Abstract
Shikimic acid is an important intermediate for the manufacture of the antiviral drug oseltamivir (Tamiflu®) and many other pharmaceutical compounds. Much of its existing supply is obtained from the seeds of Chinese star anise (Illicium verum). Nevertheless, plants cannot supply a stable source of affordable shikimate along with laborious and cost-expensive extraction and purification process. Microbial biosynthesis of shikimate through metabolic engineering and synthetic biology approaches represents a sustainable, cost-efficient, and environmentally friendly route than plant-based methods. Metabolic engineering allows elevated shikimate production titer by inactivating the competing pathways, increasing intracellular level of key precursors, and overexpressing rate-limiting enzymes. The development of synthetic and systems biology-based novel technologies have revealed a new roadmap for the construction of high shikimate-producing strains. This review elaborates the enhanced biosynthesis of shikimate by utilizing an array of traditional metabolic engineering along with novel advanced technologies. The first part of the review is focused on the mechanistic pathway for shikimate production, use of recombinant and engineered strains, improving metabolic flux through the shikimate pathway, chemically inducible chromosomal evolution, and bioprocess engineering strategies. The second part discusses a variety of industrially pertinent compounds derived from shikimate with special reference to aromatic amino acids and phenazine compound, and main engineering strategies for their production in diverse bacterial strains. Towards the end, the work is wrapped up with concluding remarks and future considerations.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Shikimic acid (3,4,5-trihydroxy-1-cyclohexene-1-carboxylic acid) is an important biochemical metabolite in plants and microorganisms. Many potent characteristics such as highly functionalized six-membered carbocyclic ring, six-carbon cyclitol with three asymmetric centers, and a functional carboxylic group render shikimate as a versatile enantiomerically pure precursor for the synthesis of different compounds with considerable biological/pharmaceutical activities, such as aromatic amino acids, alkaloid, coumarins, flavonoid, salicylic acid, and violacein (Momen and Hoshino 2000; Knaggs 2003; Rodrigues et al. 2013; Jiang and Zhang 2016). Exceptional interest in shikimate biosynthesis has been rekindled as it is a key building block for the synthesis of the antiviral drug oseltamivir, commercially known as Tamiflu® (Genentech, Inc., South San Francisco, CA, USA; Tamiflu®; Kramer et al. 2003; Ghosh et al. 2012). Given the flu pandemic impact, the limited use of vaccines against rapidly evolving flu viruses, stockpiles of effective drugs are necessary for managing a significant outbreak (Horimoto and Kawaoka 2001). Oseltamivir is effective against both type A and type B influenza, avian influenza virus H5N1, and human influenza virus H1N1, especially if administered early and also used in prophylaxis (Widmer et al. 2010). Due to this unique application, the production of shikimate from different sources has attracted significant attention and has been extensively studied over the last several years.
At present, the fruit of Chinese star anise is a major source for the shikimate supply at commercial level. However, inadequate raw feedstocks, and a multi-step, inefficient, and costly plant-based extraction process have rendered it challenging to meet the ever-increasing worldwide demand for oseltamivir (Ghosh et al. 2012; Rawat et al. 2013). Chemical-based strategies, on the other hand, are also known, but commercially unattractive due to environmental concerns. To tackle and overcome such problematic issues, the efficient use of renewable sources, e.g., glycerol for shikimate production via well-established fermentation-based strategies and metabolic engineering has been advocated as a sustainable alternative approach to meet the current market demand (Bochkov et al. 2012; Chen et al. 2014; Cui et al. 2014; Martinez et al. 2015; Gu et al. 2016, 2017; Bilal et al. 2018a).
The shikimate pathway
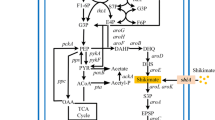
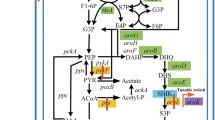
The shikimate pathway is ubiquitously found in bacteria, fungi, plants, and algae, as well as some parasitic protozoans. Figure 1 portrays that the shikimate metabolic pathway used to synthesize aromatic amino acids begins with the condensation of two compounds, namely phosphoenolpyruvate (PEP) and d-erythrose 4-phosphate (E4P) that are generated from glycerol, glucose, or other carbohydrates. PEP is obtained from the glycolysis (Embden-Meyerhof-Parnas pathway), whereas the pentose phosphate (PP) pathway yielded the E4P. Once carbohydrate metabolism has produced PEP and E4P, they are initially combined in the shikimate pathway to produce 3-deoxy-d-arabino-heptulosonate-7-phosphate (DAHP) by DAHP synthase. The resultant DAHP is then converted to shikimate through three-step enzyme-catalyzed reactions. After shikimate synthesis, the subsequent shikimate pathway reactions lead to the formation of chorismate, a common metabolite, used for the production of a variety of other aromatic end products including l-phenylalanine (l-Phe), l-tyrosine (l-Tyr), l-tryptophan (l-Trp), folic acid, etc. (Dosselaere and Vanderleyden 2001; Bongaerts et al. 2001). In plants, it is an indispensable source for the production of secondary metabolites (Eudes et al. 2018). Thanks to the key scientific advancements by biotechnologist and biochemists, all the enzymatic reactions concerning shikimate pathway have been identified and well characterized (Herrmann and Weaver 1999). Today, this pathway has fascinated intensive researchers interest, as the metabolic intermediates and by-products of the pathway can serve as starting feedstocks for producing a great variety of metabolites/compounds with novel pharmaceutical functionalities. Decades of the dedicated research efforts accompanied by the state-of-the-art engineering tools for pathway engineering have made possible for producing molecules with desired titer, yield, and productivity (Zhang et al. 2016). The overproduction of shikimate can be obtained by genetically blocking biochemical pathways consuming shikimate and overexpressing key enzymes responsible for its biosynthesis. Fermentative production using engineered microbial strains is already providing the marketable supply of shikimate (Chen et al. 2012; Chen et al. 2014). To date, several scientists have widely practiced shikimate pathway engineering in Escherichia coli (Johansson et al. 2005; Johansson and Liden 2006; Estevez and Estevez 2012; Martinez et al. 2015; Bilal et al. 2018a; Diaz-Quiroz et al. 2018). However, the PEP availability limits the induced shikimate production along with other industrially relevant aromatic compounds (Flores et al. 2002; Kogure et al. 2016). One possible reason behind this PEP-based limitation is a metabolic competition among DAHP synthase and numerous PEP-exhausting activities involved in central carbon metabolism (Flores et al. 2002). So far, metabolic engineers have been proposed numerous strategies to overcome the abovementioned issue to evidence progressive effect on induced aromatics production by enhancing the PEP pool. Markedly, metabolic engineering strategies directed the development of microbial catalysts with requisite shikimate yield (Báez-Viveros et al. 2007; Martinez et al. 2015). This review elaborates a plenty of novel metabolic engineering strategies for the elevated production of shikimate.
Metabolic engineering related to shikimate pathway for the biosynthesis of chorismate derivatives. G6P—glucose 6 phosphate; E4P—erythrose 4-phosphate; PEP—phosphoenolpyruvate; DAHP—3-deoxy-d-arobino-heptulosonate 7-phosphate; PABA—para-aminobenzoic acid; 4-HBA—4-hydroxy benzoic acid; ADIC—2-amino-2-deoxyisochorismate; PCA—phenazine-1-carboxylic acid; Phe—phenylalanine; Trp—tryptophan; Tyr—tyrosine; tktA—transketolase 1; ppsA—phosphoenolpyruvate synthase; phzC—2-keto-3-deoxy-d-arabino-heptulosonate-7-phosphate synthase; aroB—3-dehydroquinate synthase; aroD—3-dehydroquinate dehydratase; aroE—shikimate dehydrogenase; aroA—5-enolpyruvylshikimate-3-phosphate synthetase; aroC—chorismate synthase; aroK—shikimate kinase I gene; aroL—shikimate kinase II gene; aroF, aroG, aroH—DAHP synthase isoenzyme genes; pabB—aminodeoxychorismate synthase; pabC—4-amino-4-deoxychorismate lyase; folA—dihydrofolate reductase; folC—folylpolyglutamate synthase; folP—dihydropteroate synthase; ubiC—chorismate-pyruvate lyase; ubiA-H—Ubiquinone synthease genes; trpE—anthranilate synthase; trpA-F—tryptophan synthase; pheA—prephenate dehydratase; phhC—aromatic amino acid aminotransferase; tyrC—prephenate dehydrogenase; pheC—cyclohexadienyl dehydratase; phzA-G—phenazine biosynthesis proteins
Use of recombinant and engineered strains
Although extensive research has been dedicated to the development of a commercially viable/sustainable synthesis of shikimate, these methods remain expensive with several limitations. Therefore, substantial consideration has been rekindled toward alternative biotechnologically engineered bacterial strains that provide a significant and promising route for shikimate biosynthesis via fermentation strategies (Bongaerts et al. 2001; Bilal et al. 2018a). To date, most of the metabolic engineering strategies for shikimate biosynthesis has been attempted on the bacterium E. coli (Knop et al. 2001; Ahn et al. 2009; Escalante et al. 2010), but some reports in other bacteria have also been documented in the literature. Metabolic engineering approaches endeavored, over the past several years, to the improved production of shikimate in E. coli are primarily based on genetic manipulations of the central carbon metabolism (CCM) and the shikimate pathway. The fluxes of PEP and E4P can be increased or directed into shikimate pathway through genetically engineering the glycolytic pathway and the PP pathway, respectively. Knop and coworkers (Knop et al. 2001) reported that overexpression of the transketolase (tktA) led to an elevated shikimate accumulation from 38 to 52 g/L with a yield improvement of 0.12 to 0.18 mol/mol by increasing the concentration of E4P. Similarly, increased availability of PEP by engineering the glycolytic pathway has also enhanced the shikimate biosynthesis in recombinant E. coli strain. The overexpression of phosphoenolpyruvate synthase (ppsA) resulted in 66 g/L of shikimate titer with a yield of 0.23 mol/mol utilizing glucose as a sole carbon source. Inactivating phosphotransferase system (PTS) operon and overexpressing non-PTS glucose transporters like glucose facilitators (glf) and glucokinase (glk) accompanied by overexpression of the tktA gene has been shown to increase the shikimate level to 71 g/L. The shikimate titer was further enhanced to 84 g/L in the engineered E. coli strain by the supplementation of the minimal medium with yeast extract as a nitrogen source (Chandran et al. 2003). Genetic engineering techniques have also been utilized to synthesize shikimate in an evolved E. coli strain PB 12 with no PTS system. The double mutant PB 12.SA22 strain with inactivated aroK and aroL genes produced 7 g/L shikimate titer with a corresponding yield of 0.29 mol/mol (Escalante et al. 2010). Apart from engineered E. coli, genetically manipulated Bacillus subtilis (Iomantas et al. 2002) and Citrobacter freundii (Shirai et al. 2001) strains have also been exploited successfully to produce shikimate; however, the titers were not exceeded beyond 20 g/L. A shikimate kinase (aroI)-inactivated B. subtilis was shown to produce 8.5 g/L shikimate together with 9.5 g/L of dehydroshikimate (DHS) (Iomantas et al. 2002).
Since DHS can be readily converted to shikimate, many metabolic engineering studies have focused on the hyper of DHS to increase the shikimate titer. In previous studies, substantial overproduction of DHS has been achieved in the genetically engineered microbial strains (Yi et al. 2002, 2003). For instance, modified E. coli derivatives have synthesized 60 g/L DHS from glucose-based cultivation medium after 60 h (Yi et al. 2002). The allosteric enzyme DAHP synthase catalyzes the first committed step in the biosynthesis of aromatic compounds in microorganisms and plants (Bongaerts et al. 2001), and is the primary feedback regulation site of carbon flux through the shikimate pathway (Kim et al. 2000). In E. coli, the DAHP synthase isozymes encoded by aroG, aroF, and aroH contribute to the total DAHP synthase activity but are allosterically inhibited by L-Phe, L-Tyr, and L-Trp, respectively (Herrmann and Weaver 1999; Sprenger 2007a). In contrast, Corynebacterium glutamicum has only two DAHP synthase isoenzymes, AroF (encoded by aroF) and AroG (encoded by aroG), which are feedback inhibited by L-Tyr, and L-Tyr, L-Phe, prephenate, and chorismate, respectively (Liu et al. 2008b; Li et al. 2009). Many feedback-resistant variants of the DAHP synthases have been constructed with random mutagenesis, over the past few years, for elevated biosynthesis of aromatic amino acids. Zhang and coworkers (Zhang et al. 2014) identified and characterized an L-tyrosine insensitive AroF variant with a deficiency in residue Ile11 (named AroF*). After that, nine AroF variants with different truncated fragments were constructed, and overexpression of the variants AroFΔ(1–9), AroFΔ(1–10), AroFΔ(1–12), and particularly, AroFΔ(1–11) considerably enhanced the accumulation of L-Phe. By co-overexpressing AroFΔ(1–11) and PheAfbr, the L-Phe titer was increased from 2.36 to 4.29 g L−1, indicating the great potential of novel AroFΔ(1–11) variant for producing aromatic amino acids and their derivatives. The amplification and de-regulation of the catalytic activity of DAHP synthase might be a distinctive approach for hyperproduction of aromatic amino acids, shikimate, and its DHS precursors (Weaver and Herrmann 1990). Table 1 illustrates the recent overview of engineered microbial strains for enhanced shikimate biosynthesis reported in the last few years.
Increasing or diverting metabolic flux toward the shikimate pathway
One of the principle and most important challenge to engineer shikimate pathway is increasing the supply of key pathway precursors, i.e., PEP and E4P. As a metabolic intermediate in glycolysis, PEP plays a noteworthy role in transporting glucose across the membrane serving as a phosphoryl group donor. The flux towards the shikimate pathway is often constrained in the presence of glucose as the sole carbon source that might be ascribed to the PEP consumption for glucose uptake through the PTS system. In this juncture, modulation of the PTS system or exploring other glucose transport systems have been demonstrated to significantly amplify the biosynthetic proficiency of the shikimate pathway (Yi et al. 2003). On the other hand, E4P is sourced from the PP pathway, whose metabolic efficiency fluctuates depending on the genetic manipulation and cultivation conditions (Breitenbach et al. 2014). The overexpression of ppsA and tktA genes is a common approach to ameliorate the intracellular levels of PEP and E4P precursors, respectively. Besides their improvement, a balanced supply of both precursors is also crucial to directing metabolic flux into the shikimate pathway. To circumvent the inadequate PEP accessibility concerns, research efforts have also been devoted to pyruvate utilization, rather than PEP, to generate DAHP effectively (Ran and Frost 2007). Also, glycerol can serve as a potential alternative to glucose for stimulating the production of value-added compounds through the shikimate pathway (Ahn et al. 2009; Khamduang et al. 2009).
Another challenge that needs to be elucidated is the complex regulatory system of the shikimate pathway. The recent protein and metabolic engineering developments provide new and state-of-the-art technologies to significantly enhance the metabolic flux by modifying the shikimate pathway. Thanks to the key advancements, the feedback resistance (Fbr) enzymes of AroF, AroG, and AroH for DAHP production, the first rate-limiting step of the shikimate pathway, have been identified, which considerably improved the pathway efficiency by eliminating the regulation on this step (Jossek et al. 2001). The expression of the related pathway enzymes can also be increased by eliminating the pathway regulator TyrR (Lutke-Eversloh and Stephanopoulos 2007). In conclusion, the studies mentioned above offer sustainable opportunities for tailoring the shikimate pathway to produce a wide-variety of fine chemicals with potential biological or pharmaceutical applications.
Modified chemically inducible chromosomal evolution: stable gene overexpression system
The plasmid-based overexpression of rate determining enzymes, e.g., AroE, AroB, and AroG is a useful strategy to achieve a high-level accumulation of shikimate. Nevertheless, genetic instability often exhibited for plasmids presumably due to segregational and structural instability and allele segregation that results in reduced productivity of the target product (Friehs 2004). Stable maintenance of plasmids in the host cells necessitates the use of selective agents, i.e., antibiotic which consequently raises the overall production cost and provokes environmental issues. More importantly, the metabolic load or burden can be imposed as a result of duplicated plasmids competency for carbon sources, energy, and reducing equivalents within the host cell. Though several chromosomal integration approaches with single copy have been widely pursued to mitigate the plasmids instability drawbacks, little consideration so far has been given to integrating genes with multiple copies. Tyo and coworkers (Tyo et al. 2009) introduced a novel plasmid-free and high gene copy chemically induced chromosomal evolution (CIChE) expression system for the incorporation of target genes into E. coli genome. The pathway copy numbers are maintained by recA inactivation, and the tailored strain was found to be unaffected by plasmid instability issues and necessitating no selection markers. In contrast to plasmid-assisted strain, CIChE-engineered strains revealed approximately 10-folds increased genetic stability and 4-folds improved the specific productivity of a biopolymer poly-3-hydroxybutyrate (PHB). Using this technique, target genes can be directly implanted into the DNA of E. coli by the λInCh genomic integration strategy and target gene copy numbers were evolved by chemical induction. Nonetheless, the λInCh genomic integration procedure is tedious, labor-intensive, and complicated, as it comprises three steps that include two recombination steps. Chiang et al. (2008) modified the conditional-replication, integration, and modular plasmid system produced by Haldimann and Wanner (2001) and developed a replicon-free and markerless method (RMM) for the chromosomal insertion of genes. Genes of interest can be directly integrated into the bacterial attachment site of the E. coli chromosome as single copies, through transformation. However, the CIChE strains reported by Tyo et al. (2009) still have an antibiotic resistance marker (chloramphenicol resistance). To address the fundamental shortcomings of CIChE technology, Liu et al. (2012) constructed a series of triclosan induction-based integration expression plasmids, pXKF3T5bbe employed to integrate genes of interest into E. coli chromosome by transformation using replicon-free and markerless method (RMM). In a step-forward, Chen et al. (2013) developed a high-level lycopene producing an E. coli strain without carrying a plasmid or an antibiotic selection marker using triclosan-induced chromosomal evolution (Fig. 2). A gene cluster comprising deregulated aroB, aroE, aroG, and tktA was inserted into site-specific genome site of E. coli BW25113 adopting this approach. In combination with aroK and aroL knockout, incorporating additional chromosomal copies of pntAB, tktA, and nadK, the resulting engineered strain produced 3.12 g/L of shikimate with a corresponding yield of 0.33 mol/mol glucose (Cui et al. 2014).
A schematic representation of modified chemically inducible chromosomal evolution (CIChE). The CIChE DNA cassette contains a triclosan marker and target gene(s), flanked by homologous regions. By recA-mediated recombination between the leading homologous regions in one DNA strand with the trailing homologous region in another strand, one daughter cell contains two copies of the cassette will be generated. This process can be repeated when recA is present (adopted from Gu et al. 2017; an open-access article distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/)
Fluxomics and metabolomics approaches
Genetic manipulation of single or multiple genes is a most common and powerful strategy in metabolic engineering to improve the flux of target pathway. Nevertheless, inadequate supply of key precursors, excessive knocking out of competing pathways, and/or over-engineering of the pathway unnecessarily caused a metabolic burden on the host cell (Jiang and Zhang 2016). Fluxomics is a widely emerging field to measure the metabolic reaction states in different biological systems, whereas metabolomics is one of the powerful omics tools that enable the identification, exploration, and quantification of intricate biochemical changes in cells occurring as a result of different environmental stimuli, such as nutrition or stress (Wojtowicz and Piotr 2016). Combining metabolomics and fluxomics accompanied by dynamic 13C-labeling approach might provide insightful clues/evidence on the pathway activities and qualitative changes in pathway contributions (Yao et al. 2018). These can also be used to pinpoint bottleneck steps in the biosynthesis of target metabolite within the pathways and may determine the underlying pathway interactions (Nöh et al. 2007; McAtee et al. 2015; Jazmin et al. 2017). A comprehensive carbon and energy metabolism remodeling can be demonstrated in shikimate producers by advanced metabolomics and fluxomics strategies. Recently, Rodriguez and coworkers (Rodriguez et al. 2017) carried out the fluxomics and metabolomics analysis of a recombinant E. coli strain AR36 with a specific shikimate production rate of 0.20 g/g h. The global remodeling of the carbon and energy metabolism analysis revealed that a greater portion of glucose substrate was directed and assimilated into shikimate pathway while circumventing the intermediates conversion to more toxic compounds. Furthermore, the pentose phosphate pathway was found to be profoundly stimulated in engineered strain to delivering E4P and equilibrating the NADPH supplies for shikimate biosynthesis. Similarly, Yao and coworkers (Yao et al. 2016) scrutinized the fluxome profiles and regulation of glucose and glycerol-based co-metabolism in a carbon catabolite repression-negative E. coli mutant (ΔptsGglpK) to decipher its metabolic potential and matched with the native strain. Metabolic flux distribution results revealed that, in contrast to the wild-type strain, the engineered derivative repressed its carbon flux through the tricarboxylic acid (TCA) cycle and results in elevated acetate overflow. The fluxes regulation results were found to be in consonance with transcriptional analysis of several key genes concerning TCA cycle. Moreover, the redox NADPH/NADH state was considerably affected by the mutant strain that led to reduced ATP level. From these outcomes, it can be concluded that combined utilization of metabolomics and fluxomics approaches, the novel candidate targets can be efficiently discovered and reorganized for genetic modification to further tailoring the shikimate pathway for diverse valuable chemicals production.
Bioprocess engineering and fermentation strategies
Microorganisms need to constantly absorb nutrients (for example, carbon and nitrogen), growth factors, inorganic salts, and water. Microorganisms used them to synthesize new cell materials and excrete metabolites while taking energy from them. The culture medium provides essential nutrients for the growth, metabolism, and reproduction of the producing bacteria. The composition and proportion of the medium are suitable for the growth and development of the bacteria, the fermentation unit of the metabolites, the extraction process, and the quality and yield of the final product (Yang et al. 2018). As reported, altered glucose transport and modulation of PEP synthase expression could improve the shikimate pathway-derived target bioproducts titers in E. coli (Yi et al. 2002). Therefore, increasing the supply of carbon metabolites and their channeling toward the production of aromatic intermediates improves shikimate production and its aromatic precursors in E. coli (Escalante et al. 2010). The production of shikimate can be improved via fermentation optimization. In a study, Iyer et al. (2007) achieved an increasing titer of shikimate to 648.12 mg/L in shake flask fermentation by optimizing fermentation time, culture medium, and the growth conditions for the shikimate-producing bacteria.
Substrate engineering and optimization is a noteworthy approach for the high-level biosynthesis of shikimate production. The exploration and utilization of a broad variety of easily-available and inexpensive bio-based materials ensure several advantages such as overall cost-effective ratio along with long-term feasibility and sustainability of the process. For example, glycerol, a by-product of the biodiesel production process, appeared as a readily available and inexpensive feedstock for the production of industrially relevant products with substantial commercial interest such as shikimate by metabolically engineered strains. Nonetheless, carbon flux through the PP pathway might be insufficient using only a glycerol-based medium, which may lead to the inadequate availability of E4P (Chubukov et al. 2013). For induced shikimate production, the addition of glucose or other pentoses in the culture medium could increase the carbon flux through the PP pathway to supplement E4P. For example, the combination of low-cost glycerol with some other sugars such as glucose led to an accelerated shikimate production and hence be advantageous for the biosynthesis of other value-able compounds. Nevertheless, it is imperative to investigate the effect of cultivation parameters such as pH and temperature on the cell biomass and product accumulation. In a recent study, Bilal et al. (2018a) reported high shikimate titters from a genetically engineered E. coli strain with simultaneous utilization of glycerol/glucose. For this, the ptsG gene encoding EIICBglc protein followed by the shikimate kinase genes aroK and aroL were inactivated in the BW25113 to construct SA-B2 strain resulting in 42.3% improvement in shikimate titer. Subsequently, three critical genes, namely mutant aroGfbr, ppsA, and tktA, were overexpressed, and the resulting engineered SA-B7 strain accumulated 0.92 g/L shikimate in a mixed-substrate strategy, which was 0.64 g/L using the only glycerol-based medium in shake flask cultures. In another study, C. glutamicum was engineered with the capability to use glucose/pentose mixed fermentation medium concurrently, and the tailored strain was applied to anaerobic, growth-arrested, and high-density cell reaction to hyper-produce shikimic acid. A high titer of shikimate (141 g/L) with a 51%yield was achieved in the glucose-containing minimal medium after 48 h without the cell growth essential nutrients. Furthermore, equivalent shikimate yield was attained by the simultaneous assimilation of arabinose, glucose, and xylose, allowing proficient shikimate biosynthesis from lignocellulosic raw materials (Kogure et al. 2016).
Shikimate pathway based biosynthesis of value-added compounds
The engineering of shikimate pathway is a principal approach for the manufacturing of a huge variety of compounds with diverse applications in chemical, cosmetic, pharmaceutical, food, and agricultural sectors (Fig. 3). This section summarizes the generally applicable and successful strategies for the production of shikimate-derived compounds including aromatic amino acids, polyphenol, and phenazine-derived compounds.
Diversity of chorismate derived produced based on shikimate pathway. A1: resveratrol, A2: danshensu, A3: ferulate, A4: chalcones; B1: phenazine-1-carboxylic acid, B2: phenazine-1-carboxamide, B3: phenazine-1,6-dicarboxylic acid, B4: pyoluteorin; C1: phenylpropanoids, C2: chaconnes, C3: cinnamic acid, C4: ferulic acid
Aromatic amino acids
Shikimate can be used as a common precursor to produce three aromatic amino acids l-Phe, l-Tyr, and l-Trp, so the same process of aromatic amino acid synthesis can also be referred to as the shikimate pathway (Sprenger 2007b; Keseler et al. 2013). Aromatic amino acids are essential dietary constituents for humans and higher animals, therefore used as dietary supplements, and important precursors of industrial and therapeutic compounds (Lütke-Eversloh et al. 2007; Treibmann et al. 2017). Among the aromatic amino acids, the worldwide market growth of l-Tyr exceeds 14,000 tons/year, whereas the manufacturing of l-Phe is approximated to be more than 30,000 tons/year (Li et al. 2010).
In earlier studies, several metabolic approaches have been successfully exploited to strengthen the shikimate pathway for producing an array of desired metabolites particularly aromatic amino acids in E. coli strains. These strategies include enhancing the supply of direct key precursors PEP and E4P, elevating DAHP level in the shikimate pathway, improving or diverting carbon flux through the biosynthetic pathway by eliminating allosteric/transcriptional regulation, inactivating competing pathways to prevent carbon loss, determining and overexpressing enzymes involved in the rate-limiting steps, and preventing product degradation or re-internalization (Rodriguez et al. 2013). One of the important direct precursor’s PEP availability can be increased by recycling of pyruvate to PEP by overexpressing ppsA gene (Yi et al. 2002). Similarly, the overexpression of tktA gene encoding a transketolase enzyme led to an accelerated supply of E4P (Baez et al. 2001). It is important to mention that appropriately overexpressing a few genes can augment the metabolic flux towards the aromatic biosynthetic pathway. Thus, the resultants products are significantly influenced by several factors, e.g., genetic background, a combination of expression modules, and bioprocessing environments. Therefore, it is of considerable significance to design experiments to get insight about the influence of each factor to the phenotype.
4-Hydroxybenzoate and its derivatives
4-Hydroxybenzoate is a raw material for the manufacturing of antibacterial parabens, a group of compounds widely used as stabilizers in food, cosmetic, and pharmaceutical products. It is also a major building block for the manufacturing of specialty chemical, i.e., a high-performance liquid crystal polymer (LCP) that is extensively employed in the fiber and thermoplastic industry for high-strength applications (Ibeh 2011). Existing synthesis of 4-HBA is largely sourced from petrochemicals. Nevertheless, the limited petroleum resources, generation of undesired by-products, and serious environmental concerns render the chemical process comparatively expensive and environmentally unfriendly. Biotechnological production of 4-HBA from renewable resources appears an environmentally responsive and economically favorable bioprocessing approach for the enhanced biosynthesis of aromatic 4-HBA (Yao et al. 2016; Wang et al. 2018a). Barker and Frost (2001) constructed an E. coli strain with pheA tyrA4 trpE-C gene deletions and shikimate pathway genes, i.e., aroA, aroB, aroC, aroL overexpression for the biosynthesis of 4-HBA from glucose via chorismate. An elevated titer of 12 g/L 4-HBA with a yield of 13% (mol/mol) was achieved following plasmid-assisted expression of ubiC, aroFFBR, and tktA under fed-batch fermentation conditions. Increased understanding of metabolic directions derived from 4-HBA accompanied by biochemical characterization of pathway associated genes and enzymes offer novel clues for rationally engineering strains producing an array of polyphenol and polyketide-derived compounds such as resveratrol, gastrodin, etc. Resveratrol constitutes the most studied plant-based polyphenols with enormous biological activities, such as antibacterial, antioxidant, anticarcinogenic, hypolipidemic, and antimutagenic activities (Tissier et al. 2014). Lim and coworkers (Lim et al. 2011) developed a synthetic approach in E. coli to optimizing resveratrol synthesis from p-coumaric acid. The modified strain yielded an impressive level of resveratrol up to 2.3 g/L. Gastrodin is an important and active constituent of a renowned Chinese medicine gastrodiaelata B1. This drug is widely used as a sedative, antiaging, anti-inflammatory, anticonvulsant, and antimyocardial ischemia. It is usually synthesized from chemical routes and by extraction from the plant which exhibits the drawbacks of complicated manufacturing procedure and health compliances (Wang et al. 2007). Microbial transformation of p-hydroxybenzaldehyde to gastrodin might be an economical and environmentally friendly alternative to conventional extraction and chemical catalyst (Fig. 4). For example, Bai et al. (2016) constructed a non-native biosynthetic pathway in an E. coli strain and achieved 545 mg/L of gastrodin in the shake flask culture medium.
Proposed biosynthetic pathway of gastrodin. E4P: erythrose 4-phosphate; PEP: phosphoenolpyruvate: CAR: carboxylic acid reductase; Sfp: phosphopantetheinyl transferase: ADHs, alcohol dehydrogenase; UGT73B6FS: uridine sugar glycosyltransferase (Bai et al. 2016)
Phenazine and its derived compounds
Phenazines have been known for their wider-antibiotic spectrum and thus represent a large group of bacterial secondary metabolites. Phenazine and its derived compounds have been extensively used as biocontrol candidates for a range of fungal phytopathogens (Pierson and Pierson 2010; Zhao et al. 2017a, b; Yue et al. 2018). Characteristically, phenazines are pigmented heterocyclic nitrogen-containing compounds with two distinct absorption spectra in the ultraviolet and at least one in the visible range. These functional groups differ depending on the nature and position of substituents on the heterocyclic ring and are mainly accountable for distinction in their physicochemical properties, and hence, biocontrol activities. The addition of a variety of functional groups determines the solubility and redox potential of these compounds, thus influencing their biological properties (Laursen and Nielsen 2004). Most of the phenazines are simple carboxy- and hydroxyl-substituted derivatives with unique physicochemical and antibiotic activities. Evidence that phenazine producers survive longer than non-phenazine-producing species indicated that the phenazine production protects its producer’s habitat against other microbial competitors due to the antibiotic activity.
In recent years, phenazine and its derivative compounds have gained particular interest as leading molecules with potential applications such as environmental sensing/monitoring, manufacturing of microbial fuel cell, antitumor, antimalarial and antiparasitic compounds, and anticancer prodrug (Fig. 5) (Pierson and Pierson 2010; Bilal et al. 2018b; Liu et al. 2018; Wang et al. 2018b).
Reports have shown that above 6000 phenazine-containing compounds have been documented in the scientific literature in the last century. Though less than 100 are of natural origin with the same basic structure, many of these exhibit remarkable antibiotic activities toward a range of bacteria, fungi, and animal and plant tissues (Laursen and Nielsen 2004). Among the potent phenazine producers, fluorescent pseudomonads such as Pseudomonas aeruginosa, Pseudomonas chlororaphis, and Pseudomonas fluorescens constitute the best-characterized phenazine-producing bacterial strains. In vitro screening and genetic engineering experiments revealed that the phenazine biosynthetic profile of P. aeruginosa comprises pyoluteorin (PYO), phenazine 1-carboxylic acid (PCA), phenazine 1-carboxamide (PCN), 1-hydroxy phenazine (1-OH-PHZ), and Aeruginosin A and B, etc. whereas P. chlororaphis has shown to produce PCA, 2-hydroxy PCA, and 2-hydroxy phenazine (Huang et al. 2011). It is reported that the genes encoding the enzymes involved in the assemblage of the three-ringed structure of phenazine display a highly conserved set of five genes (Gross and Loper 2009).
Phenazine 1-carboxylic acid is one of the significant nitrogen-containing heterocyclic phenazine derivatives. It displays profound antifungal activity and used in both agriculture and medicine (Puopolo et al. 2013; Gorantla et al. 2014). Recently, it has been certified as a new, green, and environmentally friendly biopesticide (Shenqinmycin) and granted a pesticide production approval certificate issued by the Chinese Ministry of Agriculture to enter industrial production. Among microbial strains, Pseudomonas and Streptomyces species have been the extensively studied PCA-producing bacterial genera, as biological control agents (Geiger et al. 1988; Park et al. 2012). In the past years, PCA has been reported as a curative agent against numerous fungal-based crop diseases, for example, ginger rhizome rot, cucumber anthracnose, pepper Phytophthora blight potato scab, and wheat take-all disease, etc. (St-Onge et al. 2011; Arseneault et al. 2013; Jasim et al. 2014). Owing to other potent functionalities such as low toxicity, fungicidal efficacy, biocompatibility, and biodegradability, PCA and PCA-producing strains have attracted a substantial researcher’s interest as biocontrol agents (Daes et al. 2011). However, wild-type strains have a low industrial fermentation titer of PCA that limits its commercial-scale application feasibility due to high production costs. Therefore, it is necessary to enhance the yield of PCA for its application in agriculture. In this background, a plethora of studies have been focused, in recent years, on engineering high-yielding strains and bioprocessing conditions optimization for induced production of phenazine and its derivatives (Liu et al. 2016a; Jin et al. 2016; Hu et al. 2017; Yue et al. 2018; Peng et al. 2018). For example, Jin et al. (2015) applied a combined genetic strategy including gene, promoter, and protein engineering to tailoring the metabolic pathways for the hyperproduction of PCA in P. aeruginosa PA1201. The resulting engineered strain PA-IV produced up to 9882 mg/L PCA under the fed-batch fermentation conditions. Similarly, 2.0 g/L PCA was accumulated in the culture broth of a gacA-inactivated Pseudomonas sp. M18G derivative after 60 h of fermentation time under standardized carbon and nitrogen sources (Li et al. 2008).
Phenazine 1-carboxamide is another important phenazine derivative that possesses pronounced antagonistic activity against Fusarium oxysporum than that of PCA. Recently, novel genetic modification approaches have been endeavored to develop bacterial strains with a great capacity to produce PCN for its commercial applications (Jin et al. 2016). Also, the critical factors influencing the phenazine biosynthesis have been elucidated in Pseudomonas strains by comparative genomic and transcriptomic approaches (Chen et al. 2015). As a phenazine derivative, phenazine-1,6-dicarboxylic acid (PDC) functions as an outstanding antibiotic and anticancer agent. Interestingly, researchers have found that all the PDC producers lack the PhzA that plays an important role in phenazine production (Chin-A-Woeng et al. 2001). However, the role of phzA in relation to PDC production has not been described yet. Therefore, explicating the function of phzA in phenazine biosynthesis might provide a feasible method for the enhanced biosynthesis of PDC in Pseudomonads. Table 2 demonstrates the proposed biotechnological, in particular, agricultural applications of phenazine derivatives produced through shikimate pathway.
Biological or pharmaceutical functionalities of shikimate and its derivatives
Shikimate is an important chiral precursor for the biosynthesis of many valuable compounds with remarkable potentialities in the chemical, pharmaceutical, and cosmetic industry. Regarding biological and pharmaceutical applications, shikimate displays enormous potential as an anticoagulant, anti-inflammatory, antioxidant, antipyretic, antithrombotic, and analgesic agent. It also has a significant role in the manufacturing of compounds with pharmaceutical perspective such as anticancer and antibacterial agents, and hormonal therapy (Blanco et al. 2013). Use of shikimate as the starting material for the chemical production of oseltamivir phosphate is considered to be the most prevalent application (Bochkov et al. 2012; Rawat et al. 2013), which could considerably meet the commercial demand and trim-down the production cost of oseltamivir phosphate or other oseltamivir carboxylates (Guo and Frost 2004).
Shikimate diminishes the occurrence of neurological deficit and cerebral infarction, abates brain edema, and increases the blood coagulation time and cerebral blood flow in ischemic areas of mice (Ma et al. 2000). Several shikimate derivatives such as the triacetyl, monopalmityloxy, and isopropylidene have been synthesized and assessed for their biological activities as antithrombotic and protecting agents in brain damage triggered by cerebral ischemia. Notably, triacetyl shikimate displays potent antithrombotic and anticoagulant activities in rats (Huang et al. 2002). Monopalmityloxy shikimate can prolong the coagulation time accompanied by its antithrombotic effects (Tang et al. 2009). In addition to antithrombotic and anticoagulant activities, the 3,4-oxo-isopropylidene shikimate prevents in vitro adhesion of polymorphonuclear leukocytes to tumor necrosis factor alpha (TNF-α)-induced endothelial cells. More importantly, this shikimate derivative has shown potential to manufacture drugs for the treatment of ulcerative colitis (Xing et al. 2012). The shikimate complexes of platinum (II) are effective against in vivo B16 melanoma as well as L1210 and P388 leukemia (Farrell et al. 1991). In recent years, the synthesis of shikimate and its derivatives has gained considerable research interest for inhibitors development, as represented by the manufacturer of the influenza virus NA inhibitor (NAI) OSP, the α-glycosidase inhibitor valiolamine, and the glyoxalase I inhibitor COCT (2-crotonyloxymethyl-(4R,5R,6R)-4,5,6-trihydroxycyclohex-2-enone) (Lo et al. 2012).
Concluding remarks and future perspectives
The present review systematically summarizes a panorama of the current metabolic engineering achievements combined with newly developed technologies for elevated biosynthesis of shikimate in shikimate-producing strains. Extensive research efforts have been carried out over the last several years to produce shikimate from microorganism using different carbon sources. Nevertheless, the majority of the shikimate producers were acquired by rational metabolic engineering depending on the availability of genetic tools for target candidate. Importantly, shikimate production was almost restricted to E. coli strain because of the expensive and time-consuming development of new genetic tools for non-model microorganisms. Therefore, very scarce attempts have been made on C. glutamicum and B. subtilis for shikimate biosynthesis in the last decade. Random mutagenesis is often considered as a feasible approach to improve the characteristics of non-model strains and further engineering other high-yielding shikimate producers. Enhanced biosynthesis of shikimate requires the consideration of several important aspects: (1) appropriate level of shikimate pathway enzyme expression should be identified to prevent unnecessary metabolic burden imposed by over-engineering of the pathway, (2) eliminate excessive deletion of competing pathways even though it increase the metabolic flux through the shikimate pathway. It might decrease the production capacity by impairing the growth of the host strain. (3) Selection of a suitable carbon source and/or co-fermentation could be an effective way to improve the efficiency of the shikimate pathway.
References
Ahn J, Hong J, Park M, Lee H, Lee E, Kim C, Lee J, Choi E, Jung J, Lee H (2009) Phosphate-responsive promoter of a Pichia pastoris sodium phosphate symporter. Appl Environ Microbiol 75(11):3528–3534
Arseneault T, Goyer C, Filion M (2013) Phenazine production by Pseudomonas sp. LBUM223 contributes to the biological control of potato common scab. Phytopathology 103:995–1000
Baez JL, Bolivar F, Gosset G (2001) Determination of 3-deoxy-D-arabino-heptulosonate 7-phosphate productivity and yield from glucose in Escherichia coli devoid of the glucose phosphotransferase transport system. Biotechnol Bioeng 73:530–535
Báez-Viveros JL, Flores N, Juárez K, Castillo-España P, Bolívar F, Gosset G (2007) Metabolic transcription analysis of engineered Escherichia coli strains that overproduce L-phenylalanine. Microb Cell Factories 6:28
Bai Y, Yin H, Bi H, Zhuang Y, Liu T, Ma Y (2016) De novo biosynthesis of gastrodin in Escherichia coli. Metab Eng 35:138–147
Barker JL, Frost JW (2001) Microbial synthesis of p-hydroxybenzoic acid from glucose. Biotechnol Bioeng 76:376–390
Bilal M, Guo S, Iqbal HMN, Hu H, Wang W, Zhang X (2017) Engineering Pseudomonas for phenazine biosynthesis, regulation, and biotechnological applications: a review. World J Microbiol Biotechnol 33(10):191
Bilal M, Yue S, Hu H, Wang W, Zhang X (2018a) Systematically engineering Escherichia coli for enhanced shikimate biosynthesis co-utilizing glycerol and glucose. Biofuels Bioprod Biorefin 12(3):348–361
Bilal M, Yue S, Hu H, Wang W, Zhang X (2018b) Adsorption/desorption characteristics, separation and purification of phenazine-1-carboxylic acid from fermentation extract by macroporous adsorbing resins. J Chem Technol Biotechnol. https://doi.org/10.1002/jctb.5673
Blanco B, Prado V, Lence E, Otero JM, Garcia-Doval C, van Raaij MJ, Llamas-Saiz AL, Lamb H, Hawkins AR, Gonzalez-Bello C (2013) Mycobacterium tuberculosis shikimate kinase inhibitors: design and simulation studies of the catalytic turnover. J Am Chem Soc 135(33):12366–12376
Bochkov DV, Sysolyatin SV, Kalashnikov AI, Surmacheva IA (2012) Shikimic acid: review of its analytical, isolation, and purification techniques from plant and microbial sources. J Chem Biol 5:5–17
Bongaerts J, Krämer M, Müller U, Raeven L, Wubbolts M (2001) Metabolic engineering for microbial production of aromatic amino acids and derived compounds. Metab Eng 3(4):289–300
Breitenbach M, Rinnerthaler M, Hartl J, Stincone A, Vowinckel J, Breitenbach-Koller H, Ralser M (2014) Mitochondria in ageing: there is metabolism beyond the ROS. FEMS Yeast Res 14(1):198–212
Chandran SS, Yi J, Draths KM, von Daeniken R, Weber W, Frost JW (2003) Phosphoenolpyruvate availability and the biosynthesis of shikimic acid. Biotechnol Prog 19(3):808–814
Chen K, Dou J, Tang S, Yang Y, Wang H, Fang H, Zhou C (2012) Deletion of the aroK gene is essential for high shikimic acid accumulation through the shikimate pathway in E. coli. Bioresour Technol 119:141–147
Chen YY, Shen HJ, Cui YY, Chen SG, Weng ZM, Zhao M, Liu JZ (2013) Chromosomal evolution of Escherichia coli for the efficient production of lycopene. BMC Biol 13:6
Chen X, Li M, Zhou L, Shen W, Algasan G, Fan Y, Wang Z (2014) Metabolic engineering of Escherichia coli for improving shikimate synthesis from glucose. Bioresour Technol 166:64–71
Chen Y, Shen X, Peng H, Hu H, Wang W, Zhang X (2015) Comparative genomic analysis and phenazine production of Pseudomonas chlororaphis, a plant growth-promoting rhizobacterium. Genom Data 4:33–42
Chiang C-J, Chen PT, Chao YP (2008) Replicon-free and markerless methods for genomic insertion of DNAs in phage attachment sites and controlled expression of chromosomal genes in Escherichia coli. Biotechnol Bioeng 101:985–995
Chin-A-Woeng TFC, van den Broek D, de Voer G, van der Drift KM, Tuinman S, Thomas-Oats JE, Lugtenberg BJJ, Bloemburg GV (2001) Phenazine-1-carboxamide production in the biocontrol strain Pseudomonas chlororaphis PCL1391 is regulated by multiple factors secreted in the growth medium. Mol Plant-Microbe Interact 14:969–979
Chubukov V, Uhr M, Chat LL, Kleijn RJ, Jules M, Link H (2013) Transcriptional regulation is insufficient to explain substrate-induced flux changes in Bacillus subtilis. Mol Syst Biol 9:709
Cui YY, Chen L, Zhang YY, Jian H, Liu JZ (2014) Production of shikimic acid from Escherichia coli through chemically inducible chromosomal evolution and cofactor metabolic engineering. Microb Cell Factories 13:21
Daes J, Hua GK, De Maeyer K, Pannecoucque J, Forrez I, Ongena M, Dietrich LE, Thomashow LS, Mavrodi DV, Hofte M (2011) Biological control of Rhizoctonia root rot on bean by phenazine- and cyclic lipopeptide-producing Pseudomonas CMR12a. Phytopathology 101:996–1004
Dasgupta D, Kumar A, Mukhopadhyay B, Sengupta TK (2015) Isolation of phenazine 1,6-di-carboxylic acid from Pseudomonas aeruginosa strain HRW.1-S3 and its role in biofilm-mediated crude oil degradation and cytotoxicity against bacterial and cancer cells. Appl Microbiol Biotechnol 99:8653–8665
Delaney SM, Mavrodi DV, Bonsall RF, Thomashow LS (2001) phzO, a gene for biosynthesis of 2-hydroxylated phenazine compounds in Pseudomonas aureofaciens 30–84. J Bacteriol 183:318–327
Diaz-Quiroz DC, Cardona-Felix CS, Viveros-Ceballos JL, Reyes-Gonzalez MA, Bolivar F, Ordonez M, Escalante A (2018) Synthesis, biological activity and molecular modelling studies of shikimic acid derivatives as inhibitors of the shikimate dehydrogenase enzyme of Escherichia coli. J Enzyme Inhib Med Chem 33(1):397–404
Dosselaere F, Vanderleyden J (2001) A metabolic node in action: chorismate-utilizing enzymes in microorganisms. Crit Rev Microbiol 27:75–131
Du X, Li Y, Zhou W, Zhou Q, Liu H, Xu Y (2013) Phenazine-1-carboxylic acid production in a chromosomally non-scar triple-deleted mutant Pseudomonas aeruginosa using statistical experimental designs to optimize yield. Appl Microbiol Biotechnol 97:7767–7778
Escalante A, Calderon R, Valdivia A, de Anda R, Hernandez G, Ramirez OT, Gosset G, Bolivar F (2010) Metabolic engineering for the production of shikimic acid in an evolved Escherichia coli strain lacking the phosphoenolpyruvate: carbohydrate phosphotransferase system. Microb Cell Factories 9:21
Estevez AM, Estevez RJ (2012) A short overview on the medicinal chemistry of (-)-shikimic acid. Mini Rev Med Chem 12(14):1443–1454
Eudes A, Berthomieu R, Hao Z, Zhao N, Benites VT, Baidoo EEK, Loque D (2018) Production of muconic acid in plants. Metab Eng 46:13–19
Farrell N, Roberts JD, Hacker MP (1991) Shikimic acid complexes of platinum. Preparation, reactivity, and antitumor activity of (R,R-1,2-diaminocyclohexane) bis(shikimato) platinum(II). Evidence for a novel rearrangement involving platinum-carbon bond formation. J Inorg Biochem 42(4):237–246
Flores S, Gosset G, Flores N, de Graaf AA, Bolívar F (2002) Analysis of carbon metabolism in Escherichia coli strains with an inactive phosphotransferase system by 13C labeling and NMR spectroscopy. Metab Eng 4:124–137
Friehs K (2004) Plasmid copy number and plasmid stability. In: Scheper TH (ed) New trends and developments in biochemical engineering. Springer, Berlin, pp 47–82
Gao M, Cao M, Suastegui M, Walker JA, Rodriguez-Quiroz N, Wu Y (2017) Innovating a nonconventional yeast platform for producing shikimate as the building block of high-value aromatics. ACS Synth Biol 6:29–38
Geiger A, Keller-Schierlein W, Brandl M, Zahner H (1988) Metabolites of microorganisms. Phenazines from Streptomyces antibioticus, strain Tu 2706. J Antibiot (Tokyo) 41:1542–1551
Ghosh S, Chisti Y, Banerjee UC (2012) Production of shikimic acid. Biotechnol Adv 30:1425–1431
Gorantla JN, Kumar SN, Nisha GV, Sumandu AS, Dileep C, Sudaresan A, Kumar MM, Lankalapalli RS, Kumar BS (2014) Purification and characterization of antifungal phenazines from a fluorescent Pseudomonas strain FPO4 against medically important fungi. J Mycol Med 24:185–192
Gross H, Loper JE (2009) Genomics of secondary metabolite production by Pseudomonas spp. Nat Prod Rep 26:1408–1446
Gu P, Su T, Wang Q, Liang Q, Qi Q (2016) Tunable switch mediated shikimate biosynthesis in an engineered non-auxotrophic Escherichia coli. Sci Rep 6:29745
Gu P, Fan X, Liang Q, Qi Q, Li Q (2017) Novel technologies combined with traditional metabolic engineering strategies facilitate the construction of shikimate-producing Escherichia coli. Microb Cell Factories 16(1):167
Guo J, Frost JW (2004) Synthesis of aminoshikimic acid. Org Lett 6(10):1585–1588
Haldimann A, Wanner BL (2001) Conditional-replication, integration, excision, and retrieval plasmid-host systems for gene structure-function studies of bacteria. J Bacteriol 183:6384–6393
Herrmann KM, Weaver LM (1999) The shikimate pathway. Ann Rev Plant Physiol Plant Mol Biol 50(1):473–503
Horimoto T, Kawaoka Y (2001) Pandemic threat posed by avian influenza A viruses. Clin Microbiol Rev 14(1):129–149
Hu H, Li Y, Liu L, Zhao J, Wang W, Zhang X (2017) Production of trans-2,3-dihydro-3-hydroxyanthranilic acid by engineered Pseudomonas chlororaphis GP72. Appl Microbiol Biotechnol 101:6607–6613
Huang F, Xiu Q, Sun J, Hong E (2002) Anti-platelet and anti-thrombotic effects of triacetylshikimic acid in rats. J Cardiovasc Pharmacol 39(2):262–270
Huang L, Chen M, Wang W, Hu H, Peng H, Xu Y, Zhang X (2011) Enhanced production of 2-hydroxyphenazine in Pseudomonas chlororaphis GP72. Eur J Appl Microbiol Biotechnol 89(1):169–177
Ibeh CC (2011) Thermoplastic materials: properties, manufacturing methods, and applications. CRC Press, Boca Raton
Iomantas YAV, Abalakina EG, Polanuer BM, Yampolskaya TA, Bachina TA, Kozlov YI (2002) Method for producing shikimic acid, US
Iyer S, Pejakala V, Karabasanagouda V, Wagle S, Balaya L, Kanaka M, Hiremath M (2007) Method for obtaining shikimic acid, wo
Jasim B, Anisha C, Rohini S, Kurian JM, Jyothis M, Radhakrishnan EK (2014) Phenazine carboxylic acid production and rhizome protective effect of endophytic Pseudomonas aeruginosa isolated from Zingiber officinale. World J Microbiol Biotechnol 30:1649–1654
Jazmin LJ, Xu Y, Cheah YE, Adebiyi AO, Johnson CH, Young JD (2017) Isotopically nonstationary 13C flux analysis of cyanobacterial isobutyraldehyde production. Metab Eng 42:9–18
Jiang M, Zhang H (2016) Engineering the shikimate pathway for biosynthesis of molecules with pharmaceutical activities in E. coli. Curr Opin Biotechnol 42:1–6
Jin K, Zhou L, Jiang H, Sun S, Fang Y, Liu J, Zhang X, He YW (2015) Engineering the central biosynthetic and secondary metabolic pathways of Pseudomonas aeruginosa strain PA1201 to improve phenazine-1-carboxylic acid production. Metab Eng 32:30–38
Jin XJ, Peng HS, Hu HB, Huang XQ, Wang W, Zhang XH (2016) iTRAQ-based quantitative proteomic analysis reveals potential factors associated with the enhancement of phenazine-1-carboxamide production in Pseudomonas chlororaphis P3. Sci Rep 6:27393
Johansson L, Liden G (2006) Transcriptome analysis of a shikimic acid producing strain of Escherichia coli W3110 grown under carbon- and phosphate-limited conditions. J Biotechnol 126(4):528–545
Johansson L, Lindskog A, Silfversparre G, Cimander C, Nielsen KF, Liden G (2005) Shikimic acid production by a modified strain of E. coli (W3110 Shik 1) under phosphate-limited and carbon-limited conditions. Biotechnol Bioeng 92:541–552
Jossek R, Bongaerts J, Sprenger GA (2001) Characterization of a new feedback-resistant 3-deoxy-D-arabino-heptulosonate 7-phosphate synthase AroF of Escherichia coli. FEMS Microbiol Lett 202(1):145–148
Kerr JR, Taylor GW, Rutman A, Hoiby N, Cole PJ, Wilson R (1999) Pseudomonas aeruginosa pyocyanin and 1-hydroxyphenazine inhibit fungal growth. J Clin Pathol 52(5):385–387
Keseler IM, Mackie A, Peralta-Gil M, Santos-Zavaleta A, Gama-Castro S, Bonavides-Martinez C, Fulcher C, Huerta AM, Kothari A, Krummenacker M, Latendresse M, Muniz-Rascado L, Ong Q, Paley S, Schroder I, Shearer AG, Subhraveti P, Travers M, Weerasinghe D, Weiss V, Collado-Vides J, Gunsalus RP, Paulsen I, Karp PD (2013) EcoCyc: fusing model organism databases with systems biology. Nucleic Acids Res 41:D605–D612
Khamduang M, Packdibamrung K, Chutmanop J, Chisti Y, Srinophakun P (2009) Production of L-phenylalanine from glycerol by a recombinant Escherichia coli. J Ind Microbiol Biotechnol 36(10):1267–1274
Kim TH, Namgoong S, Kwak JH, Lee SY, Lee HS (2000) Effects of tktA, aroF FBR, and aroL expression in the tryptophan-producing Escherichia coli. J Microbiol Biotechnol 10:789–796
Knaggs AR (2003) The biosynthesis of shikimate metabolites. Nat Prod Rep 20(1):119–136
Knop DR, Draths KM, Chandran SS, Barker JL, von Daeniken R, Weber W, Frost JW (2001) Hydroaromatic equilibration during biosynthesis of shikimic acid. J Am Chem Soc 123(42):10173–10182
Kogure T, Kubota T, Suda M, Hiraga K, Inui M (2016) Metabolic engineering of Corynebacterium glutamicum for shikimate overproduction by growth-arrested cell reaction. Metab Eng 38:204–216
Kramer M, Bongaerts J, Bovenberg R, Kremer S, Muller U, Orf S, Wubbolts M, Raeven L (2003) Metabolic engineering for microbial production of shikimic acid. Metab Eng 5(4):277–283
Laursen JB, Nielsen J (2004) Phenazine natural products: biosynthesis, synthetic analogues, and biological activity. Chem Rev 104:1663–1685
Lee MY, Hung WP, Tsai SH (2017) Improvement of shikimic acid production in Escherichia coli with growth phase-dependent regulation in the biosynthetic pathway from glycerol. World J Microbiol Biotechnol 33:25
Li Y, Jiang H, Xu Y, Zhang X (2008) Optimization of nutrient components for enhanced phenazine-1-carboxylic acid production by gacA-inactivated Pseudomonas sp. M18G using response surface method. Appl Microbiol Biotechnol 77:1207–1217
Li PP, Liu YJ, Liu SJ (2009) Genetic and biochemical identification of the chorismate mutase from Corynebacterium glutamicum. Microbiology 155:3382–3391
Li Z, Ji X, Kan S, Qiao H, Jian M, Lu D, Wang J, Huang H, Jia H, Ouyuang P, Ying H (2010) Past, present and future industrial biotechnology in China. In: Tsao GT, Ouyang P, Berlin CJ (eds) Biotechnol China II Chem Energy Environ. Springer, Heidelberg, pp 1–42
Lim CG, Fowler ZL, Hueller T, Schaffer S, Koffas MAG (2011) High-yield resveratrol production in engineered Escherichia coli. Appl Environ Microbiol 77(10):3451–3460
Liu HM, Zhang XH, Huang XQ, Cao CX, Xu YQ (2008a) Rapid quantitative analysis of phenazine-1-carboxylic acid and 2-hydroxyphenazine from fermentation culture of Pseudomonas chlororaphis GP72 by capillary zone electrophoresis. Talanta 76(2):276–281
Liu YJ, Li PP, Zhao KX, Wang BJ, Jiang CY, Drake HL, Liu SJ (2008b) Corynebacterium glutamicum contains 3-deoxy-D-arabino-heptulosonate 7-phosphate synthases that display novel biochemical features. Appl Environ Microbiol 74:5497–5503
Liu JZ, Huang MT, Cui YY, Chen YY (2012) A series of expression plasmids for chromosomal integration and evolution. Chinese patent 201210060042.5
Liu K, Hu H, Wang W, Zhang X (2016a) Genetic engineering of Pseudomonas chlororaphis GP72 for the enhanced production of 2-hydroxyphenazine. Microb Cell Factories 15:131
Liu X, Lin J, Hu H, Zhou B, Zhu B (2016b) Site-specific integration and constitutive expression of key genes into Escherichia coli chromosome increases shikimic acid yields. Enzym Microb Technol 82:96–104
Liu Y, Wang Z, Bilal M, Hu H, Wang W, Huang X, Peng H, Zhang X (2018) Enhanced fluorescent siderophore biosynthesis and loss of phenazine-1-carboxamide in phenotypic variant of Pseudomonas chlororaphis HT66. Front Microbiol 9:759
Lo HJ, Chen CY, Zheng WL, Yeh SM, Yan TH (2012) A C2-symmetric pool based flexible strategy: an enantioconvergent synthesis of (+)-valiolamine and (+)-valienamine. Eur J Org Chem 2012(14):2780–2785
Lutke-Eversloh T, Stephanopoulos G (2007) L-tyrosine production by deregulated strains of Escherichia coli. Appl Microbiol Biotechnol 75(1):103–110
Lütke-Eversloh T, Santos CNS, Stephanopoulos G (2007) Perspectives of biotechnological production of L-tyrosine and its applications. Appl Microbiol Biotechnol 77:751–762
Ma Y, Sun JN, Xu QP, Guo YJ (2000) Inhibitory effects of shikimic acid on platelet aggragation and blood coagulation. Acta Pharmacol Sin 5(5):600–612
Martinez JA, Bolivar F, Escalante A (2015) Shikimic acid production in Escherichia coli: from classical metabolic engineering strategies to omics applied to improve its production. Front Bioeng Biotechnol 3:45
McAtee AG, Jazminm LJ, Young JD (2015) Application of isotope labeling experiments and 13C flux analysis to enable rational pathway engineering. Curr Opin Biotechnol 36:50–56
Momen AR, Hoshino T (2000) Biosynthesis of violacein: intact incorporation of the tryptophan molecule on the oxindole side, with intramolecular rearrangement of the indole ring on the 5-hydroxyindole side. Biosci Biotechnol Biochem 64(3):539–549
Nöh K, Grönke K, Luo B, Takors R, Oldiges M, Wiechert W (2007) Metabolic flux analysis at ultra short time scale: isotopically non-stationary 13C labeling experiments. J Biotechnol 129:249–267
Park GK, Lim JH, Kim SD, Shim SH (2012) Elucidation of antifungal metabolites produced by Pseudomonas aurantiaca IB5-10 with broad-spectrum antifungal activity. J Microbiol Biotechnol 22:326–330
Peng H, Ouyang Y, Bilal M, Wang W, Hu H, Zhang X (2018) Identification, synthesis and regulatory function of the N-acylated homoserine lactone signals produced by Pseudomonas chlororaphis HT66. Microb Cell Factories 17:9
Pierson LS, Pierson EA (2010) Metabolism and function of phenazines in bacteria: impacts on the behavior of bacteria in the environment and biotechnological processes. Appl Microbiol Biotechnol 86:1659–1670
Puopolo G, Masi M, Raio A, Andolfi A, Zoina A, Cimmino A, Evidente A (2013) Insights on the susceptibility of plant pathogenic fungi to phenazine-1-carboxylic acid and its chemical derivatives. Nat Prod Res 27:956–966
Ran N, Frost JW (2007) Directed evolution of 2-keto-3-deoxy-6-phosphogalactonate aldolase to replace 3-deoxy-D-arabino-heptulosonic acid 7-phosphate synthase. J Am Chem Soc 129(19):6130–6139
Rawat G, Tripathi P, Saxena RK (2013) Expanding horizons of shikimic acid. Recent progresses in production and its endless frontiers in application and market trends. Appl Microbiol Biotechnol 97:4277–4287
Rodrigues AL, Trachtmann N, Becker J, Lohanatha AF, Blotenberg J, Bolten CJ, Korneli C, de Souza Lima AO, Porto LM, Sprenger GA (2013) Systems metabolic engineering of Escherichia coli for production of the antitumor drugs violacein and deoxyviolacein. Metab Eng 20:29–41
Rodriguez A, Martínez JA, Báez-Viveros JL, Flores N, Hernández-Chávez G, Ramírez OT, Gosset G, Bolivar F (2013) Constitutive expression of selected genes from the pentose phosphate and aromatic pathways increases the shikimic acid yield in high-glucose batch cultures of an Escherichia coli strain lacking PTS and pykF. Microb Cell Factories 12:86
Rodriguez A, Martinez JA, Millard P, Gosset G, Portais JC, Letisse F, Bolivar F (2017) Plasmid-encoded biosynthetic genes alleviate metabolic disadvantages while increasing glucose conversion to shikimate in an engineered Escherichia coli strain. Biotechnol Bioeng 114:1319–1330
Shanmugaiah V, Mathivanan N, Varghes B (2010) Purification, crystal structure and antimicrobial activity of phenazine-1-carboxamide produced by a growth-promoting biocontrol bacterium, Pseudomonas aeruginosa MML2212. J Appl Microbiol 108:703–711
Shirai M, Miyata R, Sasaki S, Sakamoto K, Yahanda S, Shibayama K, Yonehara T, Ogawa K (2001) Microorganism belonging to the genus citrobacter and process for producing shikimic acid. European Patent, 1092766
Sprenger G (2007a) Aromatic amino acids. In Amin Acid Biosynth - Pathways, Regul Metab Eng. Wendisch VF (ed) Berlin, Heidelberg: Springer, p 418 [Microbiology Monographs, vol. 5]
Sprenger GA (2007b) From scratch to value: engineering Escherichia coli wild type cells to the production of L-phenylalanine and other fine chemicals derived from chorismate. Appl Microbiol Biotechnol 75:739–749
St-Onge R, Gadkar VJ, Arseneault T, Goyer C, Filion M (2011) The ability of Pseudomonas sp. LBUM 223 to produce phenazine-1-carboxylic acid affects the growth of Streptomyces scabies, the expression of thaxtomin biosynthesis genes and the biological control potential against common scab of potato. FEMS Microbiol Ecol 75:173–183
Tang L, Xiang H, Sun Y, Qiu L, Chen D, Deng C, Chen W (2009) Monopalmityloxy shikimic acid: enzymatic synthesis and anticoagulation activity evaluation. Appl Biochem Biotechnol 158(2):408–415
Tissier A, Ziegler J, Vogt T (2014) Specialized plant metabolites: diversity and biosynthesis. Wiley-VCH Verlag GmbH & Co KGaA. pp 14–37. https://doi.org/10.1002/9783527686063.ch2
Treibmann S, Hellwig A, Hellwig M, Henle T (2017) Lysine-derived protein-bound Heyns compounds in bakery products. J Agric Food Chem 65(48):10562–10570
Tyo KE, Ajikumar PK, Stephanopoulos G (2009) Stabilized gene duplication enables long-term selection-free heterologous pathway expression. Nat Biotechnol 27:760–765
Wang MW, Hao X, Chen K (2007) Biological screening of natural products and drug innovation in China. Philos Trans Biol Sci 362(1482):1093–1105
Wang D, Yu JM, Dorosky RJ, Pierson LS 3rd, Pierson EA (2016) The phenazine 2-hydroxy-phenazine-1-carboxylic acid promotes extracellular DNA release and has broad transcriptomic consequences in Pseudomonas chlororaphis 30–84. PLoS One 11(1):e0148003
Wang S, Bilal M, Hu H, Wang W, Zhang X (2018a) 4-Hydroxybenzoic acid—a versatile platform intermediate for value-added compounds. Appl Microbiol Biotechnol 102(8):3561–3571
Wang S, Bilal M, Zong Y, Hu H, Wang W, Zhang X (2018b) Development of a plasmid-free biosynthetic pathway for enhanced muconic acid production in Pseudomonas chlororaphis HT66. ACS Synth Biol 7(4):1131–1142
Weaver LM, Herrmann KM (1990) Cloning of an aroF allele encoding a tyrosine insensitive 3-deoxy-D-arabino-heptulosonate 7-phosphate synthase. J Bacteriol 172:6581–6584
Widmer N, Meylan P, Ivanyuk A, Aouri M, Decosterd LA, Buclin T (2010) Oseltamivir in seasonal, avian H5N1 and pandemic 2009 A/H1N1 influenza: pharmacokinetic and pharmacodynamic characteristics. Clin Pharmacokinet 49(11):741–765
Wojtowicz W, Piotr M (2016) Metabolomics and fluxomics in biotechnology: current trends. J Biotechnol Comput Biol Bionanotechnol 97(2):137–144
Xing J, Sun J, You H, Lv J, Sun J, Dong Y (2012) Anti-inflammatory effect of 3,4-oxo-isopropylidene-shikimic acid on acetic acid-induced colitis in rats. Inflammation 35(6):1872–1879
Yang X, Geng B, Zhu C, Li H, He B, Guo H (2018) Fermentation performance optimization in an ectopic fermentation system. Bioresour Technol 260:329–337
Yao R, Xiong D, Hu H, Wakayama M, Yu W, Zhang X, Shimizu K (2016) Elucidation of the co-metabolism of glycerol and glucose in Escherichia coli by genetic engineering, transcription profiling, and 13C metabolic flux analysis. Biotechnol Biofuels 9:175
Yao R, Pan K, Peng H, Feng L, Hu H, Zhang X (2018) Engineering and systems-level analysis of Pseudomonas chlororaphis for production of phenazine-1-carboxamide using glycerol as the cost-effective carbon source. Biotechnol Biofuels 11:130
Yi J, Li K, Draths KM, Frost JW (2002) Modulation of phosphoenolpyruvate synthase expression increases shikimate pathway product yields in E. coli. Biotechnol Prog 18:1141–1148
Yi J, Draths KM, Li K, Frost JW (2003) Altered glucose transport and shikimate pathway product yields in E. coli. Biotechnol Prog 19:1450–1459
Yue SJ, Bilal M, Guo SQ, Hu HB, Wang W, Zhang XH (2018) Enhanced trans-2, 3-dihydro-3-hydroxyanthranilic acid production by pH control and glycerol feeding strategies in engineered Pseudomonas chlororaphis GP72. J Chem Technol Biotechnol 93(6):1618–1626
Zhang C, Kang Z, Zhang J, Du G, Chen J, Yu X (2014) Construction and application of novel feedback-resistant 3-deoxy-D-arabino-heptulosonate-7-phosphate synthases by engineering the N-terminal domain for L-phenylalanine synthesis. FEMS Microbiol Lett 353(1):11–18
Zhang Z, Jiang Z, Shangguan W (2016) Low-temperature catalysis for VOCs removal in technology and application: a state-of-the-art review. Catal Today 264:270–278
Zhao Q, Bilal M, Yue S, Hu H, Wang W, Zhang X (2017a) Identification of biphenyl 2, 3-dioxygenase and its catabolic role for phenazine degradation in Sphingobium yanoikuyae B1. J Environ Manag 204:494–501
Zhao Q, Yue S, Bilal M, Hu H, Wang W, Zhang X (2017b) Comparative genomic analysis of 26 Sphingomonas and Sphingobium strains: dissemination of bioremediation capabilities, biodegradation potential and horizontal gene transfer. Sci Total Environ 576:646–659
Funding
This work was supported by the National Natural Science Foundation of China (No. 31670033).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no competing interest.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Rights and permissions
About this article
Cite this article
Bilal, M., Wang, S., Iqbal, H.M.N. et al. Metabolic engineering strategies for enhanced shikimate biosynthesis: current scenario and future developments. Appl Microbiol Biotechnol 102, 7759–7773 (2018). https://doi.org/10.1007/s00253-018-9222-z
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-018-9222-z