Abstract
The diagnosis of hepatocellular carcinoma relies largely on non-invasive imaging, and is well suited for radiomics analysis. Radiomics is an emerging method for quantification of tumor heterogeneity by mathematically analyzing the spatial distribution and relationships of gray levels in medical images. The published studies on radiomics analysis of HCC provide encouraging data demonstrating potential utility for prediction of tumor biology, molecular profiles, post-therapy response, and outcome. The combination of radiomics data and clinical/laboratory information provides added value in many studies. Radiomics is a multi-step process that requires optimization and standardization, the development of semi-automated or automated segmentation methods, robust data quality control, and refinement of algorithms and modeling approaches for high-throughput data analysis. While radiomics remains largely in the research setting, the strong associations of predictive models and nomograms with certain pathologic, molecular, and immune markers with tumor aggressiveness and patient outcomes, provide great potential for clinical applications to inform optimized treatment strategies and patient prognosis.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Hepatocellular carcinoma (HCC) is the third leading cause of cancer-related death in the US [1]. HCC is typically diagnosed and staged using cross-sectional imaging techniques such as magnetic resonance imaging (MRI) and computed tomography (CT) and generally has a poor prognosis, often due to advanced stage at the time of diagnosis [2]. While early and intermediate stage HCC can be treated with surgical and locoregional therapies (LRT), patients with advanced HCC may benefit from systemic treatment [3]. Tyrosine kinase (TK) inhibitor sorafenib was the first systemic therapy approved for HCC that was shown to confer a survival benefit in patients with advanced disease [4], and recent clinical trials have found that newer systemic agents, such as other TK Inhibitors and immune checkpoint inhibitors, also show improvements in outcome [3, 5].
Because HCC is a heterogeneous and therapy-resistant disease, HCC characterization at the individual patient level is urgently needed in order to stratify patients to a personalized systemic, surgical or LRT regimen to maximize their prognosis. Inaccuracy in this regard could result in unnecessary harm to patients, preclude the use of curative therapies, and impact healthcare costs [6]. While pathological, molecular, and immune classifications of HCC are promising for prognostication and prediction of post-therapy response of HCC [7, 8], these techniques require invasive tissue sampling, specialized equipment and analysis.
Radiomics is an emerging field in image analysis, which consists of the extraction of large numbers of features from the imaging data, which are hypothesized to contain information that reflects underlying tissue biology [9]. The number of publications in the field of radiomics, in particular for tumor characterization including HCC, have increased exponentially in recent years [10]. In this review, we will provide an overview of a typical HCC radiomics workflow, discuss key applications relevant to HCC, review limitations, and discuss future directions of this emerging field with emphasis on CT and MRI.
Radiomics workflow
Radiomics is defined as the quantitative extraction, analysis, and modeling of a large amount of features from medical images in relation to prediction targets, such as clinical end-points, and pathological and genomic features [11]. Radiomics analysis has numerous applications in research [9, 11, 12], but is not yet validated for clinical practice. Here, we will provide a brief overview of the typical radiomics workflow. Radiomics analysis requires in-house developed or commercial software, several of which are available as open-source. The Imaging Biomarker Standardization Initiative (IBSI) is an independent international collaboration to standardize the extraction of radiomics features across software applications and imaging platforms to allow for high-throughput radiomics studies with large patient cohorts. The IBSI manual is a useful reference for the development of robust radiomics analysis tools [13]. When using either commercial or open-source software, it is advisable to check whether the specific tool is in agreement with the IBSI guidelines.
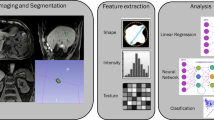
A radiomics study can generally be divided in five phases: data selection, segmentation, feature extraction, exploratory analysis, and modeling (Fig. 1).
Data selection
A radiomics study begins with the selection of imaging data (in retrospective studies) or the set-up of an imaging protocol (in prospective studies), the tissue of interest, and the prediction target (i.e., the event/characteristic that one wishes to predict). Images from any modality (e.g., MRI, CT, PET, ultrasound) can be used, while most studies have investigated CT or MRI. While standardized imaging protocols would be optimal for radiomics analysis, in clinical reality, large variability in imaging protocols across centers and platforms is common and can influence feature measurement [10]. For reproducibility and comparability of radiomics studies, it is therefore of critical importance that the used imaging protocols are extensively described in publications [11].
Segmentation
Volumes of interest (VOIs) or regions of interest (ROIs) in the tissue of interest can be segmented either manually or (semi-)automatically [10]. Manual segmentation is both time-consuming and affected by inter-observer variability. For these reasons, radiomics studies typically advise using a semi-automated approach for tissue segmentation [10]. The segmentation should be performed carefully and consistently, since variability in the segmentation may introduce bias in the radiomics quantification. For radiomics studies in tumors, such as HCC, the inclusion of non-tumor tissue in the tumor ROI should be avoided, as this would also influence the feature extraction. Nevertheless, several recent studies have shown potential utility of a separate peritumoral ROI in the liver parenchyma for improved diagnostic performance of radiomics in HCC [14,15,16].
Feature extraction
Radiomics features can be divided into semantic and agnostic features [9]. An overview of the different types of radiomics features can be found in Table 1. Semantic features include qualitative features that are typically included in radiology reports, including lesion size, shape, location, and necrosis. While semantic feature analysis is easily implemented in clinical practice, it is observer dependent and prone to inter- and intra-observer variability. Agnostic features are mathematically extracted quantitative descriptors in the tissue of interest and are not included in clinical radiological analysis of medical images or radiology reports. Agnostic features are each defined by an advanced mathematical algorithm and can be divided into morphological and statistical features [17]. Morphological features describe the shape and physical composition of the segmented volume. Statistical features can be further classified into first-order, second-order, and higher order features. First-order statistical features describe the distribution of pixel intensity values in the VOIs without taking into account spatial relationships [18]. Second-order features are most often referred as texture features. Image texture describes the spatial relationships of voxels with a certain gray level with adjacent voxels with (dis)similar gray levels (Fig. 2). Gray-level co-occurrence matrix (GLCM) features introduced by Haralick et al. are the most often analyzed texture features [19]. However, there are many texture analysis methods resulting in hundreds of possible texture features that can be included in radiomics analysis [13, 20, 21]. Higher order radiomics methods impose filters on the images to extract patterns.
Exploratory analysis and feature reduction
The large number of assessed features in radiomics comes with a considerable risk of overfitting in the developed radiomics models [12]. Datasets consisting of a minimum of 10–15 patients per assessed feature have been recommended for radiomics studies [10]. The relationships between the extracted features and the prediction target can be assessed prior to modeling to aid in feature selection, for example, using correlation or univariate logistic regression analysis. Only features that show significant diagnostic performance for assessment of the prediction target can be selected for further analysis. In addition, further feature reduction can be performed by elimination of redundant and/or non-robust features [11]. Robustness of features can be assessed by performing repeatability and reproducibility studies, including test–retest experiments and radiomics assessment by multiple observers.
Modeling
The selection of statistical methods for data analysis in radiomics is dependent upon several factors, including the sample size and whether radiomics measurements are used as the predictor or the outcome measure. There is a wide selection of models types for radiomics analysis, including a variety of statistical methods and machine learning approaches, such as random forest, linear regression, logistic regression, Cox proportional hazards regression, and nomograms [22]. The selection of the model is known to affect the performance of the radiomics model for assessment of the prediction target [23]. For small pilot studies where it is not possible to achieve a large sample size, univariate analysis may be a preferred statistical method [24]. While small pilot or retrospective studies are important for providing preliminary evidence or proof-of-concept that certain features or analytical methods warrant further investigation as imaging biomarkers and surrogates for tumor biology, it should be emphasized that feature selection bias and false-positive results can be found when the number of radiomics features assessed exceeds the number of patients [10]. It is therefore advisable to test several model types to select the best performing model.
Validation
The models can be validated by internal cross-validation, which may be used to further optimize the model to maximize its prediction performance. The optimized final model should be validated in a separate independent patient cohort that was not used for model optimization. Typically, learning set should be at least equal to or is 2–4 times the size of the independent testing set, which should be ideally prospectively collected [22, 25]. Furthermore, the clinical utility of the radiomics-derived nomograms can be assessed using decision curve analysis, in which the probabilities of net benefits are assessed at different thresholds in the training and validation sets.
HCC tumor heterogeneity
HCC is a tumor type of interest for radiomics analysis, because HCC lesions have shown to exhibit substantial intra- and inter-tumor heterogeneity on the biological level [26,27,28,29]. With respect to inter-tumor heterogeneity, genome-wide molecular profiling of HCC tissues has identified heterogeneous molecular aberrations across lesions [30]. Large-scale transcriptome analysis in 603 HCC patients revealed distinct aggressive and indolent molecular subclasses of HCC [31]. In terms of immunophenotyping, gene expression analysis in 956 patients has identified a distinct immune class, characterized by expression of markers of an inflammatory response, which consisted of two subclasses reflecting adaptive and exhausted immune responses [32]. Separate analysis of response to molecular targets in these identified distinct HCC subtypes may have significant clinical utility for the development of future clinical trials that are enriched for a certain subtype [29]. Next to the biological variability across HCC tumors, significant intra-tumoral heterogeneity has also been observed in HCC, including on the histopathological and molecular genomic levels [27, 33]. Unfortunately, HCC biopsies generally only sample a small part of the tumor, precluding analysis of intratumoral heterogeneity. Radiomics analysis is a powerful, non-invasive tool to extract numerous features on the whole lesion level which may be reflective of HCC heterogeneity and aggressiveness, and potentially inform on an optimal therapeutic approach. The value of radiomics for prediction of HCC characteristics has been studied quite extensively in the last few years, as summarized in the next sections.
Radiomics for characterization of HCC pathology
Certain histopathologic markers provide valuable information regarding HCC aggressiveness and prognosis, including tumor grade, presence of microvascular invasion (MVI), and immunohistochemical (IHC) expression of markers such as proliferation marker Ki-67 and progenitor cell marker Cytokeratin 19 (CK19). Histologic grade of HCC is determined by the combination of structural and cellular features, and according to a recent meta-analysis, poor tumor differentiation is associated with worse prognosis [34]. First-order radiomics (histogram analysis) found that minimum apparent diffusion coefficient (ADCmin) derived from diffusion-weighted imaging (DWI) [35, 36] and values on the lower end of the ADC percentile range (i.e., < 30% percentile) [36] were of value for predicting poorly differentiated HCC, which most likely reflects increased tumor cellularity (Fig. 3). Radiomics signatures based on conventional pre-contrast T1-W, post-contrast T1-W, and T2-W MRI and CE-CT imaging, either alone or in combination with clinical information [37], have also shown value for the identification of poorly differentiated HCC [37,38,39].
First-order radiomics (histogram) analysis obtained from DWI-MRI. A 54-year-old male with a well-differentiated HCC in segment 6 on arterial phase T1-W a and ADC map b and a 59-year-old male with poorly differentiated HCC in segment 2 on arterial phase T1-W d and ADC map e. Differences in histogram parameters are noted between the well-differentiated c and poorly differentiated f tumors
MVI is defined by the invasion of tumor cells into a vascular space lined by endothelium, including the microscopic vessels of the portal vein, hepatic artery, and lymphatic vessels, and is not readily detected at conventional imaging [40]. MVI has been reported as the strongest independent predictor of early tumor recurrence [41, 42] and poor prognosis [43]. Most of the studies investigating radiomics analysis for prediction of MVI were in HCC patients undergoing surgical resection, demonstrating that both semantic features and quantitative models have shown value. Radiomics scores comprised of semantic features have been validated, such as “radiogenomic venous invasion” (RVI) score, which takes into account internal arteries, hypointense halo and liver-tumor difference [44], and the “two-trait predictor of venous invasion” (TTPVI) [45], that takes into account the presence of internal arteries and hypodense halo (Fig. 4).
A 78-year-old male with chronic hepatitis B infection and pathologically confirmed microvascular invasion (MVI). A 4.9 cm HCC is present in segment 8 on arterial phase a and portal venous phase b contrast-enhanced CT. The tumor demonstrates the radiogenomic venous invasion (RVI) marker, which is comprised of internal arteries (arrows, arterial phase), “hypodense halo” as defined by a rim of hypoattenuation partially or completely circumscribing the tumor (arrowhead) and the “tumor-liver difference” as defined by focal or circumferential sharp transition in attenuation between the tumor and the adjacent liver parenchyma (dashed arrow) [44]. The findings of internal arteries and portal venous phase hypointense halos in this example also fulfill the criteria for the “two-trait predictor of venous invasion” [45]
Quantitative radiomics data, models, or nomograms have demonstrated high performance for CT [46,47,48,49,50], MRI [51,52,53], and US [54] for the prediction of MVI. These studies have investigated arterial phase (AP) and portal venous phase (PVP) for both CT and MRI and DWI. A radiomics model derived from pre-operative multiphase CT outperformed models comprised of RVI and TTPVI semantic features alone (AUC 0.76 vs. AUCs 0.49-0.53, respectively) [48]. The addition of semantic imaging features, clinical and/or laboratory data has improved quantitative model performance in several studies [15, 46, 49, 51, 54]. For example, in a study of 495 HCC patients who underwent pre-operative CT and surgical resection, a combined model yielded excellent accuracy in estimating the risk of MVI, with an AUC of 0.909 in the training/validation set and 0.889 in the test set [49].
Other pathologic markers identified at IHC with prognostic implications have also been investigated. High Ki-67 labeling index values (marker of cell proliferation) are associated with worse prognosis [55]. Histogram analysis obtained from ADC (mean, median, 5th, 25th, 75th percentiles) and AP CE-MRI (mean, median, 25th, 75th, 95th percentiles) found significant inverse correlations between parameters and high Ki-67 values [56]. A separate study also found that texture analysis of hepatobiliary phase (HBP), AP, and PVP CE-MRI using gadobenate dimeglumine was also valuable for predicting Ki-67 expression [57]. Lastly, higher measurement of progenitor cell marker cytokeratin 19 (CK19) was associated with a higher rate of tumor recurrence and metastasis [58, 59]. Texture analysis of conventional T1-W and T2-W MRI outperformed other AFP and qualitative arterial phase rim enhancement for identification of CK19 positivity in HCC, while the combination of these clinical, semantic, and texture features was not significantly better than texture features alone (p = 0.0788) [60]. Finally, a separate study evaluating multi-modal US comprised of B-mode ultrasound, shear wave elastography, and shear wave viscosity imaging found that texture analysis was valuable for prediction of three biomarkers, including programmed cell death protein 1 (PD-1), Ki-67, and MVI [61].
Radiomics for characterization of HCC molecular and immune characteristics
Certain gene signatures and immune phenotypes are associated with aggressive tumor behavior and patient outcomes [6, 62]. The majority of the radiogenomic studies to date have described semantic features obtained from CT or MRI. For example, combinations of 28 qualitative traits at CE-CT were able to reconstruct 78% of HCC global gene expression profiles [63]. In another study, only tumor size showed association with fractional allelic imbalance (FAI) rate index (p = 0.005), which is a molecular marker associated with early tumor recurrence (ER) in liver transplant patients, while there was a non-significant weak positive correlation (r = 0.264) with LI-RADS categorization [64]. In 39 HCC patients, qualitative imaging traits on CT and MRI, including infiltrative pattern, mosaic appearance, presence of macrovascular invasion, and large size, demonstrated significant associations with gene signatures of aggressive HCC phenotype (G3-Boyault, Proliferation-Chiang profiles, CK19-Villanueva, S1/S2-Hoshida), with odds ratios (OR) ranging from 4.44 to 12.73 (p < 0.045) [65]. These particular gene signatures are associated with increased cellular proliferation, vascular invasion, distant metastases, and poor prognosis. A doxorubicin response gene expression program has also been described previously [66], and was found to have significant associations with the qualitative appearance of tumor margins on AP CT (p < 0.05, q < 0.1), which may inform treatment planning for patients receiving transarterial chemoembolization (TACE) [67].
More recently, agnostic features have shown value for predicting gene signatures with prognostic and therapeutic implications. Specifically, in a study of 38 patients with CE-CT, texture features volume fraction and textural heterogeneity correlated with prognostic gene modules that were associated with overall survival (OS) [68]. Hectors et al. performed histogram analysis of parameters obtained from functional multiparametric MRI, including DWI, blood-oxygenation level-dependent (BOLD), tissue-oxygenation level-dependent (TOLD), and dynamic contrast-enhanced (DCE)-MRI and assessed the associations of these histogram metrics with molecular and immune characteristics of HCC (Fig. 5) [69]. It was found that central tendency parameters (mean and median) significantly correlated with gene expression of Wnt target GLUL, pharmacological target FGFR4, stemness markers EPCAM and KRT19, and immune checkpoint PDCD1. In the same study, central tendency parameters from functional multiparametric (mp)MRI also showed significant correlations with count of endothelial cells (CD31 monoclonal antibody), macrophages (CD68), and T-cells (CD3) quantified at multiplexed immunohistochemical analysis (all p values < 0.05) [69]. In a large-scale study of 207 HCC patients, immunoscoring, defined as the comprehensive analysis of the type, functional orientation, density, and spatial location of tumor infiltrating lymphocytes, was accurately predicted using radiomics data extracted from HCC and the peritumoral regions based on gadoxetic acid-enhanced MRI (AUC 0.904) [70]. Thus, knowledge of potential actionable treatment targets and immune phenotypes using a non-invasive radiomics approach could enable improved identification of potential responders to novel immunotherapeutic agents, especially as molecular profiling, advanced immunohistochemistry and immune profiling are plagued by the need for tissue specimens, specialized equipment and expertise, all of which limit routine use.

Adapted from Hectors et al. [69]
Multiparametric MRI histogram analysis in HCC. Example shows a 54-year-old male patient with cirrhosis secondary to chronic hepatitis B virus infection and HCC. a Representative magnified parametric maps of dynamic contrast-enhanced MRI (DCE-MRI, top row) and blood blood-oxygenation level-dependent (BOLD) and tissue-oxygenation level-dependent MRI (TOLD; bottom row) in a large (8.3 cm) HCC. Location of the tumor within the liver is indicated by the white arrow on the T2-weighted image (bottom row, right). Substantial intratumor heterogeneity was observed, in particular on the arterial, portal, and total flow (Fa, Fp, and Ft) maps of DCE-MRI and R2* maps of BOLD. b Histograms of Fa, R2* pre O2, R1 pre O2, and ADC in the same lesion. The extensive heterogeneity observed in the parameter maps is also reflected in the histograms, as illustrated by the fat tails and pronounced skewness. ADC apparent diffusion coefficient, ART arterial fraction, DV distribution volume, Fa arterial flow, Fp portal flow, Ft total flow, MTT mean transit time, R1 longitudinal relaxation rate, R2* transverse relaxation rate
Radiomics for prediction of HCC response to therapy
LRT, including transarterial chemoembolization (TACE), radioembolization (RE), ablative methods, external beam radiation, and systemic therapy, are used for patients with intermediate to advanced stage HCC or in patients who are not surgical candidates [71]. These methods which offer excellent tumor control and improved outcomes for HCC patients, however, have procedure-associated risks and can be costly. Radiomics analysis serves two major roles in patients undergoing treatment for HCC: For the prediction of therapeutic efficacy based on pre-treatment imaging and response assessment based on post-treatment imaging.
A combined model including radiomics features (surface area-to-volume ratio, kurtosis, median, GLCM contrast, and size zone variability) extracted from pre-TACE AP CT and clinical factors (Child–Pugh score, AFP, and tumor size) were found to be a stronger predictor of survival (HR 19.88; p < 0.0001) compared to either the clinical or the radiomics models alone [72]. Other studies agree with these results, and found the value for radiomics features extracted from pre-treatment CE-CT or CE-MRI for prediction of tumor response [73,74,75,76] and post-therapy response evaluation in patients undergoing TACE [73, 76]. Combinations of LRTs can be administered simultaneously. Two other studies have shown that texture analysis obtained from pre-treatment CE-MRI or IVIM-DWI in patients undergoing combined TACE and high-frequency ultrasound (HIFU) found that texture features obtained from both pre-treatment and post-treatment MRI were significant predictors of an early response [76] and patient outcome, respectively [77].
Regarding Y90 RE, volumetric histogram analysis of ADC in 22 patients demonstrated that histogram parameters obtained at 6 w and early changes in ADC compared to baseline (delta) were predictive of objective response (complete and partial response), while pre-treatment vADC histogram parameters were not [78]. These results are conflicting to a separate study in 22 patients undergoing pre-RE CT, in which histogram parameters obtained from AP CT were predictive of response at follow-up (mean follow-up period was 129 days) [79]. The differences in the results of these studies could be attributed to small sample size, different modalities/sequences analyzed, and the duration of the follow-up period (6 weeks vs. 18 weeks), which is especially relevant as the radiologic response after RE evolves over time given the longer period needed for radiation to achieve maximal cytotoxic effect on HCC [80].
Radiomics for prediction of HCC outcome
Using radiomics to predict outcome after non-surgical treatment for HCC
Quantitative radiomics analysis and models have been shown to predict outcome in patients undergoing LRT and systemic therapy in several studies. For example, in a study investigating pre-treatment PET imaging in patients with unresectable HCC undergoing Y90 RE, a whole liver radiomics score including both tumor and background liver was predictive of both progression-free survival (PFS) and overall survival (OS) [81]. In both PFS and OS models, the texture features strength (based on the neighborhood gray-level difference matrix) and variance (describes the deviation from the mean of textural feature numbers) were the most predictive [81]. For patients undergoing ablation, both histogram data obtained from MRI (the ratio of ADCmedian relative to the background liver) were the only independent predictor of tumor progression (p = 0.04) compared to other parameters [82] and radiomics models derived from PVP CT combined with clinicopathologic information yielded excellent model performance for the prediction of early recurrence (training set, C-index = 0.792; validation set, C-index = 0.755 [83] in a separate study. The radiomics geometry feature compacity extracted from non-contrast pre-treatment CT was the only variable found in Cox model that was significant for predicting 12-month survival after external beam radiotherapy (AUC 0.80, p < 0.0001) [84]. In another study in patients with advanced HCC undergoing systemic therapy (sorafenib), entropy derived from PVP CT was found to be an independent predictor of OS in the training cohort (fine filtration, HR 5.08; p = 0.0033), and then confirmed in the validation cohort (p < 0.05) [85].
Using radiomics to predict outcome after surgical treatment for HCC
Radiomics analysis has been investigated for patient outcomes after surgery, which remains a significant clinical problem as up to 40% and 31.6% of HCC patients undergoing partial hepatectomy or liver transplant, respectively, develop tumor recurrence [86, 87]. Certain semantic and agnostic radiomics features extracted from both CT and MRI have also been shown to identify patients who develop early recurrence (ER), shorter recurrence-free survival (RFS), or overall survival (OS) after surgical resection [39, 44, 88,89,90,91,92]. For example, pre-operative MRI texture features were evaluated in 100 patients undergoing hepatic resection and found that the feature entropy on AP images, a manifestation of higher tumor heterogeneity, had the greatest predictive power for ER, regardless of tumor size [91]. In another study investigating CE-CT with measurement of Gabor and Wavelet radiomics features in patients with a single HCC treated by hepatectomy, several features were associated with both OS and DFS (all p values < 0.05), while the Barcelona-Clínic Liver Cancer (BCLC) parameters were not correlated with OS (p = 0.057) [90]. Similar results in a separate study found risk scores developed from radiomics nomograms obtained from CE-CT texture data outperformed traditional clinical staging systems (BCLC, TNM, etc.) in both the training and validation cohorts for both tumor recurrence and OS [50].
The combination of clinical, laboratory and radiomics data has improved quantitative model performance for several studies [15, 16, 91, 93,94,95]. Recent studies have also included assessment of the peritumoral liver parenchyma into the analysis and models [14,15,16], which has demonstrated predictive information on treatment response and outcomes other tumor types [96, 97]. In 156 patients undergoing resection or ablation for HCC, CT-based peritumoral radiomics model improved model performance for predicting ER and showed greater efficiency compared to the tumor radiomics model alone and conventional imaging features [14]. However, in another study also using gadoxetic acid-enhanced MRI, combined clinical and radiomics models, including analysis of 3 mm border of peritumoral tissue, demonstrated equivalent performance compared to the clinicopathologic model for the prediction of ER [C-index 0.716 vs. 0.696, p = 0.788] [16].
In patients undergoing liver transplant, modeling based on the radiomics features extracted from the AP CT demonstrated superior performance compared to PVP or the combination of both of AP + PVP for the prediction of tumor recurrence [87]. In this same study, a radiomics nomogram based on a model comprised of both the radiomics signature and clinical risk factors showed good predictive performance for RFS with a C-index of 0.785 in the training dataset and 0.789 in the validation dataset [87].
Limitations
Despite the ubiquitous studies on HCC radiomics showing encouraging results on its potential utility to predict tumor biology, response, and outcome, several significant hurdles exist before radiomics analysis can be applied in the clinical setting. Radiomics requires the use of specialized software packages, which may result in additional costs and need for user training. Additional key limitations include the current lack of standardization in radiomics studies, in terms of image acquisition protocol, segmentation methods, and the radiomics tools employed for analysis, which may result in differences in radiomics feature measurement that are not attributable to biological variations [9]. In retrospective studies, using data from different imaging vendors, platforms, and protocols, pre-processing consisting of normalization and resampling is essential to reduce, yet not eliminate, the signal variation between acquisitions [98]. Interestingly, the consensus is emerging that the most optimum segmentation can be reached by using a semi-automated approach consisting of computer-aided tumor edge detection followed by manual curation [9]. As mentioned before, for the selection of the radiomics software tool, it is advisable to select a software that is in agreement with the IBSI guidelines which promotes standardization of radiomics analysis [13].
There is also a need for selection of radiomics features that are most robust in terms of repeatability and reproducibility. An increasing number of studies is being performed on this particular topic (see review in [99]), but only one in HCC [100]. The latter study assessed reproducibility and redundancy of CT radiomics features in 26 HCC patients, finding that significant variation in reproducibility across features and segmentation methods (percentage of excellent reproducible features 69–79% dependent on segmentation method) and recommended that only the most reliable and uniform radiomics features should be selected for the clinical use of radiomics features [100].
Detailed reporting of radiomics studies is also essential to advance this emerging field toward clinical translation and to improve reproducibility of study results. The radiomics quality score (RQS) has been proposed for assessment of radiomics study in terms of its compliance with best-practice procedures [11]. The RQS metrics provide useful guidance for the drafting of manuscripts on radiomics studies. Finally, explaining the associations between biologic processes and radiomics features, and especially texture feature obtained using higher order analysis, remains very challenging.
Future directions
Radiomics is still an emerging analytical method and several advances are required for widespread implementation for any clinical application, such as HCC. As described above, each step of the radiomics process will require optimization and standardization, ranging from harmonization of imaging protocols and parameters, the development of (semi-)automated segmentation methods, robust data QA/QC, and refinement of algorithms and modeling approaches for high-throughput analysis [101]. Furthermore, any radiomics study will require validation and further testing in large independent datasets in order to ensure reproducibility of results. Data sharing in the future may enable such an approach. There is also a need for well-designed, prospective, comprehensive studies to integrate radiomics with advanced histopathology, genomics/molecular classification, immune phenotyping, and patient outcomes in HCC [101]. There is growing interest in leveraging the spatial and temporal information in medical imaging to use radiomics to characterize intra-tumor heterogeneity and describe discrete tumor microenvironments that may demonstrate distinct biological behavior [101].
With the exponentially growing interest in and development of artificial intelligence (AI) applications, there are also opportunities to leverage AI in radiomics analysis. Deep learning methods, in particular convolutional neural networks (CNNs), are capable of capturing textural information in the initial convolutional layers. CNNs could thus eventually replace several current methods for radiomics analysis [102]. Recently, a new concept named “deep radiomics” was introduced, which combines radiomics and deep learning analysis by creating feature images from texture features, which are then used as input for a CNN to classify the images. Initial results in a lung CT screening study suggest that this combined deep radiomics approach outperforms both separate radiomics and deep learning analysis [103]. These promising data warrant further investigation into the combination of radiomics and deep learning.
Conclusion
Radiomics analysis is a promising method for assessment of HCC characteristics, by allowing a one-stop-shop non-invasive analysis of tumor aggressiveness. The identification of high-risk features is especially important as HCC is typically diagnosed with imaging and often then managed without obtaining tissue sampling. Given the strong association of both semantic and agnostic radiomics markers with certain pathologic, molecular, and immune markers with tumor aggressiveness and patient outcomes, radiomics analysis may aid in the development of optimized and personalized treatment strategies, resource allocation, and for prognostication. Many studies have indicated that radiomics in combination with clinical and laboratory information may provide additional added value. Future work is warranted to develop a standardized method for quantifying and analyzing multi-dimensional radiomics data to enable widespread clinical application, for future clinical trial design of novel therapeutic agents and to enable advanced analysis including the application of AI.
References
Ryerson, A.B., et al., Annual Report to the Nation on the Status of Cancer, 1975-2012, featuring the increasing incidence of liver cancer. Cancer, 2016. 122(9): p. 1312-37.
Cartier, V. and C. Aube, Diagnosis of hepatocellular carcinoma. Diagn Interv Imaging, 2014. 95(7-8): p. 709-19.
Llovet, J.M., et al., Advances in targeted therapies for hepatocellular carcinoma in the genomic era. Nat Rev Clin Oncol, 2015. 12(8): p. 436
Llovet, J.M., et al., Sorafenib in advanced hepatocellular carcinoma. N Engl J Med, 2008. 359(4): p. 378-90.
Bteich, F. and A.M. Di Bisceglie, Current and Future Systemic Therapies for Hepatocellular Carcinoma. Gastroenterol Hepatol (N Y), 2019. 15(5): p. 266-272.
Villanueva, A., et al., New strategies in hepatocellular carcinoma: genomic prognostic markers. Clin Cancer Res, 2010. 16(19): p. 4688-94.
Khemlina, G., S. Ikeda, and R. Kurzrock, The biology of Hepatocellular carcinoma: implications for genomic and immune therapies. Mol Cancer, 2017. 16(1): p. 149.
Kurebayashi, Y., et al., Landscape of immune microenvironment in hepatocellular carcinoma and its additional impact on histological and molecular classification. Hepatology, 2018. 68(3): p. 1025-1041.
Gillies, R.J., P.E. Kinahan, and H. Hricak, Radiomics: Images Are More than Pictures, They Are Data. Radiology, 2016. 278(2): p. 563-77.
Yip, S.S. and H.J. Aerts, Applications and limitations of radiomics. Phys Med Biol, 2016. 61(13): p. R150-66.
Lambin, P., et al., Radiomics: the bridge between medical imaging and personalized medicine. Nat Rev Clin Oncol, 2017. 14(12): p. 749-762.
Nougaret, S., et al., Radiomics: an Introductory Guide to What It May Foretell. Curr Oncol Rep, 2019. 21(8): p. 70.
Zwanenburg, A., et al., Imaging biomarker standardisation initiative. arXiv, 2016. 1612.07003.
Shan, Q.Y., et al., CT-based peritumoral radiomics signatures to predict early recurrence in hepatocellular carcinoma after curative tumor resection or ablation. Cancer Imaging, 2019. 19(1): p. 11.
Ahn, S.J., et al., Hepatocellular carcinoma: preoperative gadoxetic acid-enhanced MR imaging can predict early recurrence after curative resection using image features and texture analysis. Abdom Radiol (NY), 2019. 44(2): p. 539-548.
Kim, S., et al., Radiomics on Gadoxetic Acid-Enhanced Magnetic Resonance Imaging for Prediction of Postoperative Early and Late Recurrence of Single Hepatocellular Carcinoma. Clin Cancer Res, 2019. 25(13): p. 3847-3855.
Yang, F., et al., Magnetic resonance imaging (MRI)-based radiomics for prostate cancer radiotherapy. Transl Androl Urol, 2018. 7(3): p. 445-458.
Just, N., Improving tumour heterogeneity MRI assessment with histograms. Br J Cancer, 2014. 111(12): p. 2205-13.
Haralick RM, S.K., Dinstein I, Textural Features for Image Classification. IEEE Trans SystMan Cybern SMC, 1973. 3: p. 610-621.
Davnall, F., et al., Assessment of tumor heterogeneity: an emerging imaging tool for clinical practice? Insights Imaging, 2012. 3(6): p. 573-89.
Parekh, V. and M.A. Jacobs, Radiomics: a new application from established techniques. Expert Rev Precis Med Drug Dev, 2016. 1(2): p. 207-226.
Rizzo, S., et al., Radiomics: the facts and the challenges of image analysis. Eur Radiol Exp, 2018. 2(1): p. 36.
Parmar, C., et al., Machine Learning methods for Quantitative Radiomic Biomarkers. Sci Rep, 2015. 5: p. 13087.
Varghese, B.A., et al., Texture Analysis of Imaging: What Radiologists Need to Know. AJR Am J Roentgenol, 2019. 212(3): p. 520-528.
Dobbin, K.K. and R.M. Simon, Optimally splitting cases for training and testing high dimensional classifiers. BMC Med Genomics, 2011. 4: p. 31.
Fransvea, E., et al., HCC heterogeneity: molecular pathogenesis and clinical implications. Cell Oncol, 2009. 31(3): p. 227-33.
Friemel, J., et al., Intratumor heterogeneity in hepatocellular carcinoma. Clin Cancer Res, 2015. 21(8): p. 1951-61.
Lin, D.C., et al., Genomic and Epigenomic Heterogeneity of Hepatocellular Carcinoma. Cancer Res, 2017. 77(9): p. 2255-2265.
Zhu, S. and Y. Hoshida, Molecular heterogeneity in hepatocellular carcinoma. Hepat Oncol, 2018. 5(1): p. HEP10.
Goossens, N., X. Sun, and Y. Hoshida, Molecular classification of hepatocellular carcinoma: potential therapeutic implications. Hepat Oncol, 2015. 2(4): p. 371-379.
Hoshida, Y., et al., Integrative transcriptome analysis reveals common molecular subclasses of human hepatocellular carcinoma. Cancer Res, 2009. 69(18): p. 7385-92.
Sia, D., et al., Identification of an Immune-specific Class of Hepatocellular Carcinoma, Based on Molecular Features. Gastroenterology, 2017. 153(3): p. 812-826.
Liu, J., H. Dang, and X.W. Wang, The significance of intertumor and intratumor heterogeneity in liver cancer. Exp Mol Med, 2018. 50(1): p. e416.
Martins-Filho, S.N., et al., Histological Grading of Hepatocellular Carcinoma-A Systematic Review of Literature. Front Med (Lausanne), 2017. 4: p. 193.
Moriya, T., et al., 3D analysis of apparent diffusion coefficient histograms in hepatocellular carcinoma: correlation with histological grade. Cancer Imaging, 2017. 17(1): p. 1.
Xu, Y.S., et al., Whole-lesion histogram analysis metrics of the apparent diffusion coefficient: a correlation study with histological grade of hepatocellular carcinoma. Abdom Radiol (NY), 2019. 44(9): p. 3089-3098.
Wu, M., et al., Predicting the grade of hepatocellular carcinoma based on non-contrast-enhanced MRI radiomics signature. Eur Radiol, 2019. 29(6): p. 2802-2811.
Zhou, W., et al., Malignancy characterization of hepatocellular carcinomas based on texture analysis of contrast-enhanced MR images. J Magn Reson Imaging, 2017. 45(5): p. 1476-1484.
Oh, J., et al., Hepatocellular Carcinoma: Texture Analysis of Preoperative Computed Tomography Images Can Provide Markers of Tumor Grade and Disease-Free Survival. Korean J Radiol, 2019. 20(4): p. 569-579.
Gouw, A.S., et al., Markers for microvascular invasion in hepatocellular carcinoma: where do we stand? Liver Transpl, 2011. 17 Suppl 2: p. S72-80.
Roayaie, S., et al., A system of classifying microvascular invasion to predict outcome after resection in patients with hepatocellular carcinoma. Gastroenterology, 2009. 137(3): p. 850-5.
Lim, K.C., et al., Microvascular invasion is a better predictor of tumor recurrence and overall survival following surgical resection for hepatocellular carcinoma compared to the Milan criteria. Ann Surg, 2011. 254(1): p. 108-13.
Mazzaferro, V., et al., Predicting survival after liver transplantation in patients with hepatocellular carcinoma beyond the Milan criteria: a retrospective, exploratory analysis. Lancet Oncol, 2009. 10(1): p. 35-43.
Banerjee, S., et al., A computed tomography radiogenomic biomarker predicts microvascular invasion and clinical outcomes in hepatocellular carcinoma. Hepatology, 2015. 62(3): p. 792-800.
Renzulli, M., et al., Can Current Preoperative Imaging Be Used to Detect Microvascular Invasion of Hepatocellular Carcinoma? Radiology, 2016. 279(2): p. 432-42.
Peng, J., et al., A radiomics nomogram for preoperative prediction of microvascular invasion risk in hepatitis B virus-related hepatocellular carcinoma. Diagn Interv Radiol, 2018. 24(3): p. 121-127.
Ma, X., et al., Preoperative radiomics nomogram for microvascular invasion prediction in hepatocellular carcinoma using contrast-enhanced CT. Eur Radiol, 2019. 29(7): p. 3595-3605.
Bakr, S., et al., Noninvasive radiomics signature based on quantitative analysis of computed tomography images as a surrogate for microvascular invasion in hepatocellular carcinoma: a pilot study. J Med Imaging (Bellingham), 2017. 4(4): p. 041303.
Xu, X., et al., Radiomic analysis of contrast-enhanced CT predicts microvascular invasion and outcome in hepatocellular carcinoma. J Hepatol, 2019. 70(6): p. 1133-1144.
Zheng, B.H., et al., Radiomics score: a potential prognostic imaging feature for postoperative survival of solitary HCC patients. BMC Cancer, 2018. 18(1): p. 1148.
Zhu, Y.J., et al., Model-based three-dimensional texture analysis of contrast-enhanced magnetic resonance imaging as a potential tool for preoperative prediction of microvascular invasion in hepatocellular carcinoma. Oncol Lett, 2019. 18(1): p. 720-732.
Huang, Y.Q., et al., Value of MR histogram analyses for prediction of microvascular invasion of hepatocellular carcinoma. Medicine (Baltimore), 2016. 95(26): p. e4034.
Li, H., et al., Preoperative histogram analysis of intravoxel incoherent motion (IVIM) for predicting microvascular invasion in patients with single hepatocellular carcinoma. Eur J Radiol, 2018. 105: p. 65-71.
Hu, H.T., et al., Ultrasound-based radiomics score: a potential biomarker for the prediction of microvascular invasion in hepatocellular carcinoma. Eur Radiol, 2019. 29(6): p. 2890-2901.
Luo, Y., et al., Clinicopathological and prognostic significance of high Ki-67 labeling index in hepatocellular carcinoma patients: a meta-analysis. Int J Clin Exp Med, 2015. 8(7): p. 10235-47.
Hu, X.X., et al., Whole-tumor MRI histogram analyses of hepatocellular carcinoma: Correlations with Ki-67 labeling index. J Magn Reson Imaging, 2017. 46(2): p. 383-392.
Li, Y., et al., Texture analysis of multi-phase MRI images to detect expression of Ki67 in hepatocellular carcinoma. Clin Radiol, 2019.
Kim, H., et al., Human hepatocellular carcinomas with “Stemness”-related marker expression: keratin 19 expression and a poor prognosis. Hepatology, 2011. 54(5): p. 1707-17.
Tsuchiya, K., et al., Expression of keratin 19 is related to high recurrence of hepatocellular carcinoma after radiofrequency ablation. Oncology, 2011. 80(3-4): p. 278-88.
Wang, H.Q., et al., Magnetic resonance texture analysis for the identification of cytokeratin 19-positive hepatocellular carcinoma. Eur J Radiol, 2019. 117: p. 164-170.
Yao, Z., et al., Preoperative diagnosis and prediction of hepatocellular carcinoma: Radiomics analysis based on multi-modal ultrasound images. BMC Cancer, 2018. 18(1): p. 1089.
Cariani, E., et al., Immunological and molecular correlates of disease recurrence after liver resection for hepatocellular carcinoma. PLoS One, 2012. 7(3): p. e32493.
Segal, E., et al., Decoding global gene expression programs in liver cancer by noninvasive imaging. Nat Biotechnol, 2007. 25(6): p. 675-80.
Furlan, A., et al., A radiogenomic analysis of hepatocellular carcinoma: association between fractional allelic imbalance rate index and the liver imaging reporting and data system (LI-RADS) categories and features. Br J Radiol, 2018. 91(1086): p. 20170962.
Taouli, B., et al., Imaging-based surrogate markers of transcriptome subclasses and signatures in hepatocellular carcinoma: preliminary results. Eur Radiol, 2017. 27(11): p. 4472-4481.
Gyorffy, B., et al., Prediction of doxorubicin sensitivity in breast tumors based on gene expression profiles of drug-resistant cell lines correlates with patient survival. Oncogene, 2005. 24(51): p. 7542-51.
Kuo, M.D., et al., Radiogenomic analysis to identify imaging phenotypes associated with drug response gene expression programs in hepatocellular carcinoma. J Vasc Interv Radiol, 2007. 18(7): p. 821-31.
Xia, W., et al., Radiogenomics of hepatocellular carcinoma: multiregion analysis-based identification of prognostic imaging biomarkers by integrating gene data-a preliminary study. Phys Med Biol, 2018. 63(3): p. 035044.
Hectors, S.J., et al., Quantification of hepatocellular carcinoma heterogeneity with multiparametric magnetic resonance imaging. Sci Rep, 2017. 7(1): p. 2452.
Chen, S., et al., Pretreatment prediction of immunoscore in hepatocellular cancer: a radiomics-based clinical model based on Gd-EOB-DTPA-enhanced MRI imaging. Eur Radiol, 2019. 29(8): p. 4177-4187.
Bruix, J., M. Reig, and M. Sherman, Evidence-Based Diagnosis, Staging, and Treatment of Patients With Hepatocellular Carcinoma. Gastroenterology, 2016. 150(4): p. 835-53.
Kim, J., et al., Predicting Survival Using Pretreatment CT for Patients With Hepatocellular Carcinoma Treated With Transarterial Chemoembolization: Comparison of Models Using Radiomics. AJR Am J Roentgenol, 2018. 211(5): p. 1026-1034.
Kloth, C., et al., Evaluation of Texture Analysis Parameter for Response Prediction in Patients with Hepatocellular Carcinoma Undergoing Drug-eluting Bead Transarterial Chemoembolization (DEB-TACE) Using Biphasic Contrast-enhanced CT Image Data: Correlation with Liver Perfusion CT. Acad Radiol, 2017. 24(11): p. 1352-1363.
Park, H.J., et al., Prediction of Therapeutic Response of Hepatocellular Carcinoma to Transcatheter Arterial Chemoembolization Based on Pretherapeutic Dynamic CT and Textural Findings. AJR Am J Roentgenol, 2017. 209(4): p. W211-W220.
Reis, S.P., et al., Tumor Enhancement and Heterogeneity Are Associated With Treatment Response to Drug-Eluting Bead Chemoembolization for Hepatocellular Carcinoma. J Comput Assist Tomogr, 2017. 41(2): p. 289-293.
Yu, J.Y., et al., Value of texture analysis based on enhanced MRI for predicting an early therapeutic response to transcatheter arterial chemoembolisation combined with high-intensity focused ultrasound treatment in hepatocellular carcinoma. Clin Radiol, 2018. 73(8): p. 758 e9-758 e18.
Wu, L.F., et al., Pre-TACE kurtosis of ADCtotal derived from histogram analysis for diffusion-weighted imaging is the best independent predictor of prognosis in hepatocellular carcinoma. Eur Radiol, 2019. 29(1): p. 213-223.
Gordic, S., et al., Prediction of hepatocellular carcinoma response to (90)Yttrium radioembolization using volumetric ADC histogram quantification: preliminary results. Cancer Imaging, 2019. 19(1): p. 29.
Reiner, C.S., et al., Histogram Analysis of CT Perfusion of Hepatocellular Carcinoma for Predicting Response to Transarterial Radioembolization: Value of Tumor Heterogeneity Assessment. Cardiovasc Intervent Radiol, 2016. 39(3): p. 400-8.
Riaz, A., et al., Radiologic-pathologic correlation of hepatocellular carcinoma treated with internal radiation using yttrium-90 microspheres. Hepatology, 2009. 49(4): p. 1185-93.
Blanc-Durand, P., et al., Signature of survival: a (18)F-FDG PET based whole-liver radiomic analysis predicts survival after (90)Y-TARE for hepatocellular carcinoma. Oncotarget, 2018. 9(4): p. 4549-4558.
Ma, X., et al., Histogram analysis of apparent diffusion coefficient predicts response to radiofrequency ablation in hepatocellular carcinoma. Chin J Cancer Res, 2019. 31(2): p. 366-374.
Yuan, C., et al., Prediction early recurrence of hepatocellular carcinoma eligible for curative ablation using a Radiomics nomogram. Cancer Imaging, 2019. 19(1): p. 21.
Cozzi, L., et al., Radiomics based analysis to predict local control and survival in hepatocellular carcinoma patients treated with volumetric modulated arc therapy. BMC Cancer, 2017. 17(1): p. 829.
Mule, S., et al., Advanced Hepatocellular Carcinoma: Pretreatment Contrast-enhanced CT Texture Parameters as Predictive Biomarkers of Survival in Patients Treated with Sorafenib. Radiology, 2018. 288(2): p. 445-455.
Shah, S.A., et al., Recurrence after liver resection for hepatocellular carcinoma: risk factors, treatment, and outcomes. Surgery, 2007. 141(3): p. 330-9.
Guo, D., et al., Radiomics analysis enables recurrence prediction for hepatocellular carcinoma after liver transplantation. Eur J Radiol, 2019. 117: p. 33-40.
Hui, T.C.H., et al., Predicting early recurrence of hepatocellular carcinoma with texture analysis of preoperative MRI: a radiomics study. Clin Radiol, 2018. 73(12): p. 1056 e11-1056 e16.
Brenet Defour, L., et al., Hepatocellular carcinoma: CT texture analysis as a predictor of survival after surgical resection. Eur Radiol, 2019. 29(3): p. 1231-1239.
Chen, S., et al., Texture analysis of baseline multiphasic hepatic computed tomography images for the prognosis of single hepatocellular carcinoma after hepatectomy: A retrospective pilot study. Eur J Radiol, 2017. 90: p. 198-204.
Zhang, J., et al., Texture Analysis Based on Preoperative Magnetic Resonance Imaging (MRI) and Conventional MRI Features for Predicting the Early Recurrence of Single Hepatocellular Carcinoma after Hepatectomy. Acad Radiol, 2018.
Zhang, W., et al., Prognostic value of preoperative computed tomography in HBV-related hepatocellular carcinoma patients after curative resection. Onco Targets Ther, 2019. 12: p. 3791-3804.
Akai, H., et al., Predicting prognosis of resected hepatocellular carcinoma by radiomics analysis with random survival forest. Diagn Interv Imaging, 2018. 99(10): p. 643-651.
Zhang, Z., et al., Hepatocellular carcinoma: radiomics nomogram on gadoxetic acid-enhanced MR imaging for early postoperative recurrence prediction. Cancer Imaging, 2019. 19(1): p. 22.
Zhou, Y., et al., CT-based radiomics signature: a potential biomarker for preoperative prediction of early recurrence in hepatocellular carcinoma. Abdom Radiol (NY), 2017. 42(6): p. 1695-1704.
Braman, N.M., et al., Intratumoral and peritumoral radiomics for the pretreatment prediction of pathological complete response to neoadjuvant chemotherapy based on breast DCE-MRI. Breast Cancer Res, 2017. 19(1): p. 57.
Prasanna, P., et al., Radiomic features from the peritumoral brain parenchyma on treatment-naive multi-parametric MR imaging predict long versus short-term survival in glioblastoma multiforme: Preliminary findings. Eur Radiol, 2017. 27(10): p. 4188-4197.
Shafiq-Ul-Hassan, M., et al., Intrinsic dependencies of CT radiomic features on voxel size and number of gray levels. Med Phys, 2017. 44(3): p. 1050-1062.
Traverso, A., et al., Repeatability and Reproducibility of Radiomic Features: A Systematic Review. Int J Radiat Oncol Biol Phys, 2018. 102(4): p. 1143-1158.
Qiu, Q., et al., Reproducibility and non-redundancy of radiomic features extracted from arterial phase CT scans in hepatocellular carcinoma patients: impact of tumor segmentation variability. Quant Imaging Med Surg, 2019. 9(3): p. 453-464.
Sala, E., et al., Unravelling tumour heterogeneity using next-generation imaging: radiomics, radiogenomics, and habitat imaging. Clin Radiol, 2017. 72(1): p. 3-10.
Parekh, V.S. and M.A. Jacobs, Deep learning and radiomics in precision medicine. Expert Rev Precis Med Drug Dev, 2019. 4(2): p. 59-72.
Paul, R., et al., Towards deep radiomics: nodule malignancy prediction using CNNs on feature images. Proc. SPIE 10950, Medical Imaging 2019: Computer-Aided Diagnosi, 2019.
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Lewis, S., Hectors, S. & Taouli, B. Radiomics of hepatocellular carcinoma. Abdom Radiol 46, 111–123 (2021). https://doi.org/10.1007/s00261-019-02378-5
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00261-019-02378-5