Abstract
Efficient approaches for the utilization of waste sewage sludge have been widely studied. One of them is to use it for the bioenergy production, specifically methane gas which is well-known to be driven by complex bacterial interactions during the anaerobic digestion process. Therefore, it is important to understand not only microorganisms for producing methane but also those for controlling or regulating the process. In this study, azithromycin analogs belonging to macrolide, ketolide, and lincosamide groups were applied to investigate the mechanisms and dynamics of bacterial community in waste sewage sludge for methane production. The stages of anaerobic digestion process were evaluated by measuring the production of intermediate substrates, such as protease activity, organic acids, the quantification of bacteria and archaea, and its community dynamics. All azithromycin analogs used in this study achieved a high methane production compared to the control sample without any antibiotic due to the efficient hydrolysis process and the presence of important fermentative bacteria and archaea responsible in the methanogenesis stage. The key microorganisms contributing to the methane production may be Clostridia, Cladilinea, Planctomycetes, and Alphaproteobacteria as an accelerator whereas Nitrosomonadaceae and Nitrospiraceae may be suppressors for methane production. In conclusion, the utilization of antibiotic analogs of macrolide, ketolide, and lincosamide groups has a promising ability in finding the essential microorganisms and improving the methane production using waste sewage sludge.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
In contrast to a cleaning system by the biological wastewater treatment process, a large amount of waste sewage sludge (WSS) can be produced as a by-product. These wastewaters and/or WSS are now considered to be potential resources for the alternative bioenergy production through anaerobic digestion processes, which is now a globally implemented technology (Weiland 2010). Recently, a variety of approaches have been implemented for the improvement of bioenergy production using WSS. One of the approaches is to use antibiotics for the degradation of organic matters, the removal of nutrients, and the production of methane, which are handled by complicate microbes in the WSS. It has been reported that the occurrence of some macrolides from municipal wastewater ranges from 1 to 10 μg/L (Senta et al. 2017), while from pharmaceutical industry and hospital effluent produced higher concentration up to few mg/L (Liu et al. 2018). The veterinary antibiotics were detected in the range of 0.1 to 10 μg/L from animal wastewater (Wei et al. 2011). In addition, some studies were done on the effect of cephalexin in WSS (Lu et al. 2014), tetracycline and sulfamethoxydiazine for livestock wastewater (Shi et al. 2011), erythromycin for pharmaceutical wastewater (Amin et al. 2006), and tylosin in granular sludge (Sanz et al. 1996; Shimada et al. 2008). These approaches indicate a certain potential to positively or negatively change the performance of microbial activity through the addition of any antibiotic. Our previous study showed that the addition of azithromycin, a macrolide antibiotic to WSS, improves sludge degradation as well as methane production (Nguyen et al. 2014). In addition, another antibiotic tested, chloramphenicol, inhibited methane production using WSS (Mustapha et al. 2016). The difference of microbial activities in each WSS with azithromycin or chloramphenicol implicates the presence of some key microorganisms which contribute to the improvement or the suppression of methane fermentation using WSS (Mustapha et al. 2016). However, another evidence should be required to determine such key microorganisms in detail, which are related to improved methane production in WSS. According to our previous experiments in which several antibiotics including azithromycin were tested to see if the methane fermentation can be improved, so far, only azithromycin was found to be a sole antibiotic to trigger the positive impact (Nguyen et al. 2014; Mustapha et al. 2016). Therefore, the next motivation of us in the study was shifted to test the impact of azithromycin analogs to the methane fermentation using WSS.
Azithromycin is a macrolide-type antibiotic which inhibits bacterial protein synthesis by targeting the 50S ribosomal subunit (Roberts 2004). In addition, the antibiotic has been reported to be a quorum quenching compound for Pseudomonas aeruginosa (Skindersoe et al. 2008). Therefore, as an initial motivation, azithromycin was used to test the influence of quorum sensing in WSS for methane fermentation. There are three types of azithromycin analogs: (1) macrolides which are categorized as an antibiotic consisting of 14-, 15-, or 16-membered rings, (2) ketolide, and (3) lincosamides. These azithromycin analogs are structurally different as shown in Fig. 1; however, they show functionally the similar action that inhibits bacterial protein synthesis (Roberts 2004). The structural alteration has been conducted to modify the spectrum of target microbes and the efficient antimicrobial activity. Generally, they are effective against Gram-positive aerobic bacteria and Gram-positive or Gram-negative anaerobic bacteria. Specifically, these analogs have various antimicrobial spectra (Table 1); although they have a similar mode of antimicrobial action which however, the impact was still not well documented. Therefore, the objective of this study is to test the impact of several azithromycin analogs (macrolides with 14-, 15-, and 16-membered rings, ketolide, and lincosamides showing the different antimicrobial spectra) during the methane fermentation using WSS.
Chemical structures of macrolides. Azithromycin with 15-membered ring (a), roxithromycin, erythromycin and clarithromycin with 14-membered ring (b), kitasamycin and josamycin with 16-membered ring (c), ketolide; telithromycin with 14-membered ring (d), and lincosamides; lincomycin and clindamycin (e)
In this study, along with azithromycin which was used as a benchmark, other macrolide-, ketolide-, and lincosamide-type antibiotics were selected for further investigation (Fig. 1). In detail, one 15-membered ring antibiotic, azithromycin (AZM), three 14-membered ring antibiotics, erythromycin (ERM), clarithromycin (CLM), and roxithromycin (RXM), two 16-membered ring antibiotics, josamycin (JSM), and kitasamycin (KTM) were used as macrolide-group antibiotics. Then, telithromycin (TLM), a semi-synthetic erythromycin derivative was used as a ketolide-group antibiotic. Lincomycin (LCM) and clindamycin (CDM) were used as lincosamide-group antibiotics. These antibiotics were added to WSS to examine the methane production and the dynamics of complex microbial community throughout the anaerobic digestion process in each sample. Some interesting points in this study were to see if (1) among the antibiotics tested only AZM has the positive impact to improve methane fermentation using WSS, (2) the difference in antimicrobial spectra by each antibiotic influences the process of methane fermentation, and (3) the microbial activity in the WSS with each antibiotic is the same as that with azithromycin. The final goal of this study is to seek key microorganisms in WSS for controlling or regulating the methane fermentation positively or negatively based on the above viewpoints.
Materials and methods
Waste sewage sludge
Sewage sludge was obtained from the Hiagari Wastewater Treatment Plant in Kitakyushu City, Japan. The fresh raw sludge was initially centrifuged at 8000 ×g for 10 min at 4 °C, and the remaining pellet was resuspended with distilled water by shaking thoroughly. Washing steps were performed three times before adjusting the final sludge concentration to 10% (wet sludge w/w) with distilled water.
Antibiotics
Antibiotics used in this study were ERM (Nacalai Tesque), CLM, CDM, RXM (Tokyo Chemical Industry), JSM (Funakoshi), KTM, LCM (Wako), AZM (LKT Laboratories Inc.), and TLM (Bioaustralis). All the antibiotics were dissolved in ethanol as stock solutions (30 mg/mL), which were used to adjust the final concentration to 15 μg/mL of each antibiotic in WSS.
Methane production
A total volume of 30-mL WSS with 15 μg/mL of each antibiotic was put into 66-mL vials. The vials were tightly sealed and sparged with nitrogen gas for 2 min to provide anaerobic conditions prior to the incubation at 37 °C at 120 rpm for 10 days. The control WSS vial was prepared without adding any antibiotic. Each experiment was conducted at least in triplicate. Methane was measured at a certain time by injecting 50 μL of headspace gas from vials into a GC-3200 gas chromatograph (GL Science, Japan) equipped with a thermal conductive detector and Molecular Sieve 13 × 60/80 mesh column, SUS 2-m × 3-mm I.D. (GL Science, Japan). Helium gas was used as a carrier gas with a flow rate of 40 mL/min. The GC condition was set as follows: current at 100 mA, oven, injector, and detector temperature at 40, 50, and 65 °C, respectively.
Analytical methods
WSS was sampled during the fermentation for the following analyses: organic acids, pH, suspended solid, and protease activity. Samples were centrifuged at 13,000 rpm for 7 min to collect the supernatants and filtered through a 0.2-μm membrane syringe filter. Organic acids were analyzed using high-performance liquid chromatography (Shimadzu LC-10AD) as described by Mohd Yusoff et al. (2012). Each pH was measured using a compact pH meter (AS ONE, AS-211, Japan). The sludge reduction and protease activity were measured as described by Maeda et al. (2011). One unit of protease activity was calculated as the quantity of tyrosine (μmol) produced from casein by 1 mg of enzyme per min.
Total RNA extraction, cDNA synthesis, and quantitative RT-PCR
RNA extraction was conducted using fermented sludge pellets as described previously (Mohd Yasin et al. 2015) and was kept at − 70 °C. Total RNA was extracted using the RNeasy kit (Qiagen Inc., Valencia, CA), and its concentration was measured using a NanoDrop spectrophotometer (SCRUM Inc., Japan). The cDNA was synthesized from RNA using the PrimeScript RT reagent kit Perfect Real-Time (TAKARA Bio Inc., Shiga, Japan) according to the manufacturer’s protocol. The final concentration of cDNA was determined using a Qubit® 2.0 Fluorometer (Invitrogen, Qubit-IT™ dsDNA HS Assay kit). The cDNA was used for bacterial community analysis and quantitative real-time polymerase chain reaction (qRT-PCR). The qRT-PCR was performed to quantify total bacteria and archaea by a TaqMan system and StepOne Real-Time PCR System (Applied Biosystem) using the primers and probes listed in Table S1. The detailed procedures and the standard curve for universal bacteria and archaea were described in our previous paper (Mohd Yasin et al. 2015). The calculation of copy numbers based on the amount of DNA was performed as described previously (Lee et al. 2008).
High-throughput 16S rRNA sequencing
The V3–V4 region of the 16S rRNA gene was used as a target of PCR amplification for the high-throughput 16S rRNA sequencing. The forward primer was 341F (5′-CCTACGGGNGGCWGCAG-3′) and reverse primer 785R (5′-GACTACHVGGGTATCTAATCC-3′) (Klindworth et al. 2013). The KAPA HiFi HotStart ReadyMix (Kapa Biosystems, Wilmington, MA, USA) was used for the amplification with 25 cycles and according to the 16S metagenomics sequencing library preparation for Illumina MiSeq system. Briefly, the PCR amplicon was attached with dual adapter index with a unique barcode sequence (Nextera XT Index kit) to differentiate each sample. Then, the first PCR products with the index were purified by AMPure XP beads and normalized to ensure that an equal library was presented in the pooled samples. Pooled samples were denatured with sodium hydroxide, diluted with hybridization buffer, and denatured by heat prior to the MiSeq sequencing. For the sequencing, 30% spike-in of PhiX was used as an internal control. A 600-cycle V3 MiSeq reagent cartridge (Illumina) was thawed and inverted ten times to mix the reagents. Finally, pooled samples were loaded in the cartridge and then onto the MiSeq instrument along with a clean flow cell. Sequencing was performed for 301, 8, 8, and 301 cycles for forward, index 1, index 2, and reverse reads, respectively. The data obtained was demultiplexed, and the raw sequence data was deposited into the NCBI short reads archive database under the accession number SRP072534.
Processing high-throughput data
The demultiplexed raw paired-end reads sequencing data was processed using the LotuS pipeline (Hildebrand et al. 2014) by certain parameters including average sequence quality > 27, sequence length > 170 bp, no ambiguous bases, and homopolymer run < 8 bp to produce high-quality reads. These reads were further performed chimera-checking and clustered into operational taxonomic units (OTUs) at the cutoff of 97% identity with UPARSE (Edgar 2013). The α- and β-diversity indices of microbial communities were calculated after the rarefaction using Quantitative Insights into Microbial Ecology (QIIME) v1.9.0 (Caporaso et al. 2010) and further classified and taxonomically assigned using the Ribosomal Database Project (RDP) classifier with the Greengenes database v13.8 (DeSantis et al. 2006) with 80% of confidence threshold. The rarefied OTU tables were generated as the basis for calculating alpha diversity, rarefaction curves were computed using the Chao1 richness estimator, and beta diversity to determine the bacterial community structure between samples was analyzed by PCoA. Shannon index was calculated using the PAST (PAleontological STatistics) software (Hammer et al. 2001).
Statistical analysis
Different antibiotics were compared with control WSS using means from at least triplicate data (n = 3). Comparison was performed using means and standard deviations by the Student’s t test (GraphPad software) at a significance level of p < 0.05.
Results
Effect of azithromycin analogs on methane production
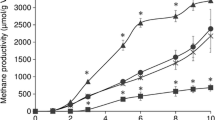
Methane production increased in all the WSS samples in the presence of each AZM analog after 6 days (Fig. 2). Besides, 14- and 16-membered ring macrolides, ketolide, and lincosamides had higher methane production than 15-membered ring AZM which is a benchmark. However, control WSS had significantly a high amount of methane at the initial incubation of day 2 compared to all the antibiotic-added WSS samples. The slower startup of methane production might be due to the impact of antibiotics to the bacterial community in WSS, which may be important in the anaerobic digestion process. In addition, the reduction of WSS was compared in all the samples based on the remaining suspended solid. The WSS reduction was similar between control and AZM which was around 25%. Also, the addition of ERM, CLM, and JSM showed slightly higher sludge reduction (26–28%) than control or AZM, although there is no significant difference. Antibiotics from ketolide (TLM) and lincosamide groups (LCM and CDM) showed lower sludge reduction than other samples which is 22 and 24%, respectively. Thus, the reason of increased methane production via antibiotics may be due to the fast digestion of WSS through changing microbial communities not due to the increased utilization of WSS.
Methane production during 10 days of anaerobic digestion from WSS with the addition of antibiotics (AZM—azithromycin, ERM—erythromycin, CLM—clarithromycin, RXM—roxithromycin, JSM—josamycin, KTM—kitasamycin, TLM—telithromycin, LCM—lincomycin, and CDM—clindamycin) or no antibiotic as control. Error bars indicate standard errors (n = 3). * Indicate the significant difference by antibiotics addition
Effect of antibiotics at the hydrolysis, acidogenesis, and acetogenesis stages during the fermentation
At first, hydrolysis process during the methane fermentation was evaluated by measuring protease activities because protein is a major composition in WSS (Maeda et al. 2009). As shown in Fig. 3a, throughout the incubation, a high protease activity was observed in the WSS in the presence of AZM in agreement with our previous result (Mustapha et al. 2016). Also, the similar trend was seen in the presence of RXM, KTM, TLM, LCM, and CDM; in particular, the protease activity of AZM and KTM was initially higher than the control, and AZM, RXM, KTM, TLM, LCM, and CDM were significantly higher than the control WSS at the end of fermentation. Despite all the macrolide-type antibiotics tested showed higher methane production than the control (Fig. 2), the protease activities in ERM, CLM, and JSM were almost the same as those in the control WSS without any antibiotic. It indicates that the mechanism to accelerate the methane production in ERM, CLM, and JSM may be different with the other macrolide, ketolide, and lincosamide antibiotics.
Protease activity at the days 2, 6, and 10 (a) and organic acid profile at the day 10 (b) from WSS with the addition of antibiotics (AZM—azithromycin, ERM—erythromycin, CLM—clarithromycin, RXM—roxithromycin, JSM—josamycin, KTM—kitasamycin, TLM—telithromycin, LCM—lincomycin, and CDM—clindamycin) or no antibiotic as control. Error bars indicate standard errors (n = 3). * Indicate the significant difference by the addition of antibiotics
The next test was to evaluate acidogenesis and acetogenesis stages during the methane fermentation using WSS, by which some precursor compounds can be produced for the methanogenesis stage. The profile of organic acids produced was determined after the 10-day incubation (Fig. 3b). As a result, formic acid was the main product in all the samples followed by acetate, propionate, isobutyrate, and butyrate, which were all increased during the incubation (data not shown). The dynamics of pH during the fermentation showed the similar trend in all the samples including the control without any antibiotic, and the final pH in all the conditions was in a range of 5.1 to 5.5. In addition, although the control WSS without any antibiotic showed the highest production of organic acids, basically the abundance ratios of organic acids were the same in all the samples except AZM. In another sense, butyric acid and propionic acid were detected remarkably, whereas formic acid was the lowest amount in the AZM compared to other samples (Fig. 3b).
Effect of antibiotics on the total bacterial and archaeal activities
Using RNA extracted from each sample, the activities of bacteria and archaea during the methane fermentation were evaluated at day 10 (Fig. 4). The total activity of archaea in the control WSS was the lowest than another WSS in the presence of each antibiotic (3.95 ± 0.07 × 108 rRNA gene copies/mL). The result is in agreement with the methane profile as the control WSS had the lowest methane production (Fig. 2). The highest archaeal activity was detected in the RXM sample which was 1.1 ± 0.2 × 109 rRNA gene copies/mL. Moreover, the total activity of bacteria was significantly low in the presence of some antibiotics (in particular, AZM, CLM, KTM, TLM, LCM, and CDM). Thus, the functions of some bacteria but not some archaea are inhibited by the antibiotics, resulting in an increased methane production. The disappearance of the bacterial species may be necessary to improve methane fermentation using WSS.
Quantity of active bacterial and archaeal populations in WSS in the presence of antibiotics (AZM—azithromycin, ERM—erythromycin, CLM—clarithromycin, RXM—roxithromycin, JSM—josamycin, KTM—kitasamycin, TLM—telithromycin, LCM—lincomycin, and CDM—clindamycin) or no antibiotic as control. Error bars indicate standard errors (n = 3). * Indicate the significant difference by antibiotics addition
Richness and diversity of microbial communities
RNA templates extracted from WSS samples with or without each antibiotic on the second day and the tenth day were used to analyze the diversity of bacterial community during the anaerobic digestion of WSS. In this study, 866–1026 OTUs were clustered at a dissimilarity level of 0.03 as stated in Table S2, and the OTUs reduced in all samples from 2 to 10 days. Chao1 indicators demonstrate the richness of bacterial community, while the Shannon index estimates the microbial population diversity. Both Chao1 and Shannon indexes showed the reduction in richness and diversity of microbial community by adding each antibiotic compared to the control WSS without any antibiotic.
Dynamics of bacterial population
RNA templates from control WSS and representative analogs from 14-, 15-, and 16-membered ring macrolides, ketolide, and lincosamides were analyze at 2 and 10 days to compare the similarity and dynamics of bacterial community. Figure 5 shows a principle coordinate analysis (PCoA) that was conducted to see the similarity of the bacterial community between samples using OTUs based on unweighted UniFrac. The bacterial community of WSS with or without each antibiotic was indicated by dark circle (at 2 days) and open circle (at 10 days), which first showed the dissimilarity at these two periods. On the second day, the bacterial communities in all antibiotics analogs were close to each other unlike that in the control WSS. However, after 10 days, the similarities were widely dispersed but still had a vertical similarity although there are some exceptions. On the other hand, ketolide (TLM) and 16-membered rings macrolide (JSM) showed more diverse community from other analogs, as well as control WSS. As expected, the control WSS without any antibiotic unlikely had a similarity in bacterial community. It is well-known that WSS contains a vast amount of different microbial community. The addition of antibiotics has changed the microbial community of WSS during the methane fermentation; for example, Proteobacteria, Firmicutes, and Bacteriodetes were the major bacterial community present in each WSS sample during the anaerobic digestion regardless of the addition of antibiotics (Fig. 6). Clostridia belongs to Firmicutes phylum which was higher in WSS with macrolide-type analogs compared to ketolide, lincosamide, and control WSS. Besides, Plantomycetes and Alphaproteobacteria were higher in the samples with antibiotics compared to antibiotic-free WSS, indicating that they may have a high resistance to a broad range of antibiotics. Caldilinea was detected at a high percentage in WSS with the addition of macrolide-, ketolide-, and lincosamide-type antibiotic when compared to AZM and control WSS as the benchmarks. In addition, the antibiotic-free WSS has a high percentage of Nitrosomonadaceae and Nitrospiraceae; which were known as nitrifiers as compared to WSS with antibiotics. These key microorganisms that present significantly in WSS with or without any antibiotic from macrolide, ketolide, and lincosamide represented by AZM, TLM and LCM, respectively, as well as with chloramphenicol from previous study as comparison were shown in Table 2. The abundant percentage of Nitrosomonadaceae was higher in control WSS and in the presence of chloramphenicol, while only Nitrospiraceae was higher in control WSS. Clostridiaceae, Caldilineaceae, Planctomycetes, and Alphaproteobacteria have high abundance percentages in antibiotic samples compared to control and WSS with chloramphenicol. The contribution of these microorganisms in anaerobic digestion stages was predicted in Fig. 7. The nitrifiers were assumed to limit the production of methane, while another four bacterial groups might be responsible for accelerating the methane production. On the other hand, methanogens was hardly detected in microbial community profile through MiSeq analysis for all WSS samples. This is due to the range of abundant percentage for methanogens was below 0.2%. Still, the percentage of methanogens in WSS in the presence of each antibiotic was higher compared to control WSS (data not shown).
Principle coordinate analysis (PCoA) in the following antibiotics (AZM—azithromycin, ERM—erythromycin, CLM—clarithromycin, RXM—roxithromycin, JSM—josamycin, KTM—kitasamycin, TLM—telithromycin, LCM—lincomycin, and CDM—clindamycin) and without antibiotic as control (CTRL) in WSS at the day 2 (black circle) and the day 10 (open circle). PCoA was conducted at the 3% cutoff OTU level
Relative abundance of the dominant microbial communities categorized at the taxonomic genus level in WSS with antibiotics (AZM—azithromycin, ERM—erythromycin, CLM—clarithromycin, RXM—roxithromycin, JSM—josamycin, KTM—kitasamycin, TLM—telithromycin, LCM—lincomycin, and CDM—clindamycin) or no antibiotic as control at day 10 of anaerobic digestion. Results were derived from high-throughput 16S rRNA sequencing. Minor classes (less than 1%) were summed up in group “other”
Key microorganisms relating to the methane production predicted by a difference in the microbial community of WSS with or without the addition of antibiotics during the anaerobic digestion process. The superscript numbers indicate the reference sources as follow; (1) NO as an oxidizing agent and O2 produced in this step are assumed to contribute to the methane oxidation by which methane can be consumed (Ettwig et al. 2012). (2) and (3) These microorganisms are well-known as nitrifiers which can oxidize ammonium to nitrite or nitrate in WSS and some studies have proved that they could be related to the methane suppression (Dunfield and Knowles 1995; Liu et al. 2017). (4) Planctomycetes also favorably present in antibiotic samples was reported to involve in sugar fermentation (Elshahed et al. 2007), which assisted in acidogenesis stage to produce volatile fatty acids, H2 and CO2. (5) Clostridia and Caldilinea of Chloroflexi pylum are responsible for the hydrolysis and acidogenesis with a high rate (Guo et al. 2015). (6) Proteobacteria is a well-known utilizer for glucose, propionate, butyrate, and acetate (Ariesyady et al. 2007)
Discussion
The application of all AZM analogs, macrolide, ketolide, and lincosamide improved the methane production using WSS. This study supported our previous study (Mustapha et al. 2016) that found that not only a macrolide antibiotic, AZM, improves methane production but also the production of methane from AZM analogs is higher than the AZM itself. Despite each analog has different structures (Fig. 1) and spectrum of activity (Table 1), the methane production was almost similar at the end of incubation for 10 days. According to Roberts (2008), the first macrolide-type antibiotic, ERM, and the analogs of lincosamides have limited antibacterial spectra on bacteria community. In addition, some macrolides were more persistent in the environment which caused the bacteria to slowly change their community and became adapted to this environment (Schlüsener and Bester 2006). LCM also has a similar characteristic as being stable during the anaerobic process (Wu et al. 2011). These conditions contributed to a high methane production even though the process slowed down initially at day 2 compared to the control sample without adding any antibiotic (Fig. 2). The methane production obtained in this study was contradicted to some other studies which inhibited methane production (Amin et al. 2006; Shimada et al. 2008; Cetecioglu et al. 2015) due to the different microbial sources, the durations and scale of experiments, and the concentration of antibiotics. The initial concentration of antibiotics used in this study was almost similar to the concentration from pharmaceutical and hospital wastewater. However, according to a previous report (Terzic et al. 2018), waste sludge has the ability to remove the macrolides through microbial biotransformation which can reduce the antibiotic activity. Besides, the anaerobic digestion strategy also improves the removal of antibiotic resistance genes, reduces the horizontal transfer of these genes, and inhibits the selection of resistant bacteria (Liu et al. 2018). Thus, anaerobic digestion technology used in this study not only may contribute to methane production but also may eliminate the antibiotic contamination from sewage sludge.
Protein is the major component in WSS rather than carbohydrates and lipids (Table S3) (Maeda et al. 2009). These compounds are degraded in a hydrolysis stage assisted by hydrolytic bacteria through secreting hydrolytic enzymes; thereby, smaller compounds can be finally converted into methane (Appels et al. 2008; Manyi-Loh et al. 2013). In this study, the addition of macrolides, ketolide, and lincosamides did not affect the hydrolytic bacteria during the anaerobic digestion of WSS as a high protease activity was detected in the samples in the presence of AZM analogs. Furthermore, the addition of antibiotics improved methane production but slightly reduced the organic acids. This was presumably due to the consumption and conversion of organic acids to acetate, CO2, and H2 by acetogenic bacteria for methane production. Clostridia and Bacteroidia, which are present in higher percentage in this study, are the main fermentative bacteria (Guo et al. 2015) in the acidogenesis stage, which convert possible WSS monomer compounds into volatile fatty acids, CO2, and H2. Later, acetogenic bacteria convert these products into acetate which can be finally utilized to methane by methanogens which belongs to archaea. Specifically, there are three methanogens which have common roles during the anaerobic digestion process (Venkiteshwaran et al. 2015). The first one is acetoclastic methanogens which convert acetate to methane and CO2; the second one, hydrogenotrophic methanogens, can use H2 or formate to convert CO2 into methane; and the third one is methylotrophic methanogens which can produce methane from methyl compounds. In addition, syntrophic and antagonistic interactions could occur between archaea and bacteria during the anaerobic process. Syntrophic interaction occurs at the acetogenic stage in which acetate is converted to H2 and CO2 by acetate-oxidizing bacteria, and the products are subsequently utilized by hydrogenotrophic methanogens for methane production (Venkiteshwaran et al. 2015). On the other hand, an antagonistic interaction occurs by sulfate-reducing bacteria (SRB) competing with methanogens and other bacteria for fermenting H2, acetate, and organic molecules. Besides, the SRB also produce hydrogen sulfide that can inhibit methanogens and several bacteria groups (Ziganshin et al. 2011). Therefore, the balance of archaea and bacteria might be important for the stable methane production.
In the control WSS, pH was initially lower and kept low; it may disturb the growth and functional abilities of methanogens to convert intermediate substrates to methane (Huser et al. 1982; Steinhaus et al. 2007) and led to the accumulation of organic acids. This situation also was proved by total archaea population quantified by qRT-PCR in the antibiotic-free WSS, which shows a low number of archaea. Moreover, according to Chen (2010), hydrogenotrophic methanogens utilize formic acid as a carbon source for methane production. Therefore, the accumulation of formic acid especially in control WSS might be due to the absence or less abundance of hydrogenotrophic methanogens. This is supported by our results which showed a low number of archaea and low methane production. However, in the AZM sample, even though the formic acid concentration was the lowest among the samples with another antibiotic, the methane production was not improved than other WSS with antibiotics. This is due to the high amount of propionic acid which can inhibit the methanogenesis process (Venkiteshwaran et al. 2015). On the other hand, antibiotics used in this study had more impact on the bacterial population as the quantity of bacteria was lower than the control WSS (Fig. 4), and the lowest bacterial population was shown in the ketolide group (TLM); a derivative of ERM. This low bacterial population might be due to the improvement in spectrum of activity of TLM that cannot be covered by ERM (Scheinfeld 2004).
Besides, richness and diversity of bacterial populations in WSS were dependent on the type of antibiotics and on the anaerobic digestion process. For example, the addition of lincosamides showed the lowest OTUs among the other antibiotics, but a reduced number of OTUs were found in the samples with the antibiotic after the anaerobic digestion process completed (Table S2). Bacterial diversity was reduced within samples presumably, because the anaerobic conditions affected the original/initial communities through the accumulation of organic acids. The small range Shannon index with or without antibiotics indicated that these samples had almost the similar diversity of bacteria, since they were originally came from the same source which is WSS.
The bacteria community present at day 10 may be important in the anaerobic digestion process. Since their abundances were high with antibiotics addition, the hydrolysis and acidogenesis/acetogenesis stages were carried out efficiently as supported by the results of protease activity, organic acids, and methane production. For example, Clostridia and Caldilinea of Chloroflexi pylum were found higher in WSS with antibiotics which are associated with a high rate of hydrolysis and acidogenesis (Guo et al. 2015; Shimada et al. 2011). The presence of these two microbes in the AZM analogs (Fig. 6) was found in our previous study using AZM (Mustapha et al. 2016). Besides, Planctomycetes also favorably present in antibiotic samples was reported to involve in sugar fermentation (Elshahed et al. 2007), which assisted in acidogenesis stage to produce volatile fatty acids, H2, and CO2. It is also assumed that in antibiotic samples, acetogenic bacteria, mainly in Alphaproteobacteria community can rapidly utilize these intermediates to produce acetate. Proteobacteria may be an important community in the anaerobic digestion process because most of the Delta-, Alpha-, Beta-, and Gammaproteobacteria are well-known utilizers for glucose, propionate, butyrate, and acetate (Ariesyady et al. 2007). In addition, Myxococcales was the major group belonging to Deltaproteobacteria at day 10 (Fig. 6). Interestingly, some bacteria belonging to this group (described as ‘micropredators’) are able to degrade other microbes by secreting hydrolytic exoenzymes (Osaka et al. 2008). It is believed that these enzymes also degrade the WSS components such as protein to smaller molecules during the hydrolysis stage. Thus, all of these communities provided sufficient soluble intermediates for methanogens for the next stage. Nitrosomonadaceae and Nitrospiraceae that abundantly present in control WSS without any antibiotic could be very sensitive to antibiotics as stated by Fan et al. (2009) that they were unable to survive at a low concentration of ERM. Also, they are well-known as nitrifiers which can oxidize ammonium to nitrite or nitrate in WSS, and some studies have proved that these microbes could contribute to the methane suppression (Dunfield and Knowles 1995; Liu et al. 2017), which explained on low methane production in WSS without antibiotic. The simple pathway was shown in Fig. 7 in which the nitrite reduced to nitric oxide, NO, nitrogen gas, N2, and oxygen, O2. The NO as an oxidizing agent and O2 produced in this step are assumed to contribute to the methane oxidation by which methane can be consumed (Ettwig et al. 2012). Regardless of the addition of antibiotic, Proteobacteria, Bacteriodetes, and Firmicutes have been reported to be common and abundant in the anaerobic digestion process (Guo et al. 2015; Shimada et al. 2011; Osaka et al. 2008; Ng et al. 2015), which play an important role in producing methanogenic precursors for methanogens. It is interesting to note that the similarity of bacterial communities by the addition of macrolides, ketolide, and lincosamides was shifted through various processes of anaerobic digestion yet maintaining the essential bacterial groups for methane production. In conclusion, all the antibiotic analogs used in this study showed a similar effect on methane production. Therefore, macrolides and another similar antibiotic of this group from ketolide and lincosamides, which have different antibacterial spectra had stimulated dynamics of the microbial community, functions, and interactions in WSS for enhancing methane production using WSS.
References
Amin MM, Zilles JL, Greiner J, Charbonneau S, Raskin L, Morgenroth E (2006) Influence of the antibiotic erythromycin on anaerobic treatment of a pharmaceutical wastewater. Environ Sci Technol 40:3971–3977
Amsden GW (1996) Erythromycin, clarithromycin, and azithromycin: are the differences real? Clin Ther 18:56–72
Appels L, Baeyens J, Degrève J, Dewil R (2008) Principles and potential of the anaerobic digestion of waste-activated sludge. Prog Energy Combust Sci 34:755–781
Ariesyady HD, Ito T, Okabe S (2007) Functional bacterial and archaeal community structures of major trophic groups in a full-scale anaerobic sludge digester. Water Res 41:1554–1568
Bryskier A (1998) Roxithromycin: review of its antimicrobial activity. J Antimicrob Chemother 41:1–21
Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Pena AG, Goodrich JK, Gordon JI, Huttley GA (2010) QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7:335–336
Cetecioglu Z, Ince B, Ince O, Orhon D (2015) Acute effect of erythromycin on metabolic transformations of volatile fatty acid mixture under anaerobic conditions. Chemosphere 124:129–135
Chantot JF, Bryskier A, Gasc JC (1986) Antibacterial activity of roxithromycin: a laboratory evaluation. J Antibiot 39:660–668
Chen Q (2010) Kinetics of anaerobic digestion of selected C1 to C4 organic acids. Dissertation, University of Missouri—Columbia
DeSantis TZ, Hugenholtz P, Larsen N, Rojas M, Brodie EL, Keller K, Huber T, Dalevi D, Hu P, Andersen GL (2006) Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl Environ Microbiol 72:5069–5072
Doons-Goossens A, Bedert R, Degreef H, Vandele M (1990) Airborne allergic contact dermatitis from kitasamycin and midecamycin. Contact Dermatitis 23:118–119
Dunfield P, Knowles R (1995) Kinetics of inhibition of methane oxidation by nitrate, nitrite, and ammonium in a humisol. Appl Environ Microbiol 61:3129–3135
Edgar RC (2013) UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat Methods 10:996–998
Elshahed MS, Youssef NH, Luo Q, Najar FZ, Roe BA, Sisk TM, Bühring SI, Hinrichs KU, Krumholz LR (2007) Phylogenetic and metabolic diversity of Planctomycetes from anaerobic, sulfide- and sulfur-rich Zodletone Spring, Oklahoma. Appl Environ Microbiol 73:4707–4716
Ettwig KF, Speth DR, Reimann J, Wu ML, Jetten MS, Keltjens JT (2012) Bacterial oxygen production in the dark. Front Microbiol 3:273
Fan C, Lee PKH, Ng WJ, Alvarez-Cohen L, Brodie EL, Andersen GL, He J (2009) Influence of trace erythromycin and erythromycin-H2O on carbon and nutrients removal and on resistance selection in sequencing batch reactors (SBRs). Appl Microbiol Biotechnol 85:185–195
Felmingham D (2001) Microbiological profile of telithromycin, the first ketolide antimicrobial. Clin Microbiol Infect 7:2–10
Goldstein EJC, Citron DM, Merriam CV, Warren Y, Tyrrel KL, Fernandez H (2003) In vitro activities of telithromycin and 10 oral agents against aerobic and anaerobic pathogens isolated from antral puncture specimens from patients with sinusitis. Antimicrob Agents Chemother 47:1963–1967
Guo J, Peng Y, Ni B-J, Han X, Fan L, Yuan Z (2015) Dissecting microbial community structure and methane-producing pathways of a full-scale anaerobic reactor digesting activated sludge from wastewater treatment by metagenomic sequencing. Microb Cell Factories 14:33
Hammer Ø, Harper DAT, Ryan PD (2001) Paleontological statistics software: package for education and data analysis. Palaeontol Electron 4
Hildebrand F, Tadeo R, Voigt AY, Bork P, Raes J (2014) LotuS: an efficient and user-friendly OTU processing pipeline. Microbiome 2:30
Huser BA, Wuhrmann K, Zehnder AJB (1982) Methanothrix soehngenii gen. nov. sp. nov., a new acetotrophic non-hydrogen-oxidizing methane bacterium. Arch Microbiol 132:1–9
Klindworth A, Pruesse E, Schweer T, Peplies J, Quast C, Horn M, Glöckner FO (2013) Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies. Nucleic Acids Res 41:1–11
Lee C, Kim J, Shin SG, Hwang S (2008) Monitoring bacterial and archaeal community shifts in a mesophilic anaerobic batch reactor treating a high-strength organic wastewater. FEMS Microbiol Ecol 65:544–554
Leigh DA (1981) Antibacterial activity and pharmacokinetics of clindamycin. J Antimicrob Chemother 7:3–9
Liu L, Xu X, Cao Y, Cai C, Cui H, Yao J (2017) Nitrate decreases methane production also by increasing methane oxidation through stimulating NC10 population in ruminal culture. AMB Express 7:76
Liu PY, Chen JR, Shao L, Tan J, Chen DJ (2018) Responses of flocculent and granular sludge in anaerobic sequencing batch reactor (ASBR) to azithromycin wastewater and its impact on microbial communities. J Chem Technol Biotechnol. https://doi.org/10.1002/jctb.5578
Lu X, Zhen G, Liu Y, Hojo T, Estrada AL, Li Y-Y (2014) Long-term effect of the antibiotic cefalexin on methane production during waste activated sludge anaerobic digestion. Bioresour Technol 169:644–651
Maeda T, Yoshimura T, García-Contreras R, Ogawa HI (2011) Purification and characterization of a serine protease secreted by Brevibacillus sp. KH3 for reducing waste activated sludge and biofilm formation. Bioresour Technol 102:10650–10656
Maeda T, Yoshimura T, Shimazu T, Shirai Y, Ogawa HI (2009) Enhanced production of lactic acid with reducing excess sludge by lactate fermentation. J Hazard Mater 168:656–663
Manyi-Loh CE, Mamphweli SN, Meyer EL, Okoh AI, Makaka G, Simon M (2013) Microbial anaerobic digestion (bio-digesters) as an approach to the decontamination of animal wastes in pollution control and the generation of renewable energy. Int J Environ Res Public Health 10:4390–4417
Mohd Yasin NH, Maeda T, Hu A, Yu C-P, Wood TK (2015) CO2 sequestration by methanogens in activated sludge for methane production. Appl Energy 142:426–434
Mohd Yusoff MZ, Maeda T, Sanchez-Torres V, Ogawa HI, Shirai Y, Hassan MA, Wood TK (2012) Uncharacterized Escherichia coli proteins YdjA and YhjY are related to biohydrogen production. Int J Hydrog Energy 37:17778–17787
Mustapha NA, Sakai K, Shirai Y, Maeda T (2016) Impact of different antibiotics on methane production using waste-activated sludge: mechanisms and microbial community dynamics. Appl Microbiol Biotechnol 100:9355–9364
Ng KK, Shi X, Ng HY (2015) Evaluation of system performance and microbial communities of a bioaugmented anaerobic membrane bioreactor treating pharmaceutical wastewater. Water Res 81:311–324
Nguyen MT, Maeda T, Mohd Yusoff MZ, Ogawa HI (2014) Effect of azithromycin on enhancement of methane production from waste activated sludge. J Ind Microbiol Biotechnol 41:1051–1059
Osaka T, Ebie Y, Tsuneda S, Inamori Y (2008) Identification of the bacterial community involved in methane-dependent denitrification in activated sludge using DNA stable-isotope probing. FEMS Microbiol Ecol 64:494–506
Peters DH, Clissold SP (1992) Clarithromycin. Drugs 44:117–164
Rakhit S, Singh K (1974) Structure activity relationship in sixteen membered macrolide antibiotics. J Antibiot 27:221–224
Roberts MC (2004) Resistance to macrolide, lincosamide, streptogramin, ketolide, and oxazolidinone antibiotics. Mol Biotechnol 28:47–62
Roberts MC (2008) Update on macrolide–lincosamide–streptogramin, ketolide, and oxazolidinone resistance genes. FEMS Microbiol Lett 282:147–159
Sanz JL, Rodriguez N, Amils ÁR (1996) The action of antibiotics on the anaerobic digestion process. Appl Microbiol Biotechnol 46:587–592
Scheinfeld N (2004) Telithromycin: a brief review of a new ketolide antibiotic. J Drugs Dermatol 3:409–413
Schlüsener MP, Bester K (2006) Persistence of antibiotics such as macrolides, tiamulin and salinomycin in soil. Environ Pollut 143:565–571
Senta I, Krizman-Matasic I, Terzic S, Ahel M (2017) Comprehensive determination of macrolide antibiotics, their synthesis intermediates and transformation products in wastewater effluents and ambient waters by liquid chromatography–tandem mass spectrometry. J Chromatogr A 1509:60–68
Shi JC, Liao XD, Wu YB, Liang JB (2011) Effect of antibiotics on methane arising from anaerobic digestion of pig manure. Anim Feed Sci Technol 166:457–463
Shimada T, Li X, Zilles JL, Morgenroth E, Raskin L (2011) Effects of the antimicrobial tylosin on the microbial community structure of an anaerobic sequencing batch reactor. Biotechnol Bioeng 108:296–305
Shimada T, Zilles JL, Morgenroth E, Raskin L (2008) Inhibitory effects of the macrolide antimicrobial tylosin on anaerobic treatment. Biotechnol Bioeng 101:73–82
Skindersoe ME, Alhede M, Phipps R, Yang L, Jensen PO, Rasmussen TB, Bjarnsholt T, Tolker-Nielsen T, Høiby N, Givskov M (2008) Effects of antibiotics on quorum sensing in Pseudomonas aeruginosa. Antimicrob Agents Chemother 52:3648–3663
Smieja M (1998) Current indications for the use of clindamycin: a critical review. Can J Infect Dis Med Microbiol 9:22–28
Steinhaus B, Garcia ML, Shen AQ, Angenent LT (2007) A portable anaerobic microbioreactor reveals optimum growth conditions for the methanogen Methanosaeta concilii. Appl Environ Microbiol 73:1653–1658
Straneo G, Scarpazza G (1990) Efficacy and safety of clarithromycin versus josamycin in the treatment of hospitalized patients with bacterial pneumonia. J Int Med Res 18:164–170
Terzic S, Udikovic-Kolic N, Jurina T, Krizman-Matasic I, Senta I, Mihaljevic I, Loncar J, Smital T, Ahel M (2018) Biotransformation of macrolide antibiotics using enriched activated sludge culture: kinetics, transformation routes and ecotoxicological evaluation. J Hazard Mater 349:143–152
Venkiteshwaran K, Bocher B, Maki J, Zitomer D (2015) Relating anaerobic digestion microbial community and process function. Microbiol Insights 8:MBI-S33593
Wei R, Ge F, Huang S, Chen M, Wang R (2011) Occurrence of veterinary antibiotics in animal wastewater and surface water around farms in Jiangsu Province, China. Chemosphere 82:1408–1414
Weiland P (2010) Biogas production: current state and perspectives. Appl Microbiol Biotechnol 85:849–860
Williams JD (1991) Spectrum of activity of azithromycin. Eur J Clin Microbiol Infect Dis 10:813–820 ratio
Wu D, Lü F, Gao H, Shao L, He P (2011) Mesophilic bio-liquefaction of lincomycin manufacturing biowaste: the influence of total solid content and inoculum to substrate. Bioresour Technol 102:5855–5862
Xu R, Peng Y, Wang M, Fan L, Li X (2014) Effects of broad-spectrum antibiotics on the metabolism and pharmacokinetics of ginsenoside Rb1: a study on rats’ gut microflora influenced by lincomycin. J Ethnopharmacol 158:338–344
Zhao Z, Jin L, Xu Y, Zhu D, Liu Y, Liu C, Lei P (2014) Synthesis and antibacterial activity of a series of novel 9-O-acetyl-4′-substituted 16-membered macrolides derived from josamycin. Bioorg Med Chem Lett 24:480–484
Ziganshin AM, Schmidt T, Scholwin F, Il’inskaya ON, Harms H, Kleinsteuber S (2011) Bacteria and archaea involved in anaerobic digestion of distillers grains with solubles. Appl Microbiol Biotechnol 89:2039–2052
Funding
The authors wish to thank the Japanese Government Scholarship (MEXT), Kitakyushu City, and Science & Technology Research Partnership for Sustainable Development Program (SATREPS) for the support of this study.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Ethical approval
This article does not contain any studies performed with human participants or with animals by any of the authors.
Conflict of interest
The authors declare that there are no conflicts of interest.
Electronic supplementary material
ESM 1
(PDF 230 kb)
Rights and permissions
About this article
Cite this article
Mustapha, N.A., Hu, A., Yu, CP. et al. Seeking key microorganisms for enhancing methane production in anaerobic digestion of waste sewage sludge. Appl Microbiol Biotechnol 102, 5323–5334 (2018). https://doi.org/10.1007/s00253-018-9003-8
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-018-9003-8