture features (r = 0.33–0.45, p < 0.049), mosaic appearance (r = 0.40, p = 0.023), and wash-in/wash-out enhancement pattern (r = 0.37, p = 0.042) [37]. Another study showed that radiomics (Rad) score of features extracted from contrast-enhanced CT positively correlated with the percentage of infiltrating CD8 + lymphocytes (r = 0.51, p < 0.0001) and could discriminate immune activated from immune-desert and immune-excluded tumors (p < 0.0001) [52]. Higher Rad scores were identified in patients with positive PD1 expression (p < 0.0001), PD-L1 immune cell expression (p < 0.0001), and PD-L1 tumor cell expression (p = 0.01498).
In a recent CT-based multi-cohort study of patients with solid organ malignancy (including HCC), a radiomics-based model based on eight variables (including one first-order and four second-order GLRLM features) successfully discriminated immune activated tumors from immune-desert tumors with an AUC of 0.76 (95%CI 0.66–0.86; p < 0.0001) [45].
Radiomics for assessment of treatment response
Assessment of treatment response to locoregional treatment
Numerous locoregional therapeutic approaches have been developed and introduced in past decades with many of them now considered standard of care [53]. They encompass various percutaneous and trans-arterial techniques aiming at tumoral cell death by direct tissue destruction (such as in case of radiofrequency and microwave ablation) or by selective trans-catheter trans-arterial delivery of embolic (trans-arterial bland embolization, TAE), chemotherapeutic (trans-arterial chemoembolization, TACE) or radioembolizing agents (trans-arterial Yttrium-90 radio-embolization, TARE). Locoregional treatments are prescribed for a wide range of indications and are used for both curative and palliative purposes [54]. Although generally less invasive than surgical resection and transplantation, these treatments can result in morbidities therefore appropriate patient selection and effective assessment of treatment response are of major clinical interest.
Current criteria for assessment of HCC treatment response such as ACR LIRADS v2018® and modified Response Evaluation Criteria in Solid Tumors (mRECIST) criteria are better suited for locoablative and TACE methods [55]. Assessment of treatment response after TARE and external beam radiation, especially in the first few months after treatment, remains challenging due to the inherent nature and time course of radiation-induced cytotoxic effects [56]. Persistent arterial enhancement and washout may be seen in the treated lesions for several months after the treatment, despite the eventual complete response [56]. Given these limitations, there is a need for having alternative methods to assess response to treatment.
Several studies have investigated role of radiomic features extracted from post-treatment CT and MRI for assessment of HCC treatment response. A prospective study of 27 patients with 56 HCCs showed high correlation (r = -0.815; p < 0.005) between the changes in CT-based textural features (heterogeneity and skewness) and perfusion indices of tumor (based on CT perfusion study) after TACE [57]. A subsequent retrospective study of 89 patients who underwent combined TACE and high intensity focused ultrasound (HIFU) showed that radiomics (uniformity, energy, entropy, and skewness) based on 1-week post-treatment MRI correlated with radiologic response on 3-month follow-up MRI [58]. A newly published retrospective study of 22 patients who underwent lobar TARE showed volumetric histogram analysis of apparent diffusion coefficient (ADC) map as a potential tool for early assessment of tumor response to TARE [59]. While the histogram-based radiomics of ADC on baseline study were not different, the ADC median/mode at 6-week post-treatment study and changes in ADC median/maximum from the baseline study were significantly higher in patients with complete or partial response (assessed based on 6-month post-treatment MRI).
While early and precise assessment of treatment response is essential in patient management, the identification of patients who may be responders to therapy before the initiation of treatment, can help with patient selection and further enhance the precision patient care. Multiple studies investigated the role of radiomics at baseline imaging as predictor of response to locoregional treatment, mainly before TACE, and showed that several radiomics features (extracted from pre-treatment CT/MRI) correlated with the future response to locoregional treatment. In the recent study by Sun et al. a model based on radiomics features extracted from pre-treatment DWI, ADC, and T2-weighted MRI had AUC of 0.8 (p < 0.001) for prediction of progressive disease after TACE [60]. Two other studies showed correlation between pre-treatment contrast-enhanced-CT-derived texture features and mid-term (at 6 month) response to drug-eluting bead TACE [57, 61]. Texture parameters (uniformity, energy, and entropy) assessed on 1-week pre-treatment MRI were also shown to correlate with response to combined treatment with TACE and HIFU [58]. In a series of 132 HCCs undergoing traditional TACE, indices of higher intra-tumoral heterogeneity (high GLCM moments and low homogeneity) correlated with complete response to treatment (odds ratios up to 6.57, p < 0.001) [62].
Assessment of treatment response to systemic therapy
Systemic therapy is increasingly used to treat patients with advanced stage HCC and the patients who are not candidates for surgical or loco-regional therapies [63]. Antiangiogenetic therapy with Sorafenib, a protein kinase inhibitor, proved to be effective in increasing overall survival in patients with advanced disease [64, 65] and is now considered as the treatment of choice in Stage C HCC [66]. New tyrosine kinase inhibitors and immune checkpoint inhibitors are shown to have superior outcomes compared to sorafenib and in patients who previously failed sorafenib therapy. Recently, a combination of atezolizumab (an anti-programmed cell death ligand 1; anti-PDL1) and bevacizumab as first-line therapy in advanced HCC patients provided objective response rate (ORR) of up to 36% and better overall and progression-free survival outcomes compared to sorafenib [67,68,69]. Currently, there are no clinical or molecular biomarkers to predict response to systemic treatment. Only few studies have been published yet to prove the ability of radiomics in assessing treatment response in patients eligible for systemic therapy. A retrospective study of 92 patients with HCC treated with sorafenib showed that entropy derived from pre-treatment portal venous phase CT was an independent predictor of survival (p < 0.05) [70]. This result could be explained by higher efficacy of anti-angiogenetic therapy in heterogeneous lesions presumably due to their higher number of unpaired arteries. Promising preliminary results have also shown that texture analysis may help in patient selection for combined treatment with TACE and Sorafenib [71].
Radiomics may also be employed for prediction of response to immune-oncologic therapy and assessment of patient outcome. For example, a recent study in solid organ malignancy including HCC found that patients treated with anti-PD-1 and PD-L1 who had a high baseline radiomic score, largely derived from GLRLM features, had a higher chance of objective response at 3 months (p = 0.049) and 6 months (p = 0.025). These patients also had superior median overall survival based on both univariate analysis (median OS 24.3 months in the high radiomic score group versus 11.5 months in the low radiomic score group) and multivariate analysis (HR 0.52, 95%CI 0.35–0.79, p = 0.0022)[45]. In a separate study, a radiomics-based biomarker using machine learning statistics was able to predict the infiltration of HCC by tumor-infiltrating CD8 + T cells. A higher score correlated with superior overall survival and disease-free survival (p = 0.012 and p = 0.0088, respectively)[52]. The radiomics scores in these studies were largely comprised of GLRLM features, which are thought to reflect the heterogeneity of an image, and GLCM features, which are thought to reflect the structural properties of images.
Radiomics for prediction of survival
Radiomics is an emerging field in clinical setting and larger and longer interval studies are needed to validate its promising results in directing patient management. A recent retrospective study analyzed the ability of radiomics analysis in predicting long-term survival by combining several clinical, radiological, and radiogenomic factors. The proposed Cox proportional hazard model predicted 3-year, 5-year, and 10-year survival in patients undergoing TACE with an AUC of 0.85, 0.90, and 0.89, respectively [72]. Another recent retrospective study of 162 patients with HCC proposed a combined model based on clinical parameters and 6 radiomics features (extracted from pre-treatment CT) to predict overall survival after TACE. The model outperformed the other clinical models (C-index of 0.73, 95%CI 0.68–0.79) [73]. Other studies also showed that combined models based on radiomics and clinical features outperformed the individual models in prediction of overall survival in patients with HCC [74,75,76]. Peritumoral radiomics features may also play a role for risk assessment and prediction of tumor recurrence after treatment [73]. A radiomics signature derived from pre-treatment FDG-PET and including both tumor and background liver was shown to be an independent predictor of overall survival and progression-free survival in patients undergoing Y90-TARE for unresectable HCC, with the strength and variance being the most predictive texture features [77].
Future directions
The majority of published literature on radiomics is based on small cohorts, and hence their proposed models could not be fully corroborated. The initial results reported herein will ultimately require independent validation in larger datasets and populations before incorporating them into decision-making algorithms and in clinical practice. Recent efforts by Image Biomarker Standardization Initiative is a promising step for standardization of radiomics workflow and for rigorous data quality control which is essential for generalization of radiomics results [27]. Application of deep learning in large-scale HCC radiomics (termed “deep radiomics”) is another exciting future direction for extracting tumor features, characterizing tumor aggressiveness, and predicting immune response and patient outcome.
Conclusion
Radiomics quantification is a powerful, noninvasive tool to extract numerous features on the whole lesion level which may be reflective of HCC heterogeneity and aggressiveness based on gene expression profiling and immunophenotyping, and could potentially inform on an optimal therapeutic approach and response to therapy. Improved identification of potential responders to novel immunotherapeutic agents could enable a personalized and cost effective patient specific treatment strategy. Radiomics has not yet reached widespread application in the clinical setting, with many existing barriers to translation, and further prospective and multi-institutional work is thus warranted to validate the results of these initial studies.
References
Gillies, R.J., P.E. Kinahan, and H. Hricak, Radiomics: Images Are More than Pictures, They Are Data. Radiology, 2016. 278(2): p. 563-77.
Khemlina, G., S. Ikeda, and R. Kurzrock, The biology of Hepatocellular carcinoma: implications for genomic and immune therapies. Mol Cancer, 2017. 16(1): p. 149.
Kurebayashi, Y., et al., Landscape of immune microenvironment in hepatocellular carcinoma and its additional impact on histological and molecular classification. Hepatology, 2018. 68(3): p. 1025-1041.
Villanueva, A., et al., New strategies in hepatocellular carcinoma: genomic prognostic markers. Clin Cancer Res, 2010. 16(19): p. 4688-94.
Cariani, E., et al., Immunological and molecular correlates of disease recurrence after liver resection for hepatocellular carcinoma. PLoS One, 2012. 7(3): p. e32493.
Lewis, S., S. Hectors, and B. Taouli, Radiomics of hepatocellular carcinoma. Abdom Radiol (NY), 2020.
Miranda Magalhaes Santos, J.M., et al., State-of-the-art in radiomics of hepatocellular carcinoma: a review of basic principles, applications, and limitations. Abdom Radiol (NY), 2020. 45(2): p. 342–353.
van Griethuysen, J.J.M., et al., Computational Radiomics System to Decode the Radiographic Phenotype. Cancer Res, 2017. 77(21): p. e104-e107.
Zhang, L., et al., IBEX: an open infrastructure software platform to facilitate collaborative work in radiomics. Med Phys, 2015. 42(3): p. 1341-53.
Apte, A.P., et al., Technical Note: Extension of CERR for computational radiomics: A comprehensive MATLAB platform for reproducible radiomics research. Med Phys, 2018.
Nioche, C., et al., LIFEx: A Freeware for Radiomic Feature Calculation in Multimodality Imaging to Accelerate Advances in the Characterization of Tumor Heterogeneity. Cancer Res, 2018. 78(16): p. 4786-4789.
Pfaehler, E., et al., RaCaT: An open source and easy to use radiomics calculator tool. PLoS One, 2019. 14(2): p. e0212223.
Chlebus, G., et al., Automatic liver tumor segmentation in CT with fully convolutional neural networks and object-based postprocessing. Scientific Reports, 2018. 8(1): p. 15497.
Fave, X., et al., Impact of image preprocessing on the volume dependence and prognostic potential of radiomics features in non-small cell lung cancer. Translational Cancer Research, 2016. 5(4): p. 349-363.
Shafiq-Ul-Hassan, M., et al., Intrinsic dependencies of CT radiomic features on voxel size and number of gray levels. Medical physics, 2017. 44(3): p. 1050-1062.
Velichko, Y.S., et al., Association Between the Size and 3D CT-Based Radiomic Features of Breast Cancer Hepatic Metastasis. Academic Radiology, 2020.
Lehmann, T.M., C. Gonner, and K. Spitzer, Addendum: B-spline interpolation in medical image processing. IEEE Transactions on Medical Imaging, 2001. 20(7): p. 660-665.
Duron, L., et al., Gray-level discretization impacts reproducible MRI radiomics texture features. PLOS ONE, 2019. 14(3): p. e0213459.
Zwanenburg, A., et al., Image biomarker standardisation initiative-feature definitions. arXiv preprint arXiv:1612.07003, 2016.
Haralick, R.M., K. Shanmugam, and I. Dinstein, Textural Features for Image Classification. IEEE Transactions on Systems, Man, and Cybernetics, 1973. SMC-3(6): p. 610–621.
Parekh, V. and M.A. Jacobs, Radiomics: a new application from established techniques. Expert review of precision medicine and drug development, 2016. 1(2): p. 207-226.
Yip, S.S. and H.J. Aerts, Applications and limitations of radiomics. Phys Med Biol, 2016. 61(13): p. R150-66.
Di Giovanni, P., et al., The biological correlates of macroscopic breast tumour structure measured using fractal analysis in patients undergoing neoadjuvant chemotherapy. Breast Cancer Research and Treatment, 2012. 133(3): p. 1199-1206.
Balboa, R.M. and N.M. Grzywacz, Power spectra and distribution of contrasts of natural images from different habitats. Vision Research, 2003. 43(24): p. 2527-2537.
Baish, J.W. and R.K. Jain, Fractals and Cancer. Cancer Research, 2000. 60(14): p. 3683-3688.
Wang, X.-H., et al., MRI-based radiomics model for preoperative prediction of 5-year survival in patients with hepatocellular carcinoma. British Journal of Cancer, 2020. 122(7): p. 978-985.
Zwanenburg, A., et al., The Image Biomarker Standardization Initiative: Standardized Quantitative Radiomics for High-Throughput Image-based Phenotyping. Radiology, 2020. 295(2): p. 328-338.
Parmar, C., et al., Machine Learning methods for Quantitative Radiomic Biomarkers. Scientific Reports, 2015. 5(1): p. 13087.
Tibshirani, R., Regression Shrinkage and Selection via the Lasso. Journal of the Royal Statistical Society. Series B (Methodological), 1996. 58(1): p. 267–288.
Clark, K., et al., The Cancer Imaging Archive (TCIA): Maintaining and Operating a Public Information Repository. Journal of Digital Imaging, 2013. 26(6): p. 1045-1057.
Llovet, J.M., et al., Molecular therapies and precision medicine for hepatocellular carcinoma. Nat Rev Clin Oncol, 2018. 15(10): p. 599-616.
Goossens, N., X. Sun, and Y. Hoshida, Molecular classification of hepatocellular carcinoma: potential therapeutic implications. Hepat Oncol, 2015. 2(4): p. 371-379.
Friemel, J., et al., Intratumor heterogeneity in hepatocellular carcinoma. Clin Cancer Res, 2015. 21(8): p. 1951-61.
Liu, J., H. Dang, and X.W. Wang, The significance of intertumor and intratumor heterogeneity in liver cancer. Exp Mol Med, 2018. 50(1): p. e416.
Segal, E., et al., Decoding global gene expression programs in liver cancer by noninvasive imaging. Nat Biotechnol, 2007. 25(6): p. 675-80.
Taouli, B., et al., Imaging-based surrogate markers of transcriptome subclasses and signatures in hepatocellular carcinoma: preliminary results. Eur Radiol, 2017. 27(11): p. 4472-4481.
Hectors, S.J., et al., MRI radiomics features predict immuno-oncological characteristics of hepatocellular carcinoma. Eur Radiol, 2020. 30(7): p. 3759-3769.
Furlan, A., et al., A radiogenomic analysis of hepatocellular carcinoma: association between fractional allelic imbalance rate index and the liver imaging reporting and data system (LI-RADS) categories and features. Br J Radiol, 2018. 91(1086): p. 20170962.
Kuo, M.D., et al., Radiogenomic analysis to identify imaging phenotypes associated with drug response gene expression programs in hepatocellular carcinoma. J Vasc Interv Radiol, 2007. 18(7): p. 821-31.
Wakabayashi, T., et al., Radiomics in hepatocellular carcinoma: a quantitative review. Hepatol Int, 2019. 13(5): p. 546-559.
Xia, W., et al., Radiogenomics of hepatocellular carcinoma: multiregion analysis-based identification of prognostic imaging biomarkers by integrating gene data-a preliminary study. Phys Med Biol, 2018. 63(3): p. 035044.
Hectors, S.J., et al., Quantification of hepatocellular carcinoma heterogeneity with multiparametric magnetic resonance imaging. Sci Rep, 2017. 7(1): p. 2452.
Zhu, A.X., et al., Ramucirumab in advanced hepatocellular carcinoma in REACH-2: the true value of alpha-fetoprotein. Lancet Oncol, 2019. 20(4): p. e191.
Sia, D., et al., Identification of an Immune-specific Class of Hepatocellular Carcinoma, Based on Molecular Features. Gastroenterology, 2017. 153(3): p. 812-826.
Sun, R., et al., A radiomics approach to assess tumour-infiltrating CD8 cells and response to anti-PD-1 or anti-PD-L1 immunotherapy: an imaging biomarker, retrospective multicohort study. Lancet Oncol, 2018. 19(9): p. 1180-1191.
Ruiz de Galarreta, M., et al., beta-Catenin Activation Promotes Immune Escape and Resistance to Anti-PD-1 Therapy in Hepatocellular Carcinoma. Cancer Discov, 2019. 9(8): p. 1124–1141.
Gabrielson, A., et al., Intratumoral CD3 and CD8 T-cell Densities Associated with Relapse-Free Survival in HCC. Cancer Immunol Res, 2016. 4(5): p. 419-30.
Sun, C., et al., The predictive value of centre tumour CD8(+) T cells in patients with hepatocellular carcinoma: comparison with Immunoscore. Oncotarget, 2015. 6(34): p. 35602-15.
Yao, Q., et al., Prognostic value of immunoscore to identify mortality outcomes in adults with HBV-related primary hepatocellular carcinoma. Medicine (Baltimore), 2017. 96(17): p. e6735.
Garnelo, M., et al., Interaction between tumour-infiltrating B cells and T cells controls the progression of hepatocellular carcinoma. Gut, 2017. 66(2): p. 342-351.
Chen, S., et al., Pretreatment prediction of immunoscore in hepatocellular cancer: a radiomics-based clinical model based on Gd-EOB-DTPA-enhanced MRI imaging. Eur Radiol, 2019. 29(8): p. 4177-4187.
Liao, H., et al., Preoperative Radiomic Approach to Evaluate Tumor-Infiltrating CD8(+) T Cells in Hepatocellular Carcinoma Patients Using Contrast-Enhanced Computed Tomography. Ann Surg Oncol, 2019. 26(13): p. 4537-4547.
Inchingolo, R., et al., Locoregional treatments for hepatocellular carcinoma: Current evidence and future directions. World J Gastroenterol, 2019. 25(32): p. 4614-4628.
Bruix, J., M. Reig, and M. Sherman, Evidence-Based Diagnosis, Staging, and Treatment of Patients With Hepatocellular Carcinoma. Gastroenterology, 2016. 150(4): p. 835-53.
Chernyak, V., et al., Liver Imaging Reporting and Data System (LI-RADS) Version 2018: Imaging of Hepatocellular Carcinoma in At-Risk Patients. Radiology, 2018. 289(3): p. 816-830.
Mendiratta-Lala, M., et al., Natural history of hepatocellular carcinoma after stereotactic body radiation therapy. Abdom Radiol (NY), 2020. 45(11): p. 3698-3708.
Kloth, C., et al., Evaluation of Texture Analysis Parameter for Response Prediction in Patients with Hepatocellular Carcinoma Undergoing Drug-eluting Bead Transarterial Chemoembolization (DEB-TACE) Using Biphasic Contrast-enhanced CT Image Data: Correlation with Liver Perfusion CT. Acad Radiol, 2017. 24(11): p. 1352-1363.
Yu, J.Y., et al., Value of texture analysis based on enhanced MRI for predicting an early therapeutic response to transcatheter arterial chemoembolisation combined with high-intensity focused ultrasound treatment in hepatocellular carcinoma. Clin Radiol, 2018. 73(8): p. 758 e9–758 e18.
Gordic, S., et al., Prediction of hepatocellular carcinoma response to (90)Yttrium radioembolization using volumetric ADC histogram quantification: preliminary results. Cancer Imaging, 2019. 19(1): p. 29.
Sun, Y., et al., Predicting the Outcome of Transcatheter Arterial Embolization Therapy for Unresectable Hepatocellular Carcinoma Based on Radiomics of Preoperative Multiparameter MRI. J Magn Reson Imaging, 2020. 52(4): p. 1083-1090.
Reis, S.P., et al., Tumor Enhancement and Heterogeneity Are Associated With Treatment Response to Drug-Eluting Bead Chemoembolization for Hepatocellular Carcinoma. J Comput Assist Tomogr, 2017. 41(2): p. 289-293.
Park, H.J., et al., Prediction of Therapeutic Response of Hepatocellular Carcinoma to Transcatheter Arterial Chemoembolization Based on Pretherapeutic Dynamic CT and Textural Findings. AJR Am J Roentgenol, 2017. 209(4): p. W211-W220.
Llovet, J.M., et al., Advances in targeted therapies for hepatocellular carcinoma in the genomic era. Nat Rev Clin Oncol, 2015. 12(8): p. 436.
Llovet, J.M., et al., Sorafenib in advanced hepatocellular carcinoma. N Engl J Med, 2008. 359(4): p. 378-90.
Cheng, A.L., et al., Efficacy and safety of sorafenib in patients in the Asia-Pacific region with advanced hepatocellular carcinoma: a phase III randomised, double-blind, placebo-controlled trial. Lancet Oncol, 2009. 10(1): p. 25-34.
European Association for the Study of the Liver. Electronic address, e.e.e. and L. European Association for the Study of the, EASL Clinical Practice Guidelines: Management of hepatocellular carcinoma. J Hepatol, 2018. 69(1): p. 182–236.
[67] Finn, R.S. and A.L. Cheng, Atezolizumab and Bevacizumab in Hepatocellular Carcinoma. Reply. N Engl J Med, 2020. 383(7): p. 695.
Lee, M.S., et al., Atezolizumab with or without bevacizumab in unresectable hepatocellular carcinoma (GO30140): an open-label, multicentre, phase 1b study. Lancet Oncol, 2020. 21(6): p. 808-820.
Bteich, F. and A.M. Di Bisceglie, Current and Future Systemic Therapies for Hepatocellular Carcinoma. Gastroenterol Hepatol (N Y), 2019. 15(5): p. 266-272.
Mule, S., et al., Advanced Hepatocellular Carcinoma: Pretreatment Contrast-enhanced CT Texture Parameters as Predictive Biomarkers of Survival in Patients Treated with Sorafenib. Radiology, 2018. 288(2): p. 445-455.
Fu, S., et al., Texture analysis of intermediate-advanced hepatocellular carcinoma: prognosis and patients' selection of transcatheter arterial chemoembolization and sorafenib. Oncotarget, 2017. 8(23): p. 37855-37865.
Liu, Q.P., et al., Prediction of prognostic risk factors in hepatocellular carcinoma with transarterial chemoembolization using multi-modal multi-task deep learning. EClinicalMedicine, 2020. 23: p. 100379.
Meng, X.P., et al., Radiomics Analysis on Multiphase Contrast-Enhanced CT: A Survival Prediction Tool in Patients With Hepatocellular Carcinoma Undergoing Transarterial Chemoembolization. Front Oncol, 2020. 10: p. 1196.
Song, W., et al., MRI-Based Radiomics: Associations With the Recurrence-Free Survival of Patients With Hepatocellular Carcinoma Treated With Conventional Transcatheter Arterial Chemoembolization. J Magn Reson Imaging, 2020. 52(2): p. 461-473.
Wang, X.H., et al., MRI-based radiomics model for preoperative prediction of 5-year survival in patients with hepatocellular carcinoma. Br J Cancer, 2020. 122(7): p. 978-985.
Kim, J., et al., Predicting Survival Using Pretreatment CT for Patients With Hepatocellular Carcinoma Treated With Transarterial Chemoembolization: Comparison of Models Using Radiomics. AJR Am J Roentgenol, 2018. 211(5): p. 1026-1034.
Blanc-Durand, P., et al., Signature of survival: a (18)F-FDG PET based whole-liver radiomic analysis predicts survival after (90)Y-TARE for hepatocellular carcinoma. Oncotarget, 2018. 9(4): p. 4549-4558.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Disclosure
The authors have nothing relevant to disclose.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Borhani, A.A., Catania, R., Velichko, Y.S. et al. Radiomics of hepatocellular carcinoma: promising roles in patient selection, prediction, and assessment of treatment response. Abdom Radiol 46, 3674–3685 (2021). https://doi.org/10.1007/s00261-021-03085-w
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00261-021-03085-w